Abstract
KAR5 is required for membrane fusion during karyogamy, the process of nuclear fusion during yeast mating. To investigate the molecular mechanism of nuclear fusion, we cloned and characterized the KAR5 gene and its product. KAR5 is a nonessential gene, and deletion mutations produce a bilateral defect in the homotypic fusion of yeast nuclei. KAR5 encodes a novel protein that shares similarity with a protein in Schizosaccharomyces pombe that may play a similar role in nuclear fusion. Kar5p is induced as part of the pheromone response pathway, suggesting that this protein uniquely plays a specific role during mating in nuclear membrane fusion. Kar5p is a membrane protein with its soluble domain entirely contained within the lumen of the endoplasmic reticulum. In pheromone-treated cells, Kar5p was localized to the vicinity of the spindle pole body, the initial site of fusion between haploid nuclei during karyogamy. We propose that Kar5p is required for the completion of nuclear membrane fusion and may play a role in the organization of the membrane fusion complex.
In the yeast Saccharomyces cerevisiae, the nuclear envelope remains intact during all stages of mating. The diploid nucleus is generated from the direct fusion of two haploid nuclei in a process called karyogamy (for review see Rose, 1996). Karyogamy proceeds in two steps: nuclear congression and nuclear fusion. Nuclear congression follows immediately after cell fusion when the haploid nuclei are drawn together in the newly formed zygote. Once the nuclei are in close proximity, the two nuclear envelopes fuse, generating a diploid nucleus.
Nuclear envelope fusion during karyogamy is a prime example of homotypic membrane fusion. Homotypic (or self) fusion involves the union of two separate membranes of the same composition and origin (for review see Rothman and Warren, 1994). Examples of self fusion in animal cells include the reassembly of the nuclear envelope (Wiese and Wilson, 1993) and the Golgi apparatus (Acharya et al., 1995; Rabouille et al., 1995) from membrane fragments during mitotic telophase. Homotypic fusion is also an inherent and essential step in secretion. Vesicle budding, like other processes of fission, requires homotypic membrane fusion. To be released from an organelle, homotypic membrane fusion is required to “pinch off” the constriction around the base of each secretory vesicle. It is likely that the study of yeast karyogamy will help identify conserved factors involved in all processes of homotypic fusion (Rothman, 1994; Rothman and Warren, 1994).
Genetic studies of yeast have identified many genes that mediate the three major pathways of membrane fusion: (a) secretory vesicles with target organelle membranes (Novick et al., 1980), (b) plasma membrane fusion during mating (McCaffrey et al., 1987; Trueheart et al., 1987; Berlin et al., 1991; Kurihara et al., 1994), and (c) nuclear envelope fusion (Conde and Fink, 1976; Polaina and Conde, 1982; Kurihara et al., 1994). The mating-specific processes of cell fusion and karyogamy are solely events of homotypic membrane fusion. For recent reviews see Rose (1996) and Marsh and Rose (1997).
During both nuclear congression and nuclear fusion, the yeast microtubule organizing center, the spindle pole body, plays a central role (Byers and Goetsch, 1975; Rose, 1991; Rose 1996). The disk-shaped spindle pole body of S. cerevisiae is embedded in the nuclear envelope and serves an analogous function to that of the centrosome in larger eukaryotes (Winey and Byers, 1993). The spindle pole body assembles both the mitotic and meiotic spindles as well as cytoplasmic microtubules. The spindle pole body plays two roles in karyogamy. During nuclear congression, the two spindle pole bodies anchor the cytoplasmic microtubules by which the two haploid nuclei are drawn together. During nuclear fusion, the spindle pole body marks the initial site of both membrane fusion and fusion of the two spindle pole bodies (Byers and Goetsch, 1975). As the two spindle pole bodies join, the two respective outer and inner nuclear envelopes fuse. Thus, nuclear fusion is a complex and coordinated series of events. It is not known whether nuclear membrane fusion proceeds by a single concerted fusion event or by a series of discrete events involving fusion of first the outer and then the inner membranes.
Genes necessary for karyogamy have been identified in genetic screens for kar mutants (Conde and Fink, 1976; Polaina and Conde, 1982; Kurihara et al., 1994). The first kar mutant screens identified mutations that caused karyogamy defect even when the mutants were mated with wild-type cells (Conde and Fink, 1976; Polaina and Conde, 1982). Such mutants were referred to as “unilateral” in that the defect was evident when a single mating partner contained the mutation. Both dominant and recessive unilateral mutants have been isolated. One explanation for the recessive unilateral mutants is that, in the zygote, the mutant nucleus cannot receive the corresponding wild-type protein from the wild-type nucleus. Presumably this results when the wild-type Kar protein is restricted to the wild-type nucleus and is not free to diffuse and rescue the mutant defect of the other nucleus before they both enter mitosis (Rose et al., 1989). In contrast, in bilateral karyogamy defects both partners in a mating must be mutant to observe the mutant phenotype. In the bilateral mutants, the gene products must fulfill one of the following criteria: (a) be diffusible, (b) be required in only one nucleus, or (c) be synthesized in the zygote such that both nuclei acquire sufficient nascent wild-type protein.
Recently, a novel screen has been used to isolate bilateral karyogamy mutants (Kurihara et al., 1994). These new mutants identified the karyogamy genes KAR4, KAR5, KAR7, KAR8, and KAR9. Based on cytological and genetic criteria, all kar mutants were grouped into two functional classes (Kurihara et al., 1994). Class I mutants exhibit a block in nuclear congression: the nuclei do not fuse and remain distant from one another in the zygote. Class II mutant zygotes exhibit closely juxtaposed but unfused nuclei, consistent with a block in nuclear membrane fusion.
Since nuclear congression is a microtubule-dependent process (Delgado and Conde, 1984; Rose and Fink, 1987; Huffaker et al., 1988; Berlin et al., 1990; Rose, 1991), it is not surprising that most Class I mutants represent genes that affect microtubule function: KAR1 encodes a component of the spindle pole body (Vallen et al., 1992; Spang et al., 1995); TUB2 encodes β-tubulin (Huffaker et al., 1987, 1988); BIK1 encodes a microtubule-associated protein (Berlin et al., 1990); KAR3 encodes a kinesin homologue (Meluh and Rose, 1990); CIK1 encodes a Kar3p- associated protein (Page and Snyder, 1992; Page et al., 1994); and KAR4 regulates the mating-induced transcription of KAR3 and CIK1 (Kurihara et al., 1996). Microtubules and associated proteins play similar roles in both karyogamy and in the condensation of vesicular ER/nuclear fragments in animals (Dabora and Sheetz, 1988). Microtubules allow membranes attached to them to be concentrated, thereby promoting their fusion.
The Class II nuclear fusion mutants are defined by the genes KAR2, KAR5, KAR7, and KAR8. Class II mutants exhibit defects in membrane fusion that are evident in vivo, as observed by electron microscopy (Kurihara et al., 1994; Beh, 1996), and in vitro, as measured in a homotypic ER/nuclear membrane fusion assay (Kurihara et al., 1994; Latterich and Schekman, 1994). The in vitro ER/nuclear membrane fusion assay indirectly measures membrane fusion by detecting the consequences of lumenal mixing. When fusion occurs, a secretory precursor protein preloaded into the lumen of one ER membrane can be processed by a glucosidase resident only in the lumen of a second ER membrane. Thus, the assay is sensitive to mutations that block both the initiation as well as the completion of membrane fusion. In contrast to the Class II mutants, ER membranes derived from the Class I mutant strains fused as efficiently as wild-type membranes in vitro.
The best characterized of the Class II genes is KAR2, which encodes an ER-localized HSP70. Kar2p is the yeast homologue of the mammalian protein BiP/GRP78 (Normington et al., 1989; Rose et al., 1989). In addition to its role in karyogamy, Kar2p is essential for the translocation of secretory precursors into the ER (Vogel et al., 1990). Given the known activities of Kar2p and its homologues, Kar2p could have several different roles in karyogamy. Kar2p may be indirectly required for nuclear fusion though its role in the translocation and/or folding of a key component of the nuclear fusion apparatus. Alternatively, Kar2p may be necessary to maintain the structure of the ER/nuclear envelope during nuclear fusion. However, a variety of genetic and biochemical data (Vogel et al., 1990; Latterich and Schekman, 1994) argue that it is more likely that KAR2 is directly involved in karyogamy. In this regard, Kar2p may be functioning as a molecular chaperone, participating in the assembly or interactions of a nuclear membrane fusogenic complex. Such a complex may be comprised of the gene products of some of the other kar genes.
In this paper, we describe the cloning and characterization of the Class II karyogamy gene KAR5. KAR5 encodes a novel protein that plays a central role in nuclear membrane fusion. KAR5 is induced during mating, and Kar5p localizes to the initial site of nuclear fusion, next to the spindle pole body. Analyses of Kar5p indicated that it is membrane bound, with most of the protein positioned within the lumen of the ER/nuclear envelope. Since it is sequestered completely within the ER, Kar5p is probably not a membrane fusogen or a receptor linking the two nuclei for docking. However, Kar5p may help determine when and where nuclear membrane fusion occurs or may be required to complete membrane fusion once nuclear contact has been made. As such, Kar5p represents a novel protein required for the regulation and activation of homotypic membrane fusion.
Materials and Methods
Microbial Techniques and Strain Constructions
Culture media and genetic manipulations were as described elsewhere (Rose et al., 1990). Cells treated with α-factor were incubated in the presence of 10 μg/ml α-factor for 2.5 h in synthetic complete (SC)1 media brought to pH 3.5 with HCl. Pheromone induction was verified microscopically when >80% of the cells formed mating projections.
Quantitative matings were performed as previously described (Rose et al., 1990). For cytoductant analysis, cyh R/rho 0 cells were obtained as previously described (Rose et al., 1990). Yeast strains used are listed in Table I.
Table I.
Yeast Strains Used
| Strain | Genotype | |
|---|---|---|
| MS52 | MATα ura3-52 leu2-3 leu2-112 trp1-Δ1 | |
| MS2685 | MATΔ::LEU2 kar5-1162 ura3-52 leu2-3 leu2-112 trp1-Δ1 his3::TRP1 [pB1311] | |
| MS2686 | MATΔ::LEU2 kar5-1162 ura3-52 leu2-3 leu2-112 trp1-Δ1 his3::TRP1 | |
| MS3258 | MAT a kar5-1162 ura3-52 ade2-101 leu2-3 leu2-112 his3-Δ200 | |
| MS3261 | MAT a kar5-486 ura3-52 ade2-101 leu2-3 leu2-112 his3-Δ200 | |
| MS3286 | MAT a kar5-Δ1::URA3 ura3-52 ade2-101 leu2-3 leu2-112 | |
| MS3289 | MATα kar5-Δ1::URA3 ura3-52 trp1-Δ1 | |
| MS3577 | MATα kar5-Δ2::LEU2 ura3-52 trp1-Δ1 leu2-3 leu2-112 | |
| MS3856 | MATα ura3-52 trp1-Δ1 leu2-3,112 cyh R rho 0 | |
| MS3857 | MATα kar5-Δ2::LEU2 ura3-52 trp1-Δ1 leu2-3 leu2-112 cyh R rho 0 | |
| MS3915 | MAT a kar5-Δ2::LEU2 ura3-52 leu2-3 leu2-112 his3-Δ200 | |
| MS3980 | MATα kar5-Δ2::LEU2 ura3-52 trp1-Δ1 leu2-3 leu2-112 [pRS426] | |
| MS3983 | MATα kar5-Δ2::LEU2 ura3-52 trp1-Δ1 leu2-3 leu2-112 [pMR3179] | |
| MS3986 | MAT a kar5-Δ2::LEU2 ura3-52 his3-Δ200 leu2-3 leu2-112 [pRS426] | |
| MS3987 | MAT a kar5-Δ2::LEU2 ura3-52 his3-Δ200 leu2-3 leu2-112 [pMR3142] | |
| MS4024 | MATα kar5-1162 ura3-52 ade2-101 his3-Δ200 lys2-801 [pMR3142] | |
| MY3264 | MAT a ura3-52 his4-519 leu2 trp1 can1-101 ste12-Δ::LEU2 | |
| MY3265 | MAT a ura3-52 his4-519 leu2 trp1 can1-101 |
All strains from the M. Rose lab collection.
KAR5 Cloning, Mapping, and Sequencing
Restriction enzymes used for cloning were obtained from New England Biolabs (Beverly, MA). PCR and sequencing primers were synthesized at the Princeton University oligonucleotide synthesis facility. Cloning techniques were performed as described in Sambrook et al. (1989). Plasmids were recovered from yeast by the method of Hoffman and Winston (1987) and transformed into Escherichia coli by electroporation.
To map KAR5 on the yeast physical map, the 2.8-kb HindIII subclone from pMR2710 was labeled by the random primer method (Sambrook et al., 1989) and hybridized to lambda prime clone filters provided by Linda Riles (Washington University School of Medicine, St. Louis, MO).
To sequence the 2.8-kb KAR5 minimal complementing region, the genomic subclone was inserted into the unique HindIII polylinker site of Bluescript KS+ and KS− (Stratagene, La Jolla, CA). Exonuclease III/mungbean nuclease digestions were performed according to Henikoff (1987). The resulting deletion clones were then sequenced using either T3 or T7 sequencing primers. Both strands of the 2.8-kb subclone were sequenced by single-strand sequencing (Sambrook et al., 1989).
Plasmid Constructions
To disrupt the KAR5 gene, two disruption alleles were generated, each with a different selectable marker for integration. The first disruption allele (pMR2741) was made through the ligation of the HindIII/KpnI and SacI/HindIII insert fragments of pMR2710 into the KpnI/SacI polylinker sites of pRS406 (Sikorski and Hieter, 1989). To integrate and disrupt the KAR5 open reading frame, pMR2741 was digested with HindIII, and the entire linearized plasmid was transformed into yeast. This disruption allele was marked with the URA3 gene and transformants were selected on SC-Ura. The second disruption plasmid (pMR2869) was constructed by ligation of the 590-bp HindIII/AvrII and 1,100-bp XhoI/HindIII fragments of pMR2710 into the XbaI/XhoI polylinker sites of pRS405, a LEU2-marked integration plasmid. Plasmid pRS405 is the equivalent of pRS305 (Sikorski and Hieter, 1989), with the polylinker in the opposite orientation with respect to the plasmid. To disrupt the KAR5 locus, pMR2869 was digested with HindIII, and yeast transformants were selected on SC-Leu. Cells bearing the kar5 disruption alleles were tested to show that CEN-based vectors bearing KAR5 [pMR2710] suppressed the kar5-Δ mating defects.
To construct a vector for the overexpression of KAR5 gene, the 2.8-kb HindIII fragment from pMR2710, bearing the KAR5 minimal complementing region, was subcloned into the HindIII sites of the 2-μm vector pRS424. This vector is equivalent to pRS306 (Sikorski and Hieter, 1989), except that the polylinker is in the opposite orientation and a segment of the yeast 2-μm plasmid has been inserted at the AatII site. At the same time, an equivalent centromeric plasmid was constructed using pRS426. Plasmid pRS426 is equivalent to pRS316 (Sikorski and Hieter, 1989), with the exception that the polylinker is in the opposite orientation. As assayed by interrupted plate matings (Rose et al., 1990), both the 2-μm plasmid, pMR3142, and the centromeric plasmid, pMR3179, suppressed the karyogamy defect of the kar5-Δ2 strain MS3577. Wild-type cells (MS52) transformed with pMR3179 or pMR3142 had no observable defects with respect to mating efficiency or viability (at 23, 30, or 37°C) and pMR3179 did not suppress the karyogamy defect of the other Class II karyogamy mutants (as tested by interrupted mating assays).
To construct a glutathione-S-transferase (GST)–Kar5p fusion protein vector for expression of Kar5p in bacteria, a fragment of KAR5 corresponding to the NH2-terminal 345 residues was amplified by PCR using primers 5′GTC-GGG-ATC-CCA-TGT-TTG-AAA-TGC-TCT-ACG3′ and 5′CCG-AGG-ATC-CTT-CCA-TTT-GTG-ATT-CAG3′. These primers had BamHI sites incorporated into the ends to allow the in-frame ligation of the amplified product into the BamHI site of the vector pGEX-1 (Smith and Johnson, 1988), thus producing the plasmid pMR2739. The corresponding transformed bacterial strain was used to make recombinant protein to generate polyclonal antisera.
RNA Analysis
For RNA-blots, total RNA was isolated, electrophoresed through 0.8% agarose formaldehyde gels, and transferred to GeneScreen hybridization transfer membrane (NEN Research Products, Boston, MA) as described elsewhere (Rose et al., 1990). Hybridization and washes were performed as follows. Blots were prehybridized for 4 h at 68°C in 5× PPiESS buffer (0.75 M NaCl, 5 mM Na4P2O7, 0.5 M Na2EDTA, 0.125 M sodium phosphate, pH 8.0) containing blocking solution (0.2% ficoll, 0.02% polyvinylpyrrolidone, 0.02% BSA), 0.1 μg/ml denatured salmon sperm DNA, and 1.0% SDS. Boiled probe was added directly to the blots in prehybridization mix and incubated overnight at 68°C. Blots were rinsed first in 0.2× PPiESS, 0.5% SDS for 15 min and then washed in the same buffer twice, each time for 20 min at 68°C.
Probes were labeled by the random primer method (Sambrook et al., 1989). The KAR5 probe used was derived from the KAR5 2.8-kb HindIII fragment isolated from pMR2710. As an internal control, levels of KAR5 mRNA were compared with levels of ACT1 transcript. The probe used for hybridization to ACT1 mRNA was derived from a 282-bp ACT1 EcoRI/ HindIII fragment isolated from pYST122 (a gift from J. Broach, Princeton University, Princeton, NJ).
Antibody Production and Purification
Antisera against bacterially expressed GST–Kar5p was generated in a New Zealand white rabbit at the Princeton University Animal Facility. GST–Kar5p was purified from inclusion bodies from four 500-ml cultures of MR2739 incubated with 0.5 mM IPTG for 4 h at 37°C. Bacterial pellets were resuspended in 1/50 culture volume of 16 mM Na2HPO4, 4 mM NaH2PO4, 150 mM NaCl, 1% Triton X-100 containing 2 μg/ml final concentration each of chymostatin, leupeptin, aprotinin, pepstatin A (Sigma Chemical Co., St. Louis, MO), and AEBSF (Calbiochem Corp., La Jolla, CA). Cells were disrupted by three 1-min pulses of a sonicator. After centrifugation at 12,000 g at 4°C, the pellet was resuspended in Laemmli sample buffer (Laemmli, 1970) and boiled for 15 min. GST–Kar5p was resolved on a 10% SDS-PAGE and transferred to nitrocellulose (Schleicher & Schuell, Inc., Keene, NH), and the band corresponding to GST–Kar5p was excised. The nitrocellulose strip, bearing ∼0.2 mg of Kar5p, was dissolved in 0.2 ml DMSO, mixed with an equal volume of sterile water, emulsified with 0.5 ml Freund's Complete Adjuvant (Sigma Chemical Co.), and injected into rabbits. Boosts were performed every month with 100 μg GST–Kar5p (either on nitrocellulose dissolved in DMSO or isolated by preparative electrophoresis [see below]) emulsified in Freund's Incomplete Adjuvant (Sigma Chemical Co.).
To affinity purify anti-Kar5p antibodies, GST–Kar5p was isolated by preparative electrophoresis and then covalently coupled to a chromatographic resin. After preparative 10% SDS-PAGE, the GST–Kar5p band was identified by negative staining with 0.3 M CuCl2. The protein was electroeluted from the excised gel slice at 100 V for 16 h in Tris-glycine buffer (25 mM Tris, 200 mM glycine) between BT1 and BT2 Elutrap membranes (Schleicher & Schuell, Inc.). 0.5–1.0 mg of the protein (as determined by Bradford assay [1976]) was conjugated to Sulfo-link gel (Pierce Chemical Co., Rockford, IL), as suggested by the manufacturer (except 1 mM DTT was used instead of 2-mercaptoethylamine). 1 ml of antisera was applied to the column, the column was washed, and anti-Kar5p–specific antibodies were eluted with 0.1 M glycine, pH 2.5.
Immunological Techniques
Yeast extracts for Western blots were prepared as described by Ohashi et al. (1982). For Western blotting, proteins were separated by SDS-PAGE and transferred in 20 mM Tris, 150 mM glycine, 20% methanol to nitrocellulose (Schleicher & Schuell, Inc.). Immunoblots were blocked in 5% nonfat milk in buffer A (10 mM Tris-HCl pH 7.4, 150 mM NaCl) for 1 h at room temperature. Blots were then incubated for at least 2 h at room temperature, with primary antibody diluted in 5% milk/buffer A. Before incubations with the secondary antibody, blots were washed four times for 15 min each in buffer A, 0.05% Tween-20 (Sigma Chemical Co.) at room temperature. HRP-conjugated anti–rabbit secondary antibody (Amersham Corp., Arlington Heights, IL), diluted in 5% milk/buffer A, was added to blots for 1 h, after which washes were performed as before.
Before use, affinity-purified anti-Kar5p antibodies were preadsorbed to kar5-Δ cells as follows: 150 μl of affinity-purified antibodies were incubated with 200 μl of packed, formaldehyde-fixed kar5-Δ2 cells (MS3980) for 2 h at 23°C. Cell walls were removed from the fixed cells by prior incubation at 30°C with 50 μl of β-mercaptoethanol and 0.5 ml 10 mg/ml Zymolyase 100,000 T in 1.2 M sorbitol. The antibody/cell mixture was then centrifuged in a microcentrifuge, and the supernatant was incubated with a second volume of fixed kar5-Δ2 cells. After centrifugation, supernatant was collected, and the preadsorbed antibody was stored at 4°C with 0.2% NaN3 as a preservative.
For Western blots, affinity-purified preadsorbed anti-Kar5p antibody was used at a 1:100 dilution, rabbit anti–Kar2p antiserum was used at a titer of 1:5,000, rabbit anti–Sec72p serum was used at a dilution of 1:1,000 (a gift of Martin Latterich, University of California, Berkeley, CA), and rabbit anti–Sec61p antibodies were used at a dilution of 1:1,000 (a gift of Vladimir Lupashin, Princeton University, NJ). Bands were visualized with a 1:3,000 dilution of the HRP-conjugated anti–rabbit secondary antibody, followed by the ECL chemiluminescent system (Amersham Corp.).
Isolation of Nuclei and Protease Protection
Nuclei were isolated as described by Aris and Blobel (1991) with minor modifications. 30,000 Klett units of cells, preincubated with 0.1 mg/ml α-factor for 2.5 h, were harvested for nuclear isolation. Cells were rinsed and pelleted twice and resuspended in 10 ml spheroplast buffer (0.1 M potassium phosphate, pH 6.5, 1.2 M sorbitol, 1 mM PMSF). To remove cell walls, cells were treated with 50 μl of β-mercaptoethanol and 0.5 ml 10 mg/ml Zymolyase 100,000 T. After 45 min of digestion, spheroplasts were recovered by centrifugation at 2.5 K rpm in a clinical centrifuge (model GLC2B; Sorvall, Newtown, CT). After two rinses with spheroplast buffer, spheroplasts were resuspended in 25 ml lysis buffer (20% Ficoll 400, 20 mM potassium phosphate, pH 6.5, 1 mM MgCl2, 1 mM PMSF). Spheroplasts were gently lysed with 20 strokes of a stainless steel Dounce homogenizer. Cellular disruption was confirmed by microscopy. After chilling 10 min at 4°C, the lysate was centrifuged 5 min at 13,000 g (9 K rpm) in a rotor (model SW27; Beckman Instruments, Fullerton, CA) at 4°C. The supernatant was recovered and poured into a second tube and centrifuged for 10 min at 13,000 g at 4°C.
For the protease protection assay, the supernatant from the 13,000-g centrifugation was diluted fivefold in ice-cold 20 mM potassium phosphate, 1 mM MgCl2, 1 mM PMSF, and membranes/nuclei were pelleted in 30-ml Corex tubes by centrifugation for 10 min at 11,725 g (9 K rpm) in a rotor (model SA600; Sorvall) at 4°C. Pellets were resuspended in 12 ml protease buffer (0.3 M mannitol, 0.1 M KCl, 50 mM Tris-HCl, pH 7.5, 1 mM EGTA) and divided into two portions. To one portion, Triton X-100 was added to a final concentration of 0.4%; both portions were incubated at 4°C with proteinase K, 0.3 mg/ml final concentration. 1-ml aliquots were removed at various times, and the aliquots were mixed with 0.1 ml 100% TCA at 4°C to terminate the digestion. After centrifugation in a microcentrifuge and washes with 90% acetone, protein pellets were resuspended in Laemmli sample buffer and boiled, and proteins were resolved by 10% SDS-PAGE.
To obtain purified nuclei, the cleared lysate was fractionated further (Aris and Blobel, 1991). The supernatant obtained from the 13,000-g spin was split and layered on top of two SW27 tubes containing step gradients of 5 ml 50, 40, and 30% Ficoll 400 (in 20 mM potassium phosphate, pH 6.5, 1 mM MgCl2, 1 mM PMSF) layered from bottom to top, respectively. The tubes were centrifuged in a rotor (model SW27; Beckman Instruments) at 18 K rpm for 1 h at 2°C. Nuclei were isolated from the 40% Ficoll layer (including the 40–50%, and 30–40% interfaces), diluted 10-fold with 20 mM potassium phosphate, pH 6.5, 1 mM MgCl2, 1 mM PMSF, and pelleted by centrifugation in 30-ml Corex tubes for 10 min at 9 K rpm in a rotor (model SA600; Sorvall) at 4°C. Nuclei were resuspended in 50 mM Tris-HCl, pH 7.0, 1 mM PMSF. Total cellular protein was precipitated by the method of Ohashi et al. (1982), and from the fractionated nuclei, proteins were precipitated with the addition of TCA to a 10% final concentration. Pellets were rinsed with 90% acetone and resuspended by boiling in Laemmli loading buffer without bromophenol blue or β-mercaptoethanol; protein concentrations were then determined by a Lowry-based protein assay (Bio-Rad DC protein assay; Bio-Rad Laboratories, Inc., Hercules, CA). Finally, β-mercaptoethanol was added (2% final concentration) with bromophenol blue and equal amounts of samples were run on a 10% SDS gel.
To determine whether Kar5p was a membrane protein, extractions were performed with 1 M NaCl, 0.1 M Na2CO3, pH 11.0, 2 M urea, 1% Triton X-100, and 1% Triton X-100 plus 0.5 M NaCl. A crude membrane fraction was isolated as described above and resuspended in 20 mM Tris-HCl, pH 7.4 with protease inhibitors (chymostatin, leupeptin, aprotinin, pepstatin A, AEBSF, and PMSF). To this fraction, NaCl, detergent, or Na2CO3 was added, and after a 1-h incubation on ice, the samples were centrifuged at 128,000 g (model TL-100 ultracentrifuge, model TLA 100.2 rotor; Beckman Instruments) for 1 h. Pellets were resuspended, and equal volumes of sample were electrophoretically separated by SDS-PAGE.
Immunofluorescence Microscopy
Indirect immunofluorescence, using paraformaldehyde fixation of intact cells, was performed with modifications of the procedure described by Roberts et al. (1991). Harvested cells were washed with 0.1 M potassium phosphate, pH 6.5, pelleted in a clinical centrifuge, and resuspended in 2 ml fresh paraformaldehyde solution (2 g paraformaldehyde, 350 μl 6 N NaOH, 50 μl 1 M MgCl2, and 0.68 g KH2PO4 in 50 ml distilled H2O) without pretreatment with formaldehyde. Cells were allowed to fix overnight at 23°C. The fixed cells were washed twice in 0.1 M potassium phosphate, pH 6.5, 1.2 M sorbitol and then resuspended in 1 ml of the same solution. To remove cell walls, fixed cells were incubated with 5 μl β-mercaptoethanol, 25 μl glusulase (DuPont Biotechnology Systems, Boston, MA), and 15 μl 10 mg/ml Zymolyase 100,000 T at 30°C for 1 h (or until they appeared dark by phase contrast microscopy). Spheroplasts were washed once with 1.2 M sorbitol and then incubated at 23°C for 10 min with 2% SDS, 1.2 M sorbitol. The cells were then washed twice in 1.2 M sorbitol. Lastly, cells were affixed to glass slides coated with polylysine.
Indirect immunofluorescence using methanol/acetone fixation of spheroplasted shmoos was performed, with minor modifications, as described by Rout and Kilmartin (1990). Spheroplasted shmoos were incubated in Wickerhams' medium for only 10 min (to avoid recovery from pheromone arrest) before being affixed onto slides. Whole cell morphology was not preserved using this method, though it was more sensitive for Kar5p visualization.
Affinity-purified, preadsorbed rabbit anti–Kar5p antibodies were used at a 1:10 dilution. Mouse anti–90-kD antibodies were used at a 1:500 dilution (a gift of John Kilmartin, Medical Research Council, Cambridge, England). Secondary antibodies were used at a 1:1,000 dilution (Boehringer Mannheim Corp., Indianapolis, IN). Secondary antibodies used for the detection of Kar5p were rhodamine-conjugated goat anti–rabbit or Cy3-conjugated goat anti–rabbit; anti–90-kD secondary antibodies were FITC-conjugated goat anti–mouse. 4′,6-diamidino-2-phenylindole (DAPI) was obtained from Accurate Chemicals and Scientific Corp. (Westbury, NY).
Results
Isolation and Mapping of KAR5
To clone the KAR5 gene, the kar5-1162 mutant strain (MS2686) was transformed with a yeast centromere vector– based genomic library (Rose et al., 1987). A total of 86,000 transformants (57 genomic equivalents) were screened by replica plating onto lawns of kar5-1162 cells of the opposite mating type (MS2685) and allowing them to mate for only 3 h before replica plating to media to select for diploids (interrupted plate matings). Several mating-proficient transformants were isolated; two of these suppressed the kar5-1162 mating defect after isolation of the plasmids in E. coli and retransformation back into kar5-1162 cells. Suppression was therefore plasmid-linked, and the genomic DNA inserts on the plasmids had related restriction maps (Fig. 1).
Figure 1.
kar5-1162–suppressing subclones. To map the two genomic library isolates containing the KAR5 gene and flanking DNA, the plasmids were analyzed by restriction mapping (A, AvrII; B, BamHI; R, EcoRI; H, HindIII; K, KpnI; S, SalI; Sc, SacI; X, XhoI; Xb, XbaI; designated to scale). To determine the minimal KAR5 complementing DNA, the indicated subclones were transformed into a kar5-1162 strain (MS2686). Subclones that suppressed the kar5-1162 karyogamy defect are indicated by black bars; open bars indicate no suppression. The minimal complementing DNA subcloned in pMR2710 was used to create the two disruption constructs pMR2741 and pMR2869. In pMR2741, the URA3 gene replaces the deleted KAR5 sequence indicated (kar5-Δ1); in pMR2869, LEU2 replaces deleted KAR5 sequence (kar5-Δ2).
To determine the minimal complementing region, several subclones were constructed, transformed into MS2686, and tested for mating proficiency in interrupted plate matings. The smallest complementing region, a 2.8-kb HindIII fragment, was identified (Fig. 1). The 2.8-kb HindIII subclone was used to construct KAR5 disruption alleles, which were viable and exhibited karyogamy defects similar to the original kar5 mutants. The phenotypes of the disruption alleles will be described below.
To demonstrate that the cloned DNA contained the authentic KAR5 gene, a kar5 disruption mutant was mated to kar5-1162 and kar5-486 mutant strains (MS3258 and MS3261, respectively). To facilitate the mating, the disruption strain contained an autonomous plasmid bearing the putative KAR5 gene (MS3983). Selected diploids were then grown on medium containing 5-fluoro-orotic acid (Boeke et al., 1987) to select for cells in which the KAR5 plasmid had been lost. After sporulation and tetrad dissection, we tested the ability of the spore clones to mate with kar5-1162 strains. From 41 tetrads, encompassing both crosses, no Kar5+ segreagants were obtained. We concluded that the cloned DNA was tightly linked to (⩽1.2 cM) and, therefore, allelic with the authentic KAR5 locus.
KAR5 Encodes a Putative Translocated Coiled-Coil Protein
To analyze the KAR5 gene, the DNA of the smallest (2.8-kb HindIII) complementing subclone (pMR2710) was sequenced. The complementing subclone contained only one complete open reading frame of 1,512 bp, encoding a protein predicted to be 504 amino acids in length (Fig. 2 B) with a molecular mass of 58.4 kD. The two disruption alleles (pMR2741 and pMR2869) delete large segments of this open reading frame (see Fig. 1), and neither complemented the kar5-1162 karyogamy defect. Using the 2.8-kb subclone as the probe, the KAR5 gene was physically mapped by hybridization with whole chromosome blots and lambda prime clone filters (Riles et al., 1993) to chromosome XIII near the ADH3 gene. The gene has also been sequenced by the Saccharomyces genome sequencing project and corresponds to hypothetical protein YMR065W. The two sequences of KAR5 concur, with the exception of 1 bp difference at residue 5. Our sequence predicts a leucine at this site, whereas the sequencing project predicts an arginine.
Figure 2.

The KAR5 sequence. (A) The DNA sequence of the 2.8-kb minimal complementing region was determined. All yeast DNA sequences on this fragment upstream of the structural gene are shown (−259 to +3). This region contained all sequence elements required for the regulation of the KAR5 gene. Within this region are two PREs at position −140 (TGTGTCA) and −122 (TGTTTTA) that match the consensus PRE (TGTTTCA) in 6/7 residues. The nucleotide numbers are indicated to the left of sequence. A potential transcriptional TATA element is underlined; the PRE homologous sequences are indicated by the reverse font. (B) The translated protein corresponding to KAR5 (YMR065W) is shown. The KAR5 structural gene began at nucleotide 260 and ended at nucleotide 1762 and potentially encodes a 504–amino acid protein. The hydrophobic regions are indicated by the reverse font, and the potential coiled-coils are indicated by double underscoring. The one difference between our DNA sequence and that produced by the Yeast Genome Sequence Consortium is indicated at residue 5, which is a leucine in our sequence. The amino acid residue number is indicated on the left.
In the smallest complementing subclone, pMR2710, 259 bp of sequence upstream of the KAR5 coding region was present together with the entire KAR5 open reading frame (Fig. 2 A). Since this subclone fully complemented the karyogamy defect of kar5 mutants, we conclude that all the sequence elements required for KAR5 function are contained within the 259-bp upstream sequence. Contained within this region are two putative pheromone response elements (PREs). PREs are the binding site for Ste12p, the transcription factor that controls genes induced in mating. Both putative PREs match the consensus sequence in seven out of eight residues.
Three regions of the Kar5p protein were predicted to be significantly hydrophobic (Figs. 2 B and 3). The first corresponds to the amino-terminal 20 residues and may therefore correspond to a secretory signal sequence. However, since no sites were found that conform to the consensus -3,-1 rule for a signal peptide cleavage site (von Heijne, 1986), if the NH2-terminal sequence does function as is a signal peptide, it may not be cleaved. Two other regions predicted to form significantly hydrophobic regions were found towards the carboxyl terminus (residues 455–470 and 485–502), and these may form transmembrane spanning domains, although no strong predictions about the orientation can be made (Hofmann and Stoffel, 1993; Rost et al., 1995). Taken together, the primary structure suggests that Kar5p enters the secretory pathway and is an integral membrane protein with one or more carboxy-terminal regions spanning the membrane.
Figure 3.
The primary sequence of Kar5p predicts a transmembrane protein with a region of coiled-coil. Kar5p has three regions of significantly hydrophobic character. Shown are (A) Hopp-Woods hydrophilicity and (B) Kyte-Doolittle hydropathy plots. The hydrophobic regions are shown as gray bars. Kar5p also has two or more regions predicted to form a coiled-coil. (C) A plot of the probability of forming a coiled-coil according to the algorithm of Lupas (1996). (D) A comparison of the predicted structures for S. cerevisiae Kar5p and the S. pombe Tht1p. See text for details.
Two other predicted regions of the protein are noteworthy. First, Kar5p is predicted to contain a region with significant probability of forming two coiled-coil domains (Lupas, 1996; Fig. 3 C). Through these two domains, Kar5p may interact with itself or other proteins. Second, the carboxy-terminal domain of Kar5p is extremely basic. The overall isoelectric point (pI) of the entire protein was predicted to be 4.59, and the pI of the first 350 residues was 4.24. In contrast, the pI of carboxy-terminal 150 residues was predicted to be 10.07. Indeed, between residues 415 and 500, there are 14 basic residues and only 1 acidic residue. The functional significance of the basic COOH-terminal domain of Kar5p is uncertain, but its potential interaction with the acidic head groups of phospholipids merits consideration.
Although KAR5 does not have close relatives in the public databases, it does share significant homology with a newly identified gene from Schizosaccharomyces pombe, tht1. (These sequence data are available from Genbank/ EMBL/DDBJ under accession number D87337.) The tht1 gene is predicted to encode a 62.9-kD protein with 578 residues. Kar5p and tht1p share three regions of significant homology (Fig. 3 D) containing 17 amino acids (33% identity, 83% similarity), 56 amino acids (28% identity, 61% similarity), and 23 amino acids (41% identity, 54% similarity). Like KAR5, the S. pombe gene encodes a potential membrane protein with as many as four hydrophobic regions that are predicted to form transmembrane spanning domains. Like Kar5p, tht1p is predicted to contain a region of α-helical coiled-coil. Finally, like Kar5p, the carboxy-terminal region of tht1p is predicted to be extremely basic; between residues 392 and 504 there are 14 basic and no acidic residues. As in Kar5p, the basic region corresponds to the putative transmembrane spanning domains. Finally, the Genbank notation for tht1 includes the statement that the gene is required for nuclear fusion in S. pombe, although no evidence for this has been published. Given their similarity of proposed structure and function, it seems likely that these two proteins are homologues.
kar5-Δ Has a Bilateral Karyogamy Defect
Karyogamy mutants can be classified as unilateral or bilateral. Unilateral mutants are defective when mated with wild-type cells; in bilateral mutants, both partners in a mating must be mutant to manifest a defect. The two original mutant alleles of KAR5, kar5-486 and kar5-1162, were identified in the screen for bilateral karyogamy mutants (Kurihara et al., 1994). To determine if the bilateral karyogamy defect reflected the true null phenotype, kar5-Δ mutants were constructed, and the zygotes were examined by microscopy and quantitative mating assays (Table II). The cloned DNA encoding the putative KAR5 gene was used to disrupt the corresponding open reading frame (Fig. 1). The disruption, marked with the LEU2 gene (kar5-Δ2) and integrated into a wild-type haploid strain, had no effect on cell viability at any temperature. The percentage of mutant zygotes was determined by microscopy (Table II) for matings involving kar5-Δ1 and kar5-486 strains. In bilateral or self-matings, the kar5-Δ1 mutants exhibited a pronounced karyogamy defect, similar to if not greater than that of kar5-486. In crosses with a wild-type strain (unilateral matings), both kar5-Δ1 and kar5-486 manifested slight karyogamy defects, indicating that wild-type Kar5p from one parent is usually sufficient to effect nuclear fusion. As was found for kar5-486 and kar5-1162, the nuclei were closely apposed in all of the kar5-Δ1 zygotes exhibiting the Kar− defect. These results confirm that Kar5p has no function in nuclear congression.
Table II.
Quantitative Mating Experiments
| Allele | X self | X wild type | ||
|---|---|---|---|---|
| Percentage of Kar− zygotes | ||||
| kar5-486 | 87% (47/54) | 20% (17/86) | ||
| kar5-Δ1 | 95% (54/57) | 20% (10/50) | ||
| KAR5 | <5% | |||
| Percentage of diploid formation | ||||
| kar5-Δ2 | 8.7% | 51% | ||
| KAR5 | 100% | |||
| C/D ratio | ||||
| kar5-Δ2 | 1.8 | 0.035 | ||
| KAR5 | 0.0005 |
Quantitative mating experiments. For each mutant, mating frequencies were determined for bilateral (self) matings and for unilateral (crosses with wild-type cells) matings. The percentage of Kar− zygotes was determined by DAPI staining and fluorescence microscopy. The number of the zygotes counted are shown in parentheses. The percent diploids formed is measured as the number of prototrophic diplids formed per total viable cells after mating. The ratio of cytoductants to total diploids (C/D) is an index of the failure of nuclear fusion. The strains used for this analysis were MS3286, MS3289, MS3856, MS3857, and MS3915.
Measurement of the formation of cytoductants provides a genetic assay for the failure in nuclear fusion. Cytoductants are the aberrant progeny that arise from Kar− zygotes. They contain the haploid nucleus of one mating parent but cytoplasmic elements (e.g., mitochondria) from the other parent. The cytoductants arising from the cross can be measured using a combined selection for the functional mitochondria coming from one parent and for the recessive drug resistance marker (cycloheximide resistance) from the nucleus of the other parent. The calculated fraction of cytoductants to the total number of diploids provides a sensitive measure of a karyogamy defect (Conde and Fink, 1976). Compared to the wild type, kar5-Δ2 strains displayed about an 11-fold decrease in diploid formation from bilateral matings (Table II). This corresponded to a roughly 3,600-fold increase in the frequency of cytoductants relative to the isogenic wild type (Table II). In unilateral matings, kar5-Δ2 strains formed cytoductants ∼70-fold more frequently than the wild-type strains. However, this amounts to a failure in nuclear fusion occurring in <5% of the matings. In contrast, viable cytoductants are formed almost twice as frequently as diploids in the bilateral matings. Because viable cytoductants are not formed in every case of a failure in nuclear fusion (Rose, 1991), it is likely that the failure rate is actually higher, similar to that seen by direct microscopy of the zygotes. The kar5-Δ2 bilateral defect was comparable to the defects observed for the original unilateral KAR mutants (Conde and Fink, 1976; Polaina and Conde, 1982). However, this defect is not as severe as that observed in kar3-Δ bilateral matings (Meluh and Rose, 1990), suggesting that a secondary pathway for nuclear envelope fusion may exist.
KAR5 Expression Is Induced by α-Factor
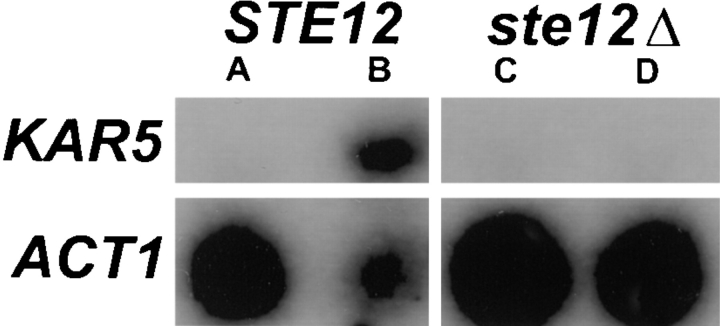
The PRE sequence motif found upstream of most pheromone-inducible genes mediates transcriptional induction in response to activation of the pheromone-activated protein kinase pathway by mating factor. Because of the PREs observed upstream of KAR5, we tested directly whether the expression of KAR5 was induced upon pheromone treatment. Total RNA was isolated from strains before and after α-factor induction, and equal amounts were analyzed by RNA-blot hybridization (Fig. 4). The observed size of the KAR5 mRNA, 2.1 kb, was consistent with the predicted length of the KAR5 open reading frame. KAR5 mRNA was present at low levels in vegetatively growing cells (more readily apparent after longer exposures of the autoradiograph) and was markedly induced after α-factor treatment (Fig. 4, lane B). In a ste12-Δ strain, the level of KAR5 mRNA did not increase, indicating that induction required the STE12 transcription factor and the mating-response signal transduction pathway. The vegetative level of KAR5 transcription was not higher in diploids and was not present in kar5-Δ strains (data not shown). Induction by mating pheromone is consistent with a specific role for Kar5p in nuclear fusion during mating.
Figure 4.
KAR5 is a pheromone-induced gene. Northern blots of mRNA from (A) a vegetatively growing wild-type strain (MY3265)—a low level of KAR5 message could be detected; (B) MY3265 after addition of α-factor—a dramatic induction of KAR5 transcription was observed; (C) a vegetatively growing ste12-Δ strain (MY3264); and (D) the ste12-Δ strain after addition of pheromone. In the ste12-Δ strain, the levels of KAR5 transcripts were the same before and after addition of pheromone.
Kar5p Is a Nuclear Envelope Protein
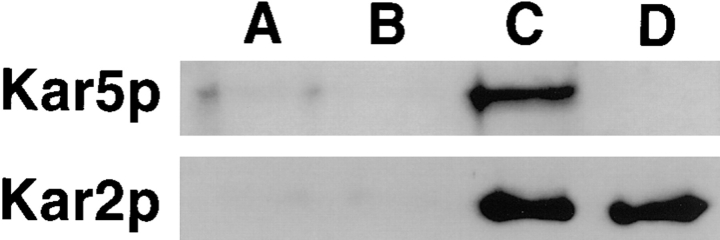
To characterize the Kar5 protein, we used GST–Kar5p expressed in E. coli to produce polyclonal antibodies that were then affinity-purified (see Materials and Methods). To detect Kar5p, total protein was extracted from cells overexpressing KAR5 from a multicopy plasmid, with and without pheromone induction. On protein blots, the Kar5p antibodies recognized a 58-kD protein after pheromone induction, corresponding to the predicted size of unmodified Kar5p (58,396 D). In contrast, the 58-kD protein was not detectable after pheromone induction in a strain bearing a deletion of the KAR5 gene (MS3986) (Fig. 5). Taken together, these data demonstrate that the 58-kD protein was Kar5p. In some experiments (e.g., Fig. 7), the Kar5p band was resolved into a doublet. The molecular nature of the two bands has not been determined. Overexpression of KAR5 had no detectable adverse phenotype (data not shown). In addition, both a low molecular mass protein (Fig. 5) and a slightly higher molecular mass protein (Fig. 7) were sometimes detected with the antibody. Since these proteins were observed in the kar5-Δ strain, they are not derived from Kar5p.
Figure 5.

Expression of Kar5p in pheromone-treated cells. Equal amounts of cellular proteins were loaded and run on a 10% polyacrylamide gel, transferred, and probed with affinity purified anti-Kar5p antibodies. (A) The kar5-Δ2 strain MS3986 treated with α-factor. (B) MS3987 without pheromone treatment, (C) MS3987 after 2.5 h pheromone treatment. The band corresponding to Kar5p is indicated by an arrow. Uninduced levels of Kar5p were not detectable by Western blot. Kar5p corresponds to a 58-kD protein that was expressed at substantially greater levels in pheromone-treated cells. A contaminating band, indicated with an asterisk (*), corresponded to a cell membrane protein (as determined by fractionation), which could be removed with successive preadsorption in other experiments (see Materials and Methods). This cross-reacting protein did not contribute to staining in immunofluorescence experiments (Fig. 9).
Figure 7.
Kar5p extraction from nuclei. To determine whether Kar5p was a lumenal, integral, or peripheral membrane protein, a purified membrane fraction (derived from MS3987 after 2.5-h α-factor induction) was subjected to various extraction conditions (see Materials and Methods): 1 M NaCl; 0.1 M Na2CO3, pH 11.0; 2 M urea; 1% Triton X-100; 1% Triton X-100, 0.5 M NaCl; and buffer alone. As a control, an equivalent membrane fraction from the kar5-Δ strain, MS3986, was used to confirm the identity of the protein species. After incubation for 1 h at 4°C, the samples were centrifuged at 128,000 g. The supernatant was collected, and the pellet was resuspended in the original volume. Equal volumes of supernatant (S) and pellet (P) were analyzed by Western blot. Blots were probed with anti-Sec61p, anti-Kar2p, anti-Sec72p, and anti-Kar5p antibodies.
To determine if Kar5p was enriched in nuclear membranes, nuclei were isolated by the method of Aris and Blobel (1991). The enriched nuclear proteins were electrophoretically separated by SDS-PAGE and analyzed by protein blot. Kar5p was found to be highly enriched in the purified nuclei (Fig. 6). As a control, we also examined the ER/nuclear envelope protein Kar2p and found it to be enriched in the nuclear fraction.
Figure 6.
Kar5p is enriched in isolated nuclei. Equal amounts of protein (20 μg), isolated from whole cells (A and B) or purified nuclei (C and D) were loaded onto 10% SDS polyacrylamide gels, transferred, and probed with anti-Kar5p and anti-Kar2p antibodies (1/50 of the protein was loaded for the Kar2p blot). In lanes A and C, protein extracts were derived from MS3987 after 2.5 h of α-factor induction. In lanes B and D, protein extracts were derived from the kar5-Δ2 strain MS3986, which had also been treated with α-factor for 2.5 h. Both Kar2p and Kar5p were highly enriched in the nuclear fractions.
The sequence data suggested that Kar5p was an integral membrane protein. After low-speed (10,000 g) centrifugation of crude yeast lysates, all detectable Kar5p was found in the pellet fraction (not shown). These results were consistent with Kar5p being a nuclear protein and suggested that Kar5p might be a membrane protein. To determine directly whether Kar5p was a membrane protein, we attempted to extract Kar5p from whole nuclei using a variety of treatments (Fig. 7). For controls, we also followed the extraction properties of Sec72p, a peripheral nuclear envelope protein; Kar2p, a lumenal protein; and Sec61p, an integral membrane protein. As previously reported, Sec72p was not released under conditions of high salt (1 M NaCl) or high pH (0.1 M Na2CO3, pH 11.0) but was partially released by detergent (1% Triton X-100) (Feldheim and Schekman, 1994). Sec72p was completely released by the combination of detergent and high salt (1% Triton X-100 + 0.5 M NaCl). Kar2p was partially released under most conditions but was extensively released by treatments that disrupt membrane integrity, such as high pH and detergent. The most extensive release was observed with the combination of detergent and high salt. Sec61p was slightly released with detergent treatment and released completely in the presence of 0.5 M NaCl (Stirling et al., 1992). Kar5p was not extracted by conditions that released the lumenal membrane proteins nor was it extracted by detergent alone. However, Kar5p was partially solubilized by the presence of detergent and high salt. Taken together, these results indicate that Kar5p is most likely an integral membrane protein. The relatively poor extraction of Kar5p by detergent suggests that Kar5p may be complexed with other proteins in the nuclear envelope.
To determine the orientation of Kar5p with respect to the ER membrane/nuclear envelope, the sensitivity of Kar5p to protease was examined. Proteins exposed on the surface of the membrane are sensitive to exogenous proteases. In contrast, proteins contained within the lumen between the inner and outer nuclear membranes are resistant to the addition of proteases. Lumenal proteins would become sensitive to proteases upon addition of detergent to disrupt the membrane. Sec72p, a peripheral ER/nuclear envelope protein (Feldheim and Schekman, 1994), was sensitive to proteinase K (Fig. 8) regardless of the presence of detergent. In contrast, Kar2p, an ER-lumenal protein (Normington et al., 1989; Rose et al., 1989), was resistant to proteinase K in the absence of detergent but sensitive in the presence of detergent. Like Kar2p, Kar5p was resistant to protease in the absence of detergent but sensitive in its presence. The detergent-dependent protease sensitivity of Kar5p indicates that the protein was not inherently protease resistant. Furthermore, the fact that Kar5p was not significantly trimmed by protease in the absence of detergent suggests that very little if any of the protein is exposed on the cytoplasmic side of the membrane. This is consistent with the observation that the putative transmembrane domains are present at the extreme carboxyl terminus of the protein. Taken together, these results demonstrate that Kar5p is a nuclear envelope/ER membrane protein, anchored in the membrane by one or more hydrophobic transmembrane domains and topologically oriented such that most or all of the hydrophilic portion of the protein is situated within the ER/nuclear envelope lumen.
Figure 8.
Kar5p was contained within the ER/nuclear envelope lumen. Protease protection was used to determined if Kar5p was exposed to the cytoplasm or sequestered within the ER between the inner and outer nuclear envelopes. Membrane fractions from pheromone-induced cells (MS3987) were either treated with proteinase K or proteinase K with Triton X-100 for 0, 1, 2, 5, 10, and 20 min. After protease treatment, proteins were separated by SDS gel electrophoresis, transferred to nitrocellulose, and probed with anti-Kar5p, anti-Kar2p, or anti-Sec72p antibodies. Sec72p is an ER protein exposed to the cytoplasm (Feldheim and Schekman, 1994), and Kar2p is an ER lumenal protein (Rose et al., 1989). Sec72p was nearly entirely digested by protease after 20 min regardless of whether Triton X-100 was added. Kar2p and Kar5p were protease protected in the absence of detergent for >20 min. Kar2p and Kar5p were not inherently protease resistant since neither was resistant to proteinase K when Triton X-100 was added; the solubilization of the nuclear membrane allowed the protease access to lumen proteins. Thus, both Kar2p and Kar5p are ER/nuclear envelope lumenal proteins.
Kar5p Is Associated with the Spindle Pole Body during Shmoo Formation
To better define the cellular location of Kar5p, cells were stained by indirect immunofluorescence using the affinity-purified anti-Kar5p antibodies. Since Kar5p staining was found to be sensitive to formaldehyde fixation, cells were either fixed in paraformaldehyde or processed using the methanol-acetone method of Rout and Kilmartin (1990; see Materials and Methods). Preadsorbed affinity-purified anti-Kar5p antibody was used for indirect immunofluorescence on wild-type, kar5-Δ, and Kar5p overexpressing strains. DAPI staining was used to visualize the nucleus. Kar5p was detectable at wild-type levels in pheromone-treated cells fixed in methanol-acetone. However, by this method the overall cellular morphology was poorly preserved. Paraformaldehyde fixation followed by SDS extraction resulted in better preservation of the cellular morphology, but Kar5p was then detectable only in pheromone-treated cells containing KAR5 on a multicopy plasmid. Kar5p was not detected in vegetative cells or in zygotes by either method.
In wild-type cells, and in cells in which Kar5p was overexpressed, Kar5p localization was confined to a spot on the periphery of the nucleus (Figs. 9 and 10). Most cells had a single large spot of localization, but some cells had one or more additional smaller spots along the nuclear periphery (71% had one spot, 19% had two spots, and 10% had three spots, out of 105 cells counted). The minor spots were largely observed in cells using methanol-acetone fixation and only rarely in cells fixed with paraformaldehyde. The minor spots may therefore result from inadequate fixation.
Figure 9.

Kar5p was localized at or in the vicinity of the spindle pole body after pheromone induction. To determine whether Kar5p was localized to the spindle pole body, the fixation method of Rout and Kilmartin (1990) was used. Before fixation and staining, cells were induced with pheromone until >80% of the cells had formed shmoos. Indirect immunofluorescence using antibodies against Kar5p and the 90-kD SPB antigen were performed on wild-type (A–C), KAR5 overexpressing (D–F), and kar5-Δ2 (G–I) strains. The nucleus was identified by DAPI staining shown in A, D, and G. Localization of the 90-kD protein is shown in C, F, and I and indicates the position of the SPB on the nucleus. Localization of Kar5p is shown in B and E. Kar5p largely colocalized with the 90-kD protein and was not detected in the kar5-Δ2 control strain (H, MS3986). Of 48 cells counted, 77% (37/48) exhibited Kar5p staining and 87% (41/48) exhibited 90-kD staining.
Figure 10.
Kar5p was localized to the nuclear periphery near the shmoo projection. In each series, the leftmost panel shows whole cell (shmoo) morphology by DIC, the middle shows the Kar5p indirect immunofluorescence, and the rightmost panel shows nuclei by DAPI staining. Kar5p was observed as a spot on the nuclear periphery in (A) Kar5+ shmoos (MS3987) and in (C) kar5-1162 shmoos (MS4024), but Kar5p staining was not visible in (B) kar5-Δ2 shmoos (MS3986). Cells were treated with α-factor for 2–2.5 h, after which time 80% or more of the cells were morphologically distinct shmoos.
To identify the location of the Kar5p spot, we performed colocalization experiments using a monoclonal antibody specific for the 90-kD protein, a major protein of the spindle pole body (SPB). For this experiment, detection of the 90-kD antigen requires use of acetone-methanol fixation. In all cells, a Kar5p spot colocalized with the 90-kD SPB marker (Fig. 9, compare B with C and E with F). In 74% of cells (50 of 68 shmoos counted), the major Kar5p spot and the 90-kD protein colocalized. In the remaining cells, one of the minor Kar5p spots colocalized with the 90-kD protein. To demonstrate that Kar5p staining was specific and not due to bleedthrough fluorescence from the 90-kD staining, the colocalization experiment was performed on kar5-Δ2 cells. Kar5p was not detected in kar5-Δ2 cells (Fig. 9 C). Furthermore, Kar5p/SPB staining was not observed in the absence of anti-Kar5p antibody (data not shown).
To examine the location of the Kar5p spot with relation to the overall cell morphology, paraformaldehyde-fixed shmoos were stained with the preadsorbed/affinity-purified anti-Kar5p antibodies (Fig. 10). By this method, 75% of all wild-type shmoos (85/113) stained with the anti-Kar5p antibody. Kar5p was present as a spot or bar on the nuclear periphery (Fig. 10, middle). Consistent with SPB localization (Byers and Goetsch, 1974; 1975), the Kar5p spot was observed on the face of the nucleus pointed towards the shmoo tip in 77% of wild-type shmoos examined (57 stained cells). To test the specificity of the anti-Kar5p staining under these conditions, immunofluorescence was performed on kar5-Δ2 shmoos (Fig. 10 B, middle); as before, in kar5-Δ2 cells, the Kar5p spot was not detected. Thus, by two different fixation and staining techniques, Kar5p was observed as a spot on the nuclear periphery. Based upon these localization data, we conclude that Kar5p is associated with the SPB during mating, where it would be in position to participate in nuclear membrane fusion during karyogamy.
Discussion
KAR5 Is a Mating-induced Gene Required for Nuclear Membrane Fusion
KAR5 is a gene required for the membrane fusion between haploid nuclei during mating in Saccharomyces cerevisiae (Kurihara et al., 1994). In this paper, we report the cloning and sequencing of the KAR5 gene, the phenotype of KAR5 gene disruptions, and the subcellular localization of the Kar5 protein. KAR5 is a nonessential gene encoding a novel 58.4-kD membrane protein with predicted coiled-coil domains.
The properties of KAR5 and its encoded protein indicate that Kar5p plays a central role in nuclear envelope fusion during karyogamy: (a) Strains bearing a disruption of the KAR5 gene manifested severe karyogamy defects in bilateral or “self” matings. As for the original KAR5 mutants (Kurihara et al., 1994), the two nuclei were found closely apposed but unfused in zygotes formed during the mating of kar5-Δ haploids. (b) Expression of KAR5 was induced upon addition of pheromone consistent with a specific role in nuclear membrane fusion during karyogamy, and induction was dependent upon the STE12 transcriptional activator. Thus, KAR5 joins a growing list of genes including FUS1, FUS2, MFA1, CIK1, KAR3, and KAR4 that are downstream targets of the mating-specific MAP kinase–activated transcriptional regulon (for recent reviews see Sprague and Thorner, 1992; Rose, 1996). Indeed, KAR5 has been identified recently as FIG3 in a screen for genes that are specifically induced by mating pheromone (Erdman, S., and M. Snyder, personal communication) (c) Kar5p was found to be a nuclear membrane protein. Thus, Kar5p is associated with the membrane whose fusion it helps mediate. (d) The dependence of nuclear membrane fusion on Kar5p has been recapitulated in vitro. It was previously shown that the original kar5 mutants are defective for membrane fusion in vitro (Kurihara et al., 1994). Recently, it has been shown that, like the original kar5 mutants, the kar5-Δ strains are also defective in the in vitro ER homotypic fusion assay (Latterich, M., personal communication). The fusion defect could not be rescued by the addition of wild-type cytosol, and anti-Kar5p antisera did not inhibit the fusion of wild-type membranes in vitro (Latterich, M., personal communication). These observations are consistent with our findings that Kar5p sedimented with membranes and was not present in cytosolic fractions. Moreover, Kar5p was localized within the ER/nuclear envelope lumen, shielded from exogenously added proteases and potentially inhibitory antibodies. (e) Lastly, in shmoos Kar5p was localized to the vicinity of the SPB, the initial site of membrane fusion between zygotic nuclei. This localization at the SPB in shmoos was observed before cell fusion or karyogamy began. Based on these results, it is clear that Kar5p plays a critical function in membrane fusion during karyogamy.
kar5 Is a Bilateral Karyogamy Mutation Affecting a “Nondiffusible” Protein
Mutant alleles of KAR5 were identified in a genetic screen specifically tailored to identify bilateral karyogamy mutants (Kurihara et al., 1994). The previously isolated kar5-486 and kar5-1162 alleles, as well as kar5-Δ alleles, only manifested karyogamy defects when both partners in the mating were mutant. The bilateral defect of some KAR mutants may be a consequence of the special conditions that arise during karyogamy. After cell fusion but before nuclear fusion, both haploid nuclei briefly coexist in the same zygotic cytoplasm. In the common milieu of the zygote, gene products are free to diffuse between nuclei. In a bilateral mutant, the karyogamy defect may not be manifested in matings with wild-type cells either because the corresponding wild-type factor is free to diffuse to the mutant nucleus, where it can suppress the defect, or because it is required in only one of the two nuclei to promote nuclear fusion. In contrast, recessive unilateral mutants, like those represented by some of the initially identified KAR genes (Conde and Fink, 1976; Polaina and Conde, 1982), may be the result of compartmentalization or membrane attachment of corresponding gene products. For example, Kar2p is not free to diffuse between nuclei because it is restricted to the ER lumen (Normington et al., 1989; Rose et al., 1989), and Kar1p is anchored to the spindle pole body (Vallen et al., 1992). However, if Kar5p is compartmentalized within the ER lumen, why are KAR5 mutants bilaterally defective? It seems unlikely Kar5p is required in only one of the two nuclei to promote nuclear fusion because it was required in both nuclei for in vitro homotypic membrane fusion (Kurihara et al., 1994). It also seems unlikely that mature Kar5p is diffusible between the nuclei. However, it is likely that KAR5 mRNA and nascent Kar5p are freely transferred. During karyogamy, Kar5p levels are substantially increased as a result of STE12-mediated transcriptional induction. As a result, in the common milieu of the zygote, the induced KAR5 mRNA from one haploid nucleus can be translated on the ER associated with either nucleus, effectively transferring Kar5p to the mutant nucleus. In contrast, neither KAR1 and KAR2 are induced by mating pheromone. Accordingly, we propose that for KAR1 and KAR2, the levels of mRNA expressed during mating would be insufficient to deliver enough wild-type gene product to the mutant nucleus to correct the defect in unilateral matings.
Possible Functions of KAR5 during Nuclear Membrane Fusion
Assuming that nuclear envelope fusion is similar to other membrane fusion systems, there must be several steps to the overall process (for review see White, 1990). First, microtubule-dependent congression would serve to bring the two nuclei close together. Second, there must be a docking/recognition event in which the fusogen proteins in the two membranes are brought into close apposition. Third, the initiation of fusion between the outer membrane leaflets would result in “hemifusion intermediates.” Fourth, the opening of the pore and its dilation would complete the outer membrane fusion event. The subsequent events of nuclear envelope fusion are less clear. Two different models have been proposed that differ in their requirements for additional inner nuclear membrane fusion events to produce the single diploid nucleus (Rose, 1996).
In keeping with other membrane fusion reactions, we presume that the proteins mediating the initial steps of nuclear envelope fusion, docking and initiation, would extend out of the nuclear envelope into the cytoplasm. However, as determined by protease protection, Kar5p was confined within the lumen of the ER/nuclear envelope, with no significant exposed cytoplasmic domain. As such, it is unlikely that Kar5p could directly mediate the recognition, docking, or initiation of membrane fusion. More likely is that Kar5p is required for the completion of membrane fusion once initial contact between nuclei has been made. Consistent with this hypothesis is the observation that electron micrographs of kar5 mutant zygotes showed small lipid bridges spanning the cytoplasm between the two nuclear envelopes (Kurihara et al., 1994; Beh, 1996). According to these studies, the two nuclei made several limited contacts in the mutant zygotes but, as judged by both biochemical and genetic assays, nevertheless failed to complete fusion. In this view, Kar5p may be a factor required to dilate or expand the initial fusion pore. One possibility is that Kar5p could directly promote fusion by destabilizing the membrane bilayer from the underside. Such an activity has been demonstrated for amphipathic peptides, which can aid in the conversion of hemifusion intermediates (Melikyan et al., 1997).
Other aspects of Kar5p's function in karyogamy are suggested by its regulation and localization. After pheromone induction, Kar5p accumulated at the spindle pole body in shmoos. Therefore, Kar5p can localize to the eventual site of membrane fusion before cell or nuclear fusion. Indeed, to date Kar5p is the only protein required for nuclear membrane fusion that is pheromone induced. Furthermore, none of the other proteins required for nuclear membrane fusion that have been subcellularly localized are specifically localized to the spindle pole body. Therefore, Kar5p might regulate nuclear fusion by virtue of its transcriptional regulation and subcellular localization to designate the timing and location of membrane fusion. In this view, Kar5p might also serve as a scaffold for the localization of the other proteins required for fusion to the vicinity of the spindle pole body.
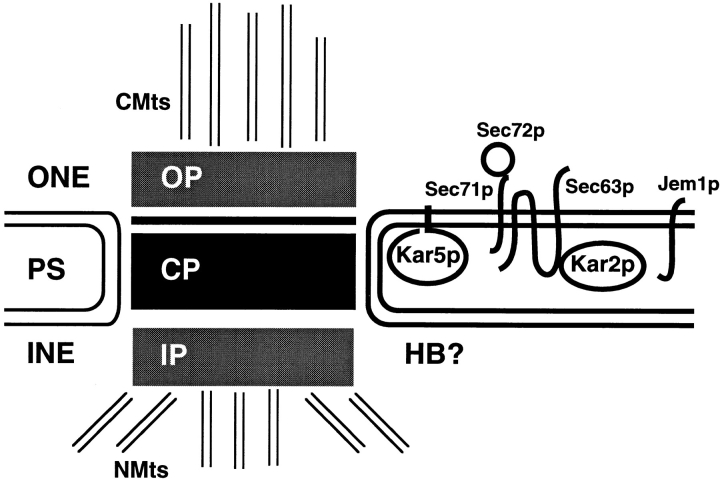
As diagrammed in Fig. 11, the SPB is a multiprotein complex of at least three layers; a dense central plaque is sandwiched between two plaques of lesser density (Winey and Byers, 1993). In pheromone-arrested cells, one edge of the central plaque abuts the “half-bridge,” a region of darkly staining nuclear envelope. Nuclear envelope fusion initiates along one edge of the half-bridge and continues along the SPB such that two SPBs fuse to a form a single enlarged SPB (Byers and Goetsch, 1975). A crease marking the edge of fusion can be observed in the zygotic SPB.
Figure 11.
Diagram of the spindle pole body with membrane fusion Kar proteins. In yeast, the disk-shaped spindle pole body assembles both the cytoplasmic and nuclear microtubules. In pheromone-arrested cells, on one edge of the central plaque lies the “half-bridge,” which sits within the nuclear envelope, and on the furthest end of the half-bridge is the “satellite,” an amorphous region of high electron density (not shown). The half-bridge and the satellite are the initial sites of nuclear and spindle pole body fusion (Byers and Goetsch, 1975). Since Kar5p was associated with the spindle pole body during mating, Kar5p may facilitate nuclear membrane fusion at the half-bridge or satellite. The other characterized fusion proteins, Kar2p, Jem1p, Sec63p, Sec72p, and Sec71p/Kar7p, are localized over the entire nuclear periphery. To date, Kar5p is the only identified Kar protein specifically localized to the spindle pole body. OP, outer plaque; CP, central plaque; HB, half bridge; IP, inner plaque; NMts, nuclear microtubules; CMts, cytoplasmic microtubules; ONE, outer nuclear envelope; INE, inner nuclear envelope; PS, periplasmic space/ER lumen.
Kar5p was clearly localized to the region of the SPB by immunofluorescence. However, Kar5p was more diffusely localized than the 90-kD SPB component, suggesting that Kar5p was associated with but not actually a central component of the SPB. A similar SPB-associated localization pattern has been described for the Nuf2 protein (Osborne et al., 1994). Given this pattern, Kar5p may be associated with the half-bridge and nuclear membrane adjacent to the SPB. Determination of the precise localization of Kar5p awaits future studies by immunoelectron microscopy.
Interactions between Membrane Fusion-specific KAR Genes
A number of genes have been reported to be required in vivo for the membrane fusion step of karyogamy, including JEM1, KAR2, KAR5, KAR7, KAR8, SEC63, SEC71, and SEC72 (Polaina and Conde, 1982; Kurihara et al., 1994; Ng and Walter, 1996; Nishikawa and Endo, 1997). All of the characterized protein components required for nuclear membrane fusion are depicted in Fig. 11. Several of the genes were first identified because of their roles in the translocation of proteins into the lumen of the ER or because their cognate proteins interact with the translocation machinery. SEC63, SEC71, and SEC72 were originally identified in screens for translocation-defective mutants (Toyn et al., 1988; Green et al., 1992). Sec63p is a DnaJ homologue resident in the ER/nuclear envelope (Sadler et al., 1989). Both genetic and biochemical experiments demonstrate that Sec63p and Kar2p interact (Brodsky and Schekman, 1993; Scidmore et al., 1993). SEC71 was also identified as a high copy suppressor of a mutation in SEC63 (Kurihara and Silver, 1993) and biochemically as a Sec63p interacting protein (Feldheim et al., 1993). KAR7 has recently been discovered to be identical to SEC71 (Brizzio, V., and M.D. Rose, unpublished data). Sec71p, Sec72p, and Kar2p copurify with Sec63p as a complex (Brodsky and Schekman, 1993). Finally, Jem1p was recently identified as another DnaJ homologue resident in the nuclear envelope (Nishikawa and Endo, 1997). The jem1 mutant results in a strong defect in nuclear fusion but has little effect on vegetative growth.
The requirement of several of the nuclear fusion proteins for both translocation and nuclear fusion raises the question of whether they act directly in nuclear fusion or are required only for the translocation of a protein like Kar5p into the ER. Several genetic and biochemical results suggest that these proteins play a role in fusion that is distinct from their role in translocation. First, there exist mutant alleles of KAR2 that have strong defects in nuclear fusion that have little if any defects in translocation (Vogel et al., 1990). Second, Kar2p and Kar7p/Sec71p are required in vitro for homotypic membrane fusion, under conditions in which protein translocation is not occurring (Latterich and Schekman, 1994). Indeed, in this assay temperature-sensitive kar2 and sec71/kar7 mutations cause temperature-sensitive defects in vitro (Kurihara et al., 1994; Latterich and Schekman, 1994; Brizzio, V., M. Latterich, M.D. Rose, unpublished data). These data suggest that both Kar2p and Sec71p directly affect nuclear fusion and are not simply required for the translocation of fusion proteins into the nuclear envelope/ER. One reasonable suggestion is that the Kar2p, Sec63p, Sec71p, and Sec72p are components of a protein complex with a primary role in translocation and a secondary role in membrane fusion (Ng and Walter, 1996).
With respect to nuclear fusion, mutations in KAR5 and KAR7 showed complex genetic behaviors that suggested that their cognate proteins may interact or be functionally related. For example, the mutations kar7-1039 and kar5-1162 exhibited unlinked noncomplementation, whereas kar7-1039 and kar5-468 showed normal complementation behavior (Kurihara et al., 1994). Furthermore, although neither kar7 nor kar5 mutants show strong nuclear fusion defects when crossed to wild-type strains (the bilateral mutant phenotype), they exhibit a strong defect when crossed against one another (Brizzio, V., and M.D. Rose, unpublished). One explanation for these genetic interactions is that the functions of Kar5p and Kar7p in nuclear fusion are in some way interdependent. For example, one attractive hypothesis is that Kar5p and Kar7p/Sec71p are components of the same nuclear fusion protein complex. Kar5p might serve to localize and concentrate a subset of Sec63p/Kar2p/ Sec71p/Sec72p complexes to the vicinity of the SPB where nuclear fusion will take place.
The location, regulation and mutant phenotypes of Kar5p demonstrate that it is a unique protein required specifically for nuclear membrane fusion during mating. Future directions for research will include elucidating its interactions with other components in the ER/nuclear envelope as well as its specific role in membrane fusion during mating and mitosis.
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- GST
glutathione-S-transferase
- PRE
pheromone response element
- SC
synthetic complete
- SPB
spindle pole body
Footnotes
Thanks to Jasper Rine, Sue Biggins, and Nancy Hawkins for critical reading of this manuscript and to Jim Broach, Gerry Waters, Scott Ederman, Mike Snyder, and Martin Latterich for reagents, useful comments, and communicating unpublished results. We also thank Don Huddler and especially Laurie Jo Kurihara for invaluable suggestions and comments.
This research was supported by a National Institutes of Health (NIH) grant (GM37739) to M.D. Rose. C.T. Beh and V. Brizzio were partially supported by institutional NIH Genetics and Cell and Molecular Biology Training grants.
Address all correspondence to Mark D. Rose, Department of Molecular Biology, Princeton University, Princeton, NJ 08544-1014. Tel.: (609) 258-2804. Fax: (609) 258-6175. E-mail: mrose@watson.princeton.edu
Christopher Beh's current address is Department of Molecular and Cellular Biology, 401 Barker Hall, University of California, Berkeley, CA 94720.
References
- Acharya U, Jacobs R, Peters J-M, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- Beh, C.T. 1996. Functions of the yeast endoplasmic reticulum. Ph.D. Thesis, Princeton University, Princeton, NJ. 178pp.
- Berlin V, Styles CA, Fink GA. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V, Brill JA, Trueheart J, Boeke JD, Fink GR. Genetic screens and selections for cell and nuclear fusion mutants. Methods Enzymol. 1991;194:774–792. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A dye binding assay for protein. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 59–96.
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation in Saccharomyces cerevisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiaedefective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabora SL, Sheetz MP. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell. 1988;54:27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Conde J. Benomyl prevents nuclear fusion in Saccharomyces cerevisiae. . Mol Gen Genet. 1984;193:188–189. doi: 10.1007/BF00327435. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Yoshimura K, Admon A, Schekman R. Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol Biol Cell. 1993;4:931–939. doi: 10.1091/mbc.4.9.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N, Fang H, Walter P. Mutants in three novel complementation groups inhibit membrane protein insertion into and soluble protein translocation across the endoplasmic reticulum membrane of Saccharomyces cerevisiae. . J Cell Biol. 1992;116:597–604. doi: 10.1083/jcb.116.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis . Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. . Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMBASE—a database of membrane spanning protein segments. Biol Chem. 1993;374:166. [Google Scholar]
- Hopp TP. Use of hydrophilicity plotting procedures to identify protein antigenic segments and other interaction sites. Methods Enzymol. 1989;178:571–585. doi: 10.1016/0076-6879(89)78040-x. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Hoyt MA, Botstein D. Genetic analysis of the yeast cytoskeleton. Annu Rev Genet. 1987;21:259–284. doi: 10.1146/annurev.ge.21.120187.001355. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Silver P. Suppression of a sec63mutation identifies a novel component of the yeast endoplasmic reticulum translocation apparatus. Mol Biol Cell. 1993;4:919–930. doi: 10.1091/mbc.4.9.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose M. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Stewart B, Gammie A, Rose MD. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol Cell Biol. 1996;16:3990–4002. doi: 10.1128/mcb.16.8.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli EK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Latterich M, Schekman R. The karyogamy gene KAR2and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Marsh, L., and M.D. Rose. 1997. The pathway of cell and nuclear fusion during mating in Saccharomyces cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces, Vol. 3, Cell Cycle and Cell Biology. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 827–888.
- McCaffrey G, Clay FJ, Kelsay K, Sprague GF. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. . Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Brener SCA, Ok DC, Cohen FS. Inner but not outer membrane leaflets control the transition from gylosylphosphatidylinositol-anchores influenza hemagglutinin-induced hemifusion to full fusion. J Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Ng DT, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiaeencodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- Osborne MA, Schlenstedt G, Jinks T, Silver PA. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. . Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the KAR3 kinesin heavy chain-like protein requires the CIK1interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaina J, Conde J. Genes involved in the control of nuclear fusion during the sexual cycle of Saccharomyces cerevisiae. . Mol Gen Genet. 1982;186:253–258. doi: 10.1007/BF00331858. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters J-M, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Riles L, Dutchik JE, Baktha A, McCauley BK, Thayer EC, Leckie MP, Braden VV, Depke JE, Olson MV. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiaeat a resolution of 2.6 kilobase pairs. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in yeast. Annu Rev Microbiol. 1991;45:539–567. doi: 10.1146/annurev.mi.45.100191.002543. [DOI] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. . Annu Rev Cell Dev Biol. 1996;12:663–695. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homologue of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods of Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Prot Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1917. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coliheat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scidmore MA, Okomura H, Rose MD. Genetic interaction between KAR2 and SEC63, eukaryotic homologues of HSP70 and DNAJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J Cell Biol. 1995;128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, G.F., and J. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In The Molecular Biology of the Yeast Saccharomyces, 2nd ed. J.R. Broach, J.R. Pringle, and E.W. Jones, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 657–744.
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn J, Hibbs AR, Sanz P, Crowe J, Meyer DI. In vivo and in vitro analysis of ptl1, a yeast ts mutant with a membrane-associated defect in protein translocation. EMBO (Eur Mol Biol Organ) J. 1988;7:4347–4353. doi: 10.1002/j.1460-2075.1988.tb03333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Misra L, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilson KL. Nuclear membrane dynamics. Curr Opin Cell Biol. 1993;5:387–394. doi: 10.1016/0955-0674(93)90002-8. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]