Abstract
Previously we isolated tub2-423, a cold-sensitive allele of the Saccharomyces cerevisiae gene encoding β-tubulin that confers a defect in mitotic spindle function. In an attempt to identify additional proteins that are important for spindle function, we screened for suppressors of the cold sensitivity of tub2-423 and obtained two alleles of a novel gene, STU2. STU2 is an essential gene and encodes a protein whose sequence is similar to proteins identified in a variety of organisms. Stu2p localizes primarily to the spindle pole body (SPB) and to a lesser extent along spindle microtubules. Localization to the SPB is not dependent on the presence of microtubules, indicating that Stu2p is an integral component of the SPB. Stu2p also binds microtubules in vitro. We have localized the microtubule-binding domain of Stu2p to a highly basic 100-amino acid region. This region contains two imperfect repeats; both repeats appear to contribute to microtubule binding to similar extents. These results suggest that Stu2p may play a role in the attachment, organization, and/or dynamics of microtubule ends at the SPB.
The microtubule-organizing center (MTOC)1 of eukaryotic cells controls the number, polarity, and organization of cellular microtubules. In the yeast Saccharomyces cerevisiae, the MTOC is the spindle pole body (SPB), which is embedded in the nuclear envelope throughout the cell cycle and nucleates microtubules that extend into the nucleus and cytoplasm. The SPB is a trilaminar disk consisting of a central plaque and flanking inner and outer plaques on the nuclear and cytoplasmic surfaces, respectively (Byers, 1981). The inner and outer plaques are the sites of microtubule association in vivo and are likely to be involved in the nucleation of microtubule polymerization and in anchoring microtubules to the SPB.
The nucleation of microtubules is thought to be accomplished through the action of the γ-tubulin complex. γ-Tubulin is localized to the MTOC in animal cells (Stearns et al., 1991; Zheng et al., 1991) and fungi (Oakley et al., 1990; Horio et al., 1991; Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996). γ-Tubulin is also present in the cytoplasm of animal cells as a component of a large, ring-shaped structure that is capable of nucleating microtubules (Stearns and Kirschner, 1994; Zheng et al., 1995). A similar γ-tubulin–containing ring structure has been identified in MTOCs at the base of nucleated microtubules (Moritz et al., 1995). γ-Tubulin is required for centrosome function in vivo (Joshi et al., 1992) and in vitro (Felix et al., 1994; Stearns and Kirschner, 1994) and is essential for the viability of fungi (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996). The phenotypes of S. cerevisiae γ-tubulin mutants are consistent with a role for this protein in microtubule nucleation (Marschall et al., 1996; Spang et al., 1996).
The relationship between microtubule nucleation and microtubule anchoring to the MTOC is not clear. Microtubules are dynamic polymers during mitosis. The observation of poleward microtubule flux in mitotic spindles (Mitchison, 1989; Sawin and Mitchison, 1991) implies that the attachment of microtubules to MTOCs must be arranged in a way that allows the exchange of tubulin subunits at the microtubule ends. One way for the MTOC to accomplish this task would be for it to make lateral attachments to microtubules. Such attachments could anchor microtubules at the MTOC and simultaneously allow subunit exchange at microtubule ends. In this report we describe a protein, Stu2p, that may play a role in anchoring and organizing microtubules at the S. cerevisiae MTOC. Stu2p is an integral component of the SPB and is capable of binding laterally to microtubules.
Materials and Methods
Yeast Strains, Media, and Plasmids
The yeast strains used in this study are listed in Table I. Yeast growth media were prepared as described by Sherman (1991). To depolymerize microtubules, cells were grown in 20 μg/ml nocodazole for 1 h at 30°C and then 1 h at 4°C.
Table I.
Yeast Strains
| Strain | Genotype | |
|---|---|---|
| CUY26 | MATα his3-Δ200 leu2-3,112 ura3-52 | |
| CUY30 | MATα his3-Δ200 leu2-3,112 ura3-52 ade2 lys2-801 | |
| CUY502 | MATa tub2-423::URA3 ACT1::HIS3 TUB1::LEU2::TUB1 met2 his3-Δ200 leu2-Δ1 lys2-801 ura3-52 | |
| CUY546 | MATa/MATα ade2/ADE2 his3/Δ200/his3-Δ200 leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | |
| CUY696 | MATα tub2-423::URA3 ade2-101 his3-Δ200 leu2-Δ1 lys2-801 ura3-52 | |
| CUY935 | MATα tub2-423::URA3 STU1::HIS3 ade2-101 his3-Δ200 leu2-Δ1 lys2-801 ura3-52 | |
| CUY936 | MATa tub2-423::URA3 STU1::HIS3 ade2-101 his3-Δ200 leu2-Δ1 lys2-801 ura3-52 | |
| CUY1042 | MATa tub2-423::URA3 ACT1::HIS3 stu2-1 ade2-101 his3 leu2 lys2-801 ura3-52 | |
| CUY1045 | MATa tub2-423::URA3 stu2-2 ade2-101 his3 leu2 lys2-801 ura3-52 | |
| CUY1046 | MATa/MATα stu2-Δ1::HIS3/STU2 ade2/ADE2 his3-Δ200/his3-Δ200 leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | |
| CUY1047 | MATa/MATα tub2-423::URA3/tub2-423::URA3 stu2-1/STU2 ACT1::HIS3/ACT1 TUB1::LEU2::TUB1/TUB1 | |
| met2/MET2 leu2/leu2 his3/his3 lys2-801/lys2-801 ura3-52/ura3-52 | ||
| CUY1060 | MATa/MATα stu2-Δ1::HIS3/stu2-Δ1::HIS3 leu2-Δ1/leu2-Δ1 ura3-52/ura3-52 ade2-101/ADE2 | |
| his3-Δ200/his3-Δ200 (pWP89 pWP90) | ||
| CUY1065 | MATa/MATα stu2-Δ1::HIS3/stu2-Δ1::HIS3 leu2-Δ1/leu2-Δ1 ura3-52/ura3-52 ade2-101/ADE2 | |
| his3-Δ200/his3-Δ200 (pWP90) | ||
| CUY1066 | MATa/MATα stu2-Δ1::HIS3/stu2-Δ1::HIS3 leu2-Δ1/leu2-Δ1 ura3-52/ura3-52 ade2-101/ADE2 | |
| his3-Δ200/his3-Δ200 (pWP89) | ||
| CUY1067 | MATa stu2-Δ1::HIS3 his3-Δ200 leu2-3,112 ura3-52 (pWP70) | |
| CUY1068 | MATa STU2-GFP::URA3 his3-Δ200 ura3-52 leu2-3, 112 | |
| CUY1069 | MATa/MATα stu2-Δ2::HIS3/STU2 ade2/ADE2 his3-Δ200/his3-Δ200 leu2-3, 112/leu2-3,112 ura3-52/ura3-52 |
Selected plasmids are listed in Table II.
Table II.
Plasmids
| Plasmids | Relevant markers | |
|---|---|---|
| pS2 | stu2-1 LEU2 CEN6 ARSH4 | |
| pWP44 | stu2-2 LEU2 CEN6 ARSH4 | |
| pWP45 | STU2 LEU2 CEN6 ARSH4 | |
| pWP70 | STU2-HA3 LEU2 CEN6 ARSH4 | |
| pWP89 | STU2-GFP LEU2 CEN6 ARSH4 | |
| pWP90 | STU2-GFP URA3 2 μm origin |
Isolation of Spontaneous Suppressors of tub2-423
100 individual colonies (∼107 cells/colony) of strain CUY696 were each resuspended in 100 μl of sterile water and spread onto separate YPD plates. Colonies that arose after incubation at 16°C for 8 d were retested for growth at 16°C. Those that retested were mated to CUY502 to determine whether the suppression was dominant or recessive. The resulting diploids were then sporulated, and tetrads were dissected to determine whether suppression is due to a mutation in a single locus. These crosses were also used to establish whether the mutations are linked to any of the tubulin genes (TUB1, TUB2, or TUB3) because the marked ACT1 locus is closely linked to the TUB2 locus and the marked TUB1 locus is linked to the TUB3 locus. The extragenic suppressors were put into linkage groups by crossing them in pairwise combinations. A suppressor strain from each linkage group was also crossed to CUY935 or CUY936 to determine whether each mutation is linked to the STU1 locus (Pasqualone and Huffaker, 1994).
Cloning and Sequencing the stu2-1 Allele
A centromere-based yeast genomic library was made from CUY1042 genomic DNA by the protocol of Rose and colleagues (Rose et al., 1987; Rose and Broach, 1991) with the following modifications. High molecular weight genomic DNA was isolated and partially digested with Sau3A. DNA fragments of 6 to 9 kb were recovered from a preparative agarose gel by electroelution and ligated into the BamHI site of pRS315 (Sikorski and Hieter, 1989). The ligated DNA was transformed into ElectroMAX DH10B™ competent cells (GIBCO BRL, Gaithersburg, MD) by electroporation. 66,000 transformants were recovered and pooled. 12 out of 15 plasmids examined had inserts ranging in size from 5.5 to 15 kb with an average size of 8.2 kb. The total genomic DNA content of this library is ∼31 times as large as yeast genome.
The stu2-1 allele was cloned by complementation of the cold-sensitive phenotype of the tub2-423 mutation. The yeast genomic library described above was transformed into CUY696. After 2 d at 30°C, 11,500 transformants were replica plated onto YPD plates and incubated at 16°C for 3 d. Then, these cells were replica plated again onto YPD plates and incubated at 16°C for 5 d. Three transformants were able to grow at 16°C. Plasmids recovered from these strains were able to suppress the tub2-423 phenotype after being retransformed into CUY696. The three plasmids, referred to as pS2, pS20, and pS28, contained 6.7-, 6.5-, and 10.5-kb DNA inserts, respectively, and had a 5-kb overlapping fragment, as shown by restriction mapping.
To verify that the authentic stu2-1 locus has been cloned, the 6.7-kb XhoI–XbaI fragment of pS2 (see Fig. 2) was subcloned into the integrating plasmid pRS305 (Sikorski and Hieter, 1989) to make plasmid pWP5. pWP5 was then linearized with BamHI, which cuts once within the insert, and transformed into CUY696 cells. Leu+ transformants were crossed to CUY1042 and the resulting diploids sporulated. In all tetrads dissected, only parental genotypes were observed; four spores in each tetrad grew at 16°C.
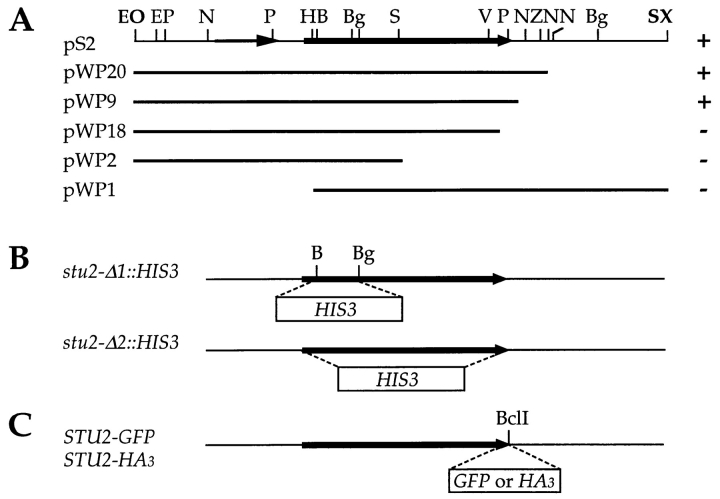
Figure 2.
STU2 subclones and plasmid constructs. (A) The top bar represents the 6.7-kb genomic insert in pS2 that contains stu2-1. The stu2-1 open reading frame is indicated by the wide arrow. A smaller ORF, YLR406c, upstream of stu2-1 is indicated by the thin arrow. Subclones of the 6.7-kb insert were made by digestion with Exonuclease III and Nuclease S1. Right column indicates whether each construct could (+) or could not (−) suppress the cold sensitivity of tub2-423. (B) The stu2-Δ1::HIS3 allele as created by replacing the 541-bp BamHI–BglII fragment with the 1.7-kb HIS3 gene. The stu2-Δ2::HIS3 allele was made by replacing the entire STU2 ORF with HIS3. (C) A BclI restriction site was introduced just before the stop codon STU2. A 120-bp fragment encoding the HA3 or a 739-bp fragment encoding GFP was cloned into this site. Abbreviations of the restriction sites: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; N, NsiI; O, XhoI; P, PstI; S, SpeI; V, EcoRV; X, XbaI; Z, SphI. Sites in bold are in the vector.
To physically map the STU2 gene, a 3-kb PstI–PstI fragment from the genomic insert of pS2 was used to probe λ phage containing clones of yeast genomic DNA (Riles et al., 1993). The probe hybridized to two clones, American Type Culture Collection (Rockville, MD) numbers 6122 and 4488. These two clones are ∼2 kb apart from each other on chromosome XII near PDC1.
To localize the stu2-1 gene within the cloned DNA fragment, a series of nested deletions starting from both ends of the pS2 insert was created using the combination of exonuclease III and nuclease S1 (Heinrich, 1991). Each of the resulting deletions was transformed into CUY696 to test its ability to suppress the cold-sensitive phenotype of tub2-423. The deletion constructs determined to span the stu2-1 locus were sequenced.
Cloning and Sequencing the STU2 and stu2-2 Alleles
Both the STU2 and stu2-2 alleles were recovered by plasmid gap repair (Rothstein, 1991). To prepare the gapped plasmid, pS2 was digested with NsiI to delete a 4.3-kb fragment containing the entire stu2-1 coding sequence, ∼1.2 kb of upstream sequence and ∼450 bp of downstream sequence. The remaining fragment was religated to create pWP42. pWP42 was linearized with NsiI, dephosphorylated, gel purified, and transformed into CUY1045 (stu2-2) and a diploid strain CUY1046 (STU2/stu2-Δ1:: HIS3) for the recovery of the stu2-2 and STU2 alleles, respectively. Plasmids were isolated from Leu+ transformants, transformed into Escherichia coli DH10BTM cells by electroporation, and analyzed by restriction mapping. Both STU2 (pWP45) and stu2-2 (pWP44) alleles were obtained. As expected, the stu2-Δ1::HIS3 allele was also recovered from CUY1046 cells.
To narrow down the location of mutations in the suppressor alleles, we interchanged the 3.3-kb SpeI fragment between pS2 (stu2-1) and pWP45 (STU2). This fragment contains 1.4 kb of the carboxy-terminal coding sequence of STU2. The same sequence-swapping experiment was done between pWP44 (stu2-2) and pWP45 (STU2). The resulting hybrid plasmids were transformed into CUY696 and tested for their ability to suppress the tub2-423 phenotype. This experiment showed that, for both stu2-1 and stu2-2 alleles, the suppressor mutations reside in the 3.3-kb SpeI fragment. The 1.4-kb carboxy-terminal coding regions of both STU2 and stu2-2 were sequenced from both strands with synthetic sequencing primers. The complete wild-type STU2 sequence has been submitted to EMBL/GenBank/ database (accession number U35247).
Disruption of STU2
The STU2 gene was disrupted by the one-step gene replacement method (Rothstein, 1991). pWP2 (see Fig. 2) was cut with BamHI and BglII, dephosphorylated and gel purified. A 1.7-kb BamHI fragment from pCU34 containing the HIS3 gene was ligated into the pWP2 backbone. In the resulting plasmid, pWP39, a 541-bp segment of stu2-1 encoding amino acids 57–235 is replaced by the 1.7-kb fragment containing the HIS3 gene (see Fig. 2). A 4.3-kb DNA fragment was liberated from pWP39 by digestion with both EcoRI and XbaI, gel purified, and transformed into the diploid strain CUY546. This disruption allele is referred to as stu2-Δ1::HIS3.
An exact deletion of STU2 open reading frame (ORF) was made by the replacement with the HIS3 gene. The two primers used for PCR amplification of HIS3 were 5′-TTACAGTGTAAAGTATTTTTGAGTTTTTATTAAAGAGTTTGAAGTTGACTGAGCTTGGTGAGCGCTAGG AG -3′ and 5′-AGTTGAAGACTATATATTTTATTGAGTTTATGTTATGGGGAGGCTACCCTCTCGTTCAGAATGACACGAT-3′. Full length primers were obtained by HPLC purification. The resulting 1.2-kb PCR product has 50-bp sequences at each end that are identical to the sequences immediately before and after the STU2 ORF, respectively, and therefore can direct homologous recombination of the HIS3 marker with the STU2 locus. His+ transformants were obtained by transforming the diploid strain CUY546 with the PCR product. The resulting disruption allele is referred to as stu2-Δ2::HIS3 (see Fig. 2).
Tagging Stu2p with Green Fluorescent Protein and the Influenza Hemagglutinin Epitope
A BclI site was engineered just before the stop codon in STU2 by PCR with the following two primers: 5′-ACGATTTCATCATACTCC-3′ and 5′-GGAGGCTACCCTTTATTGATCAGTCCTGGTTGTCCC-3′ (the stop codon is shown with boldface, and the inserted BclI site is underlined). The 384-bp fragment amplified from pWP45 with the above primers was subcloned into the TA cloning vector pCR™II (Invitrogen, Carlsbad, CA) to give rise to pWP59. The 171-bp EcoRV–EcoRV fragment of pWP59 was subcloned into the EcoRV site of pWP58 which contains the 2.6-kb BamHI–SphI fragment of pWP45 in the pTZ18U plasmid (Bio Rad, Richmond, CA), resulting in pWP61 with the right insert orientation.
To make the STU2–GFP fusion construct, the S65T mutant version of green fluorescent protein (GFP) was released from pRSETB-S65T (Heim et al., 1995) by digestion with BamHI and cloned into the engineered BclI site of pWP61 giving rise to pWP87. The 3.3-kb BamHI–SphI fragment of pWP87 was used to replace the 2.6-kb BamHI–SphI fragment of pWP45 and pWP81. pWP81 has the same stu2-1 insert as pS2 in the YEp vector pRS426 (Sikorski and Hieter, 1989). The resulting plasmids containing the STU2–GFP fusions are referred to as pWP89 (YCp) and pWP90 (YEp). STU2–GFP was also used to replace the chromosomal copy of STU2. A 1.2-kb fragment containing the URA3 gene was cloned into the SphI site of pWP89 to produce pXC277. The 5.5-kb SpeI–SpeI fragment of pXC277 was transformed into a wild-type haploid strain.
DNA encoding three tandem copies of the hemagglutinin (HA) epitope was amplified by PCR from the plasmid GTEPI (a gift of B. Futcher, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) with the following two primers: 5′-GAAGATCTTGGCCGCATCTTTTACCCA-3′ and 5′-GAAGATCTCCGCACTGAGCAGCGTAAT-3′. The PCR-amplified HA3 DNA has a BglII site on each end (underlined). The 120-bp BglII-restricted PCR product was ligated into the BclI site of pWP61 to produce pWP67. The 1.81-kb SpeI–SphI fragment of pWP67 was subcloned into the SpeI–SphI site of pWP20. The resulting YCp plasmid containing STU2-HA3 is designated pWP70.
To address whether the tagged Stu2p constructs are functional, pWP70, pWP89, and pWP90 were transformed independently into CUY1046 (STU2/stu2-Δ1::HIS3). Leu+ transformants were sporulated and tetrads dissected. In each case, viable His+ spores were always Leu+, demonstrating that the tagged STU2 constructs complement the stu2-Δ1::HIS3 disruption.
Fluorescence Microscopy
GFP was visualized either in living cells or cells that had been fixed with formaldehyde as described below. The green fluorescence of Stu2p–GFP is sensitive to fixation but recovers fluorescence at a reduced intensity after removal of formaldehyde. Cells were grown in SD medium in the presence of 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 2 to 4 h to visualize nuclear DNA in living cells.
Immunofluorescent staining of yeast cells was performed as described previously (Pasqualone and Huffaker, 1994). To visualize microtubules alone, cells were fixed by adding formaldehyde to a final concentration of 3.7% and incubating at 30°C for 1 h. To visualize the HA- or GFP-tagged Stu2p or γ-tubulin, cells were fixed for 25 min. Rat monoclonal anti–yeast α-tubulin antibody, YOL1/34 (Kilmartin et al., 1982), was a gift from J. Kilmartin (Medical Research Council, Cambridge, UK). Mouse monoclonal antibody 12CA5, which recognizes the HA epitope, was purchased from Berkeley Antibody Co.(Berkeley, CA). Rabbit anti–yeast γ-tubulin antibody, TUB4-1-4 (Marschall et al., 1996), was a gift from T. Stearns (Stanford University, Stanford, CA). Rhodamine-conjugated goat anti– rabbit and goat anti–rat and fluorescein-conjugated goat anti–mouse secondary antibodies were obtained from Cappel Research Products (Durham, NC). Before using them to visualize the HA tag, both 12CA5 and fluorescein-conjugated goat anti–mouse antibodies were preabsorbed with wild-type fixed yeast spheroplasts overnight at 4°C.
Plasmid Constructions for In Vitro Transcription and Translation
The entire STU2 coding sequence was amplified by PCR with primers SP10 and SP11. SP10: 5′-GGGTCTAGAACCATGTCAGGAGAAGAAGAAGTA-3′; SP11: 5′-GGGGTCGAC TTACGTCCTGGTTGTGCCTTCC-3′. SP10 introduces an XbaI site (underlined) and a Kozak consensus context for the start codon, which is critical for optimal translation (Kozak, 1986). SP11 introduces an SalI site (underlined) after the stop codon (bold). The PCR product was cloned into a TA cloning vector pCRTMII (Invitrogen). A plasmid with STU2 oriented so that it is under the control of T7 promoter was identified and called pWP79. The ATG codons lying between the T7 promoter and the STU2 start codon were eliminated by removing the 59-bp XbaI fragment and religating to generate pWP82. The carboxy-terminal truncation constructs C505 and C466 were made by digesting pWP82 with KpnI and SpeI, respectively, and self ligation of the resulting large fragment. All the other carboxy-terminal truncations were made by PCR with SP10 as the upstream primer and specific downstream primers containing in-frame stop codons and then subcloned into either pCR™II or pCR™2.1 (Invitrogen). Those clones with STU2 oriented under control of the T7 promoter were chosen for in vitro transcription and translation. Similarly, all of the amino-terminal truncation constructs were made using specific upstream primers and SP11 as the downstream primer. All of the upstream primers were designed to put the initiation codon (either endogenous or exogenous ATG) in a consensus or nearly consensus Kozak context.
Synthesis of Radiolabeled Stu2p by In Vitro Transcription and Translation
In vitro transcription and translation of all STU2 constructs were carried out using the TNT T7 coupled reticulocyte lysate system kit (Promega, Madison, WI). The reaction was done by combining the following reagents: 25 μl TNT rabbit reticulocyte lysate, 2 μl TNT reaction buffer, 1 μl TNT T7 RNA polymerase, 1 μl TNT amino acid mixture lacking methionine, 1 μl (40 U) RNasin ribonuclease inhibitor (Promega), 5 μl (50 μCi) of “cell-labeling” grade [35S]methionine (Amersham Intl., Arlington Heights, IL), 15 μl H2O and 1 μl (1 μg) plasmid DNA. The reaction mixture was incubated at 30°C for 60 to 80 min. Depending upon the transcription and translation efficiency, 2–10 vol of PEM-TDTG buffer (0.1 M Pipes, 2 mM EGTA, 1 mM MgSO4, 0.1% Triton X-100, 4 mM DTT, 20 μM Taxol, 1 mM GTP) was added to each reaction, and the reactions were clarified by centrifugation at 57,000 g for 20 min at 30°C. The supernatants were removed and used for the microtubule cosedimentation assay.
In Vitro Microtubule Cosedimentation Assay
Taxol-stabilized bovine brain microtubules were prepared according to Vallee (1986). Tubulin concentration was determined using protein assay kit (Bio Rad) with BSA as the reference protein. To generate more microtubule ends while keeping tubulin concentration constant, microtubules were sheared by repetitive passage through a 26-gauge hypodermic needle attached to a 1-ml syringe. Microtubule lengths were determined by pelleting them onto a 12-mm-round glass coverslip and visualizing by indirect immunofluorescence using DM1A (Blose et al., 1984) as the primary antibody. Random fields were photographed and polymer lengths measured on the negatives and converted to actual lengths (Mitchison and Kirschner, 1984).
The in vitro microtubule cosedimentation assay was performed as described by Yang et al. (1989) with minor modifications. Triton X-100 was added to taxol-stabilized microtubules to 0.1%. A constant amount of clarified Stu2p (3 μl) was incubated at 30°C for 20 min with increasing amounts of microtubules in a 40-μl reaction volume supplemented with PEM-TDTG buffer when necessary. Microtubules were pelleted by centrifugation through a 40-μl 60% glycerol PEM-TDTG cushion at 40,000 g for 20 min at 30°C. Both the supernatant and the glycerol cushion were removed and mixed with 20 μl 5× SDS sample buffer without glycerol (Laemmli, 1970). The microtubule pellet was resuspended in 100 μl SDS sample buffer. All samples were then boiled for 3 min, and 10 μl of each was subjected to SDS-PAGE analysis. After electrophoresis, the gel was fixed, dried, and quantitated using a PhosphoImager (Molecular Dynamics Inc., Sunnyvale, CA).
Results
Identification of the STU2 Gene
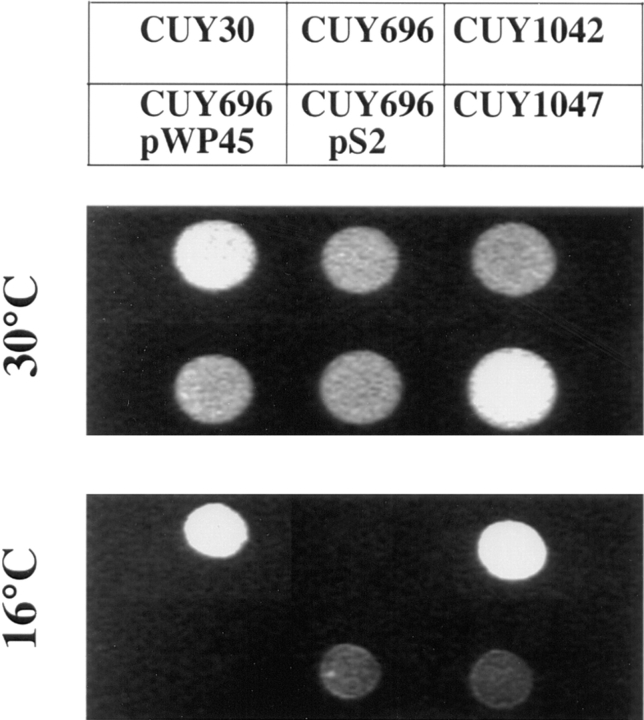
tub2-423 is a cold-sensitive allele of the yeast gene encoding β-tubulin and causes a specific defect in spindle function at 16°C (Reijo et al., 1994). Spontaneous suppressors of this mutation were isolated by plating tub2-423 cells at 16°C. Cold-resistant colonies arose at a frequency of ∼10−7. We obtained 29 independent mutants that grew at wild-type rates at 16°C. For 15 of these mutants, suppression segregated as a single gene mutation in backcrosses to a tub2-423 strain. 10 of these latter mutations were tightly linked to TUB2 and assumed to be intragenic suppressors. Three of the five extragenic suppressors were tightly linked to the STU1 locus (Pasqualone and Huffaker, 1994). The remaining two mutations were tightly linked to each other and dominant for suppression (Fig. 1). We have called the gene identified by these two mutations STU2 (suppressor of tubulin). Neither stu2-1 nor stu2-2 confers a conditional-lethal phenotype in a tub2-423 or TUB2 background.
Figure 1.
Suppression of tub2-423 by stu2-1. Yeast cells were spotted on YPD plates and incubated at 30°C for 2 d and 16°C for 5 d. The relevant genotypes of strains are as follows: CUY30, TUB2 STU2; CUY696, tub2-423 STU2; CUY1042, tub2-423 stu2-1; CUY696/pWP45, tub2-423 STU2 (STU2 on YCp plasmid); CUY696/pS2, tub2-423 STU2 (stu2-1 on YCp plasmid); CUY1047, tub2-423/tub2-423 stu2-1/ STU2.
To test for genetic interactions between the stu2sup alleles and other tub2 alleles, we constructed haploid strains containing stu2-1 and one of nine cold-sensitive tub2 alleles (120, 209, and 401–407) and haploid strains containing stu2-2 and one of seven different cold-sensitive tub2 alleles (418, 421, 429, 434, 438, 445, and 451). We observed that stu2-2 also suppressed the cold sensitivity of tub2-418, but none of the other tested tub2 alleles was suppressed by either of the stu2 suppressors. Therefore, suppression by stu2-1 and stu2-2 is allele specific. In addition, the double mutant strain containing stu2-1 and tub2-404 was inviable at 30°C. At this temperature, yeast strains carrying either one of these mutations grows at wild-type rates. Thus, lethality results from the combination of two unlinked mutations, a phenomenon referred to as synthetic lethality (Huffaker et al., 1987).
STU2 Encodes a Novel and Essential Gene
We cloned the dominant suppressor allele, stu2-1, by its ability to suppress the tub2-423 mutation. We obtained three independent clones that contain a ∼5 kb overlapping fragment. Genomic DNA from the insert of one of these plasmids directed the integration of a LEU2 marker to the STU2 locus, demonstrating that the plasmid contains the stu2-1 allele. The STU2 gene maps to chromosome XII, near PDC1 (see Materials and Methods).
The smallest fragment of DNA tested that still contains suppression activity is the 4.8-kb insert carried by plasmid pWP9 (Fig. 2). Sequence analysis showed that this fragment contains a 2,664-bp ORF. pWP9 also contains a 812-bp ORF, YLR406c, upstream of the larger ORF. Because pWP2, which contains the entire YLR406c gene, does not suppress the tub2-423 allele, we conclude that the larger ORF must encode Stu2p. Both the wild-type STU2 gene and the stu2-2 allele were cloned through plasmid gap repair and sequenced. STU2 encodes an 888-amino acid, 101-kD basic protein (isoelectric point = 8.6). Stu2p is predicted to have a coiled-coil region (Lupas et al., 1991) from residues 658 to 764. Two potential phosphorylation sites for Cdc28 kinase (S/TP[X]R/K consensus sequence) are found in a highly basic region at positions 603 and 645.
Stu2p shows a modest level of similarity to several other proteins (Fig. 3). Stu2p is 22% identical to the S. pombe p93dis1 (Nabeshima et al., 1995). It is also similar to the human ch-TOG (Charrasse et al., 1995) and the C. elegans ZYG-9 (Matthews, 1997) proteins. Both of these proteins are about twice the size of Stu2p. Stu2p is similar to the amino-terminal halves of ch-TOG and ZYG-9 (22% and 27% respectively). The S. cerevisiae genome does not contain any sequence that could encode a protein with significant similarity to the carboxy-terminal halves of these proteins.
Figure 3.

Alignments of the S. cerevisiae Stu2p, S. pombe p93dis1, and human ch-TOG. Note that only the amino-terminal half of ch-TOG is shown in this figure; the full length protein is 1,972 amino acids. The sequence of STU2 is available from GenBank/ EMBL/DDBJ under accession number U35247.
Sequence comparison between STU2 and the two suppressor alleles revealed two base pair differences in both stu2-1 (A1540G, C2561T) and stu2-2 (G1537T, C2561T). The appearance of T at position 2561 in stu2-1 and stu2-2 alleles suggested that this nucleotide difference might be due to a polymorphism between CUY696 from which both suppressor mutations were derived and CUY1046 from which the STU2 was cloned. We sequenced the STU2 gene from CUY696 and found that this strain does contain T at position 2561. Therefore, the mutations responsible for the suppressing activity of stu2-1 and stu2-2 are A1540G and G1537T, respectively. These mutations produce a T514A amino acid substitution in stu2-1 and a D513Y amino acid substitution in stu2-2.
To determine whether STU2 is required for mitotic growth, we replaced the stu2-1 sequence encoding amino acids 57-235 with a DNA fragment bearing the HIS3 marker (Fig. 2 B, stu2-Δ1::HIS3). The resulting construct was transformed into a wild-type His− diploid. A His+ transformant was shown by PCR to contain one copy of the disrupted STU2 gene (data not shown). The His+ transformant was sporulated and tetrads were dissected. Each of 20 tetrads contained two His− spores that were able to form colonies and two spores that were unable to form colonies, demonstrating that the STU2 gene is essential for viability. Similar results were obtained using a precise deletion of the entire STU2 coding region (Fig. 2 B, stu2-Δ2::HIS3). The stu2-Δ1::HIS3 spores germinated on YPD medium and each (n = 6) gave rise to an average of 12 progeny cells. The ability of stu2-Δ1::HIS3 cells to undergo three or four cell divisions may be attributable to the supply of the wild-type Stu2p inherited from their diploid parent.
Stu2p Is a Component of the SPB
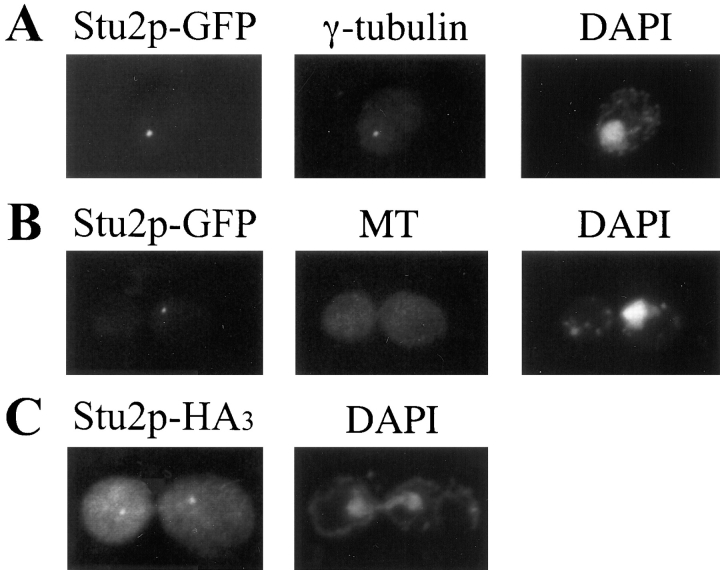
A fusion between STU2 and the coding region of GFP was constructed to allow visualization of Stu2p. This construct on a YCp plasmid, YEp plasmid, or integrated in single copy into the yeast genome was able to complement a STU2 deletion. The GFP-tagged Stu2p was visualized in stu2-Δ1::HIS3 cells carrying plasmid-borne STU2–GFP or in cells with the chromosomal copy of STU2 replaced by STU2–GFP. The fluorescence pattern observed in each of these situations was similar, but the staining intensity was somewhat greater with STU2–GFP on plasmids. In living cells, one bright fluorescent dot was observed in unbudded cells, and two dots were generally observed in budded cells (Fig. 4). In small budded cells, the two dots were either adjacent or separated by ∼1 μm. In large budded cells, the two fluorescent dots were often segregated into each cell body. The intensity of staining did not appear to change over the course of the cell cycle. When cells were grown in the presence of DAPI, we observed that the Stu2p–GFP dots resided at the periphery of the nuclear DNA.
Figure 4.
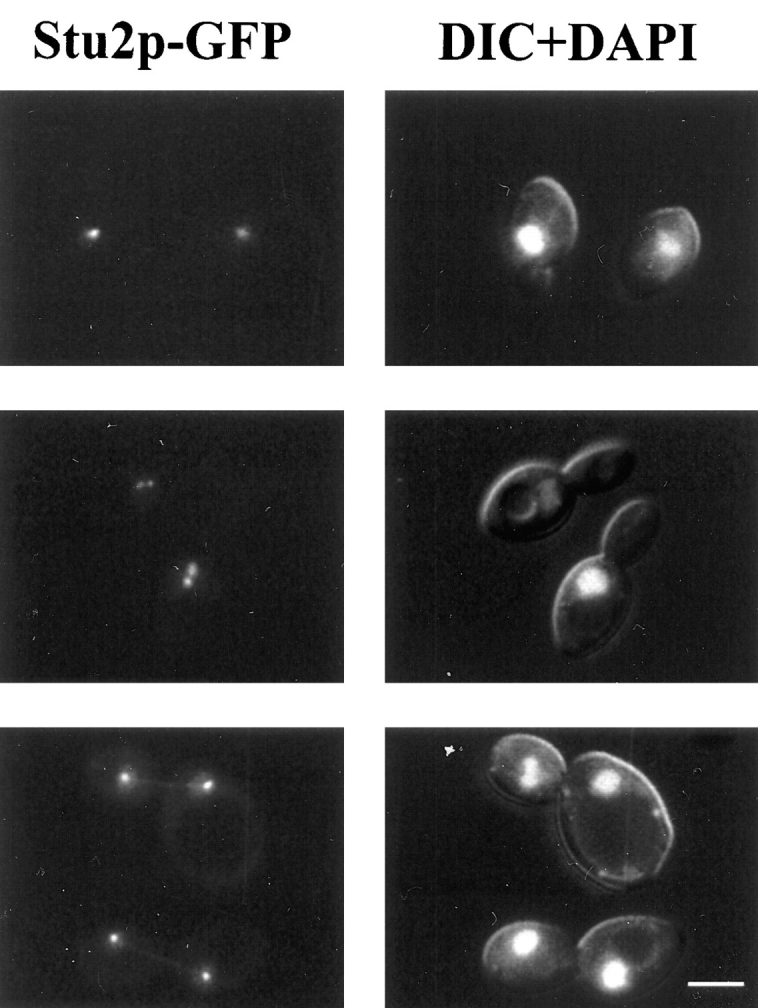
Localization of Stu2p–GFP fusion protein. GFP fluorescence was visualized in diploid cells, homozygous for the stu2-Δ1::HIS3 disruption, that contain a plasmid carrying STU2-GFP (CUY1065). Cells were grown in the presence of 1 μg/ml DAPI for 2 h in SD media. Left column shows the fluorescence of Stu2p-GFP; right column shows both DNA staining with DAPI and differential interference contrast image of cells. Bar, 5 μm.
We also examined the localization of an HA3-tagged Stu2p. This construct was able to complement a STU2 deletion, and Stu2p-HA3 was visualized in stu2-Δ1::HIS3 cells carrying plasmid-borne STU2-HA3. The localization pattern of the Stu2p-HA3 was the same as that observed for Stu2p–GFP; it resided at one or two dots on the nuclear periphery of the cells (Fig. 5 C). This localization pattern suggested that Stu2p was likely a component of the SPB. To confirm this hypothesis, we compared the Stu2p–GFP localization with that of γ-tubulin, a known component of the SPB (Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996). We observed that Stu2p–GFP colocalized with γ-tubulin (Fig. 5 A).
Figure 5.
(A) Colocalization of Stu2p-GFP with γ-tubulin. CUY1060 cells were stained with anti-Tub4p antibody (middle) and DAPI (right). The left frame shows the fluorescence of Stu2p–GFP in the same cell. (B) CUY1060 cells were treated with nocodazole to depolymerize microtubules and stained with anti-tubulin antibody (middle) and DAPI (right). The left frame shows the fluorescence of Stu2p–GFP in the same cell. (C) Localization of Stu2p-HA3. CUY1066 cells were stained with anti- hemaglutinin antibody (left) and DAPI (right).
In addition to its presence at the SPBs, minor amounts of Stu2p also appeared to localize along microtubules. Weak Stu2p–GFP staining was often observed as a straight line between SPBs in anaphase cells, indicating some Stu2p along spindle microtubules (Fig. 4, bottom row). Before anaphase, Stu2p also appeared to extend away from the SPBs toward the center of the spindle, but this was difficult to distinguish from SPB staining alone because of the relatively large size of the fluorescent dot at the SPBs and the relatively short distance between the poles. Finally, we occasionally observed weak Stu2p staining along cytoplasmic fibers that extended from the SPBs, consistent with some Stu2p localized along cytoplasmic microtubules. However, the vast majority Stu2p appeared to be associated with the SPB throughout the cell cycle.
To determine whether the association of Stu2p with the SPB depends upon the presence of microtubules, cells containing STU2–GFP were treated with the microtubule depolymerizing drug, nocodazole. After this treatment, 90% of the cells contained no microtubules that could be detected by immunofluorescence. Nocodazole treatment did not affect the number of cells containing Stu2p–GFP staining (>95%) or noticeably change the intensity of staining compared to untreated cells (Fig. 5 B).
Stu2p Binds to Microtubules In Vitro
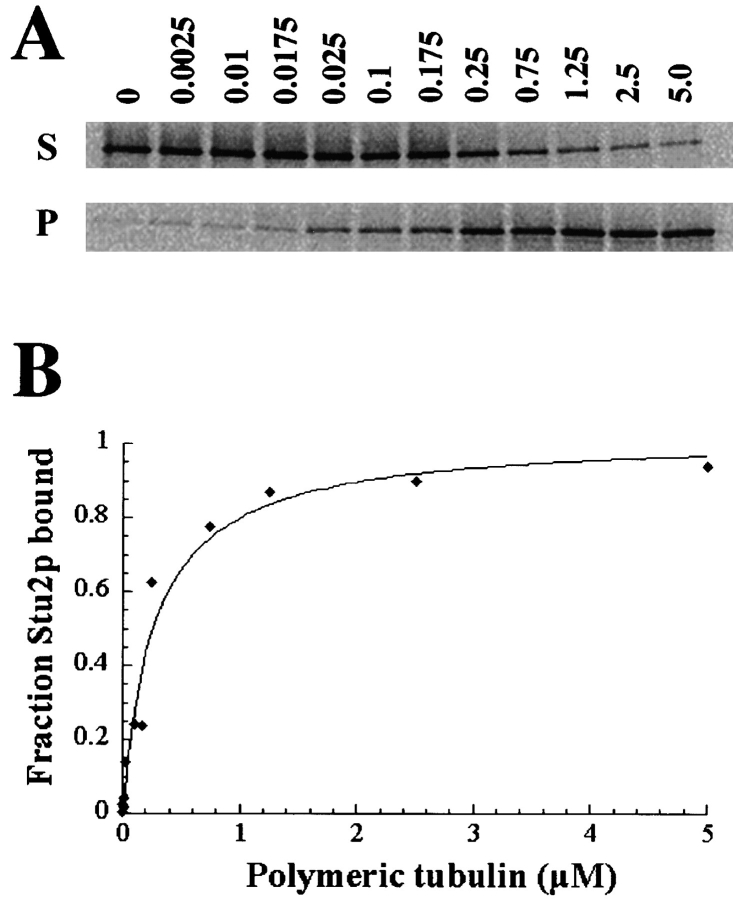
Two lines of evidence suggested that Stu2p may directly interact with microtubules. First, mutations in STU2 display allele-specific suppression of the tub2-423; second, Stu2p localizes to the SPB where the microtubule ends reside and to a lesser extent along the length of microtubules. To determine whether Stu2p binds to microtubules directly, we performed an in vitro microtubule-cosedimentation assay. 35S-labeled Stu2p was synthesized by in vitro transcription and translation and incubated with a large excess of taxol-stabilized bovine brain microtubules (tubulin/Stu2p was >100:1). When microtubules were then pelleted by centrifugation, almost all the Stu2p was pelleted with microtubules (Fig. 6 A, last lane). On the other hand, most Stu2p remained in the supernatant in the absence of microtubules (Fig. 6 A, first lane). The binding activity could be abolished by treatment with 0.4 M NaCl (data not shown).
Figure 6.
Measurement of the dissociation constant (Kd) for full length Stu2p. A constant amount of 35S-labeled in vitro translated Stu2p (∼10−11 M final concentration) was incubated with variable amounts of taxol-stabilized bovine brain microtubules. Microtubules were pelleted by centrifugation, and both bound Stu2p (in pellets) and unbound Stu2p (in supernatants) were subjected to SDS-PAGE analysis. Band intensities were quantitated using the phosphoimager software. (A) Autoradiographs of Stu2p in the supernatants (S) and in the corresponding pellets (P). Microtubule concentration ranging from 0 to 5 μM is shown above each lane. (B) The binding curve was generated from the above data. The Kd is equal to the concentration of polymerized tubulin required to cosediment 50% of the Stu2p in the reaction.
To determine the microtubule-binding affinity of Stu2p, a constant amount of 35S-labeled Stu2p was mixed with variable amounts of microtubules. As seen in Fig. 6, A and B, the binding of Stu2p to microtubules is concentration dependent and saturable. The apparent dissociation constant, Kd, equal to the concentration of polymerized tubulin required to cosediment half of the Stu2p in the reaction is 5.4 × 10−7 M.
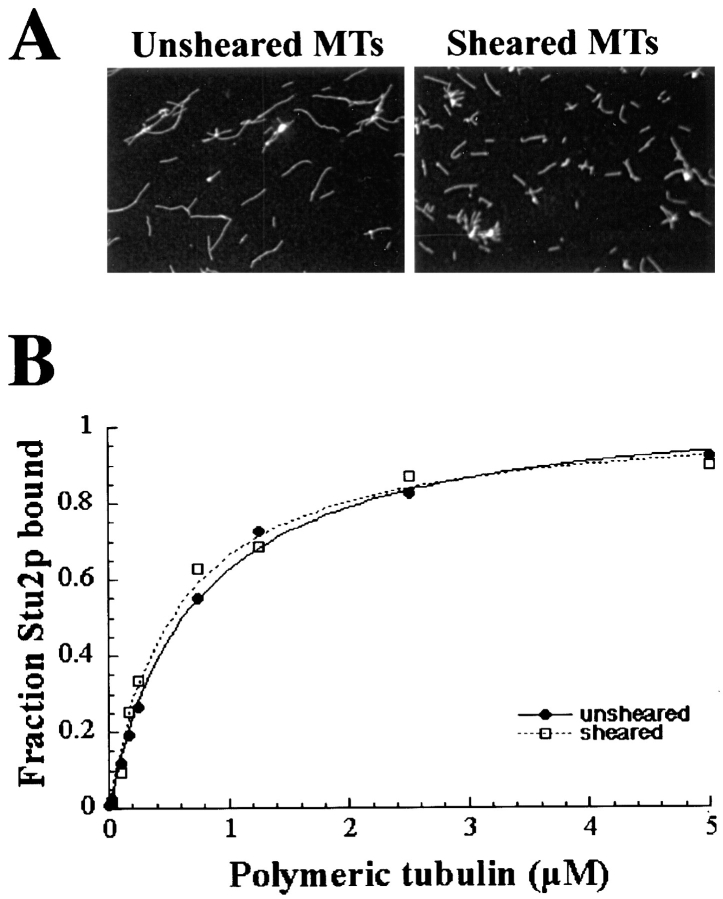
The localization of Stu2p to the SPB raises the prospect that Stu2p might bind to the ends of microtubules as γ-tubulin does. To examine this possibility, we sheared microtubules to generate more polymer ends for a given tubulin concentration. The sheared microtubules with an average length of 3.4 ± 1.2 μm were nearly three times shorter than unsheared microtubules with an average length of 9.3 ± 5.2 μm (Fig. 7 A). If Stu2p binds exclusively to the ends of microtubules, we would expect about a threefold decrease in the Kd value measured with sheared microtubules. As shown in Fig. 7 B, the Kd value measured with sheared microtubules (0.56 ± 0.07 μM) was nearly identical to that with unsheared microtubules (0.54 ± 0.19 μM). Thus, the affinity of Stu2p for microtubules does not depend on the concentration of microtubule ends, indicating that Stu2p binds laterally to microtubules.
Figure 7.
(A) Microtubules before (left) and after (right) shearing visualized by immunofluorescence. (B) The binding curves for Stu2p to both sheared and unsheared microtubules.
The Microtubule-binding Domain of Stu2p
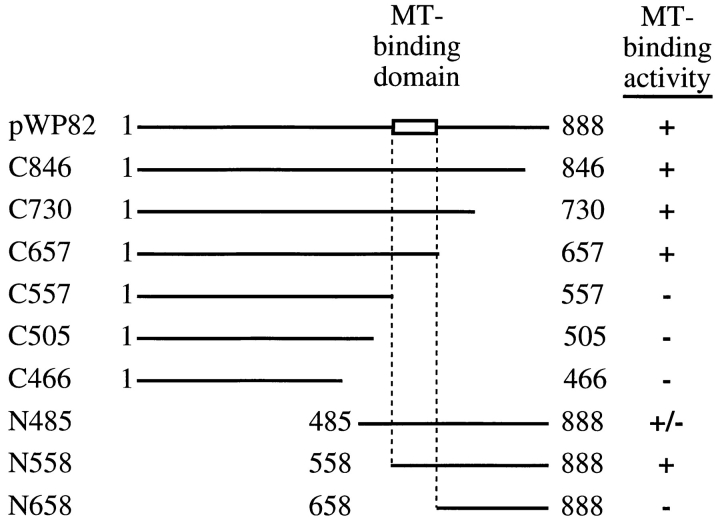
To define the microtubule-binding domain of Stu2p, we measured the relative microtubule-binding affinities of a series of amino- and carboxy-terminal truncation constructs (Fig. 8). Initially, we measured the fraction of Stu2p that cosedimented with microtubules at a single microtubule concentration (16.5 μM tubulin). Truncations that lack up to 231 amino acids from the carboxy terminus of Stu2p (Fig. 8, C846, C730, and C657) bind to microtubules nearly as well as full length Stu2p. Further deletion of 100 amino acids to residue 557 (C557) abolishes most of the binding activity. Truncations that lack up to 557 amino acids from the amino terminus of Stu2p (N485 and N558) have microtubule-binding activities comparable to that of full length Stu2p. However, a construct lacking an additional 100 amino acids from the amino terminus (N658) has only residual microtubule-binding activity. These results localize the microtubule-binding domain of Stu2p to the 100 amino acid region between amino acids 557 and 658.
Figure 8.
Identification of Stu2p microtubule-binding domain. Various truncated Stu2p polypeptides were translated in vitro and tested for microtubule-binding activity by the cosedimentation assay using a constant amount of microtubules (16.5 μM tubulin). The percentage of Stu2p that pelleted with microtubule is indicated by (+), >90%; (+/−), 70–90%; (−), <30%. The numbers adjacent to the endpoints represent the corresponding amino acid positions.
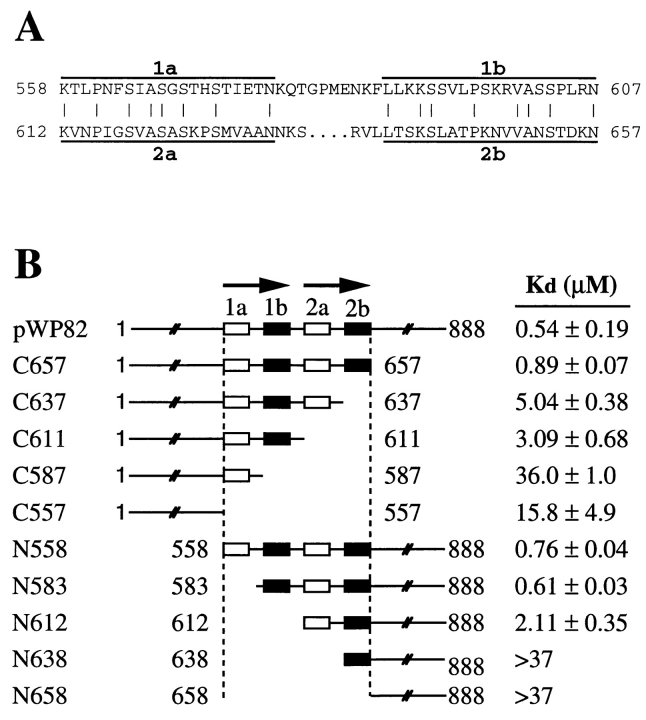
To obtain more quantitative binding data for some of these constructs, we measured the fraction of polypeptide bound to microtubules over a range of microtubule concentrations and calculated the apparent Kd as described for the full length Stu2p above (Fig. 9 B). The Kd's for the C657 and N558 polypeptides are only 1.6- and 1.4-fold higher, respectively, than the Kd for the full length protein confirming that the 231 carboxy-terminal and 557 amino-terminal amino acids do not contribute significantly to the microtubule-binding affinity of Stu2p. However, the Kd's for the C557 and N658 polypeptides are ∼30-fold and >50-fold higher than that of the full length protein.
Figure 9.
Dissection of the microtubule-binding domain of Stu2p. (A) Sequences of the two bipartite repeats in the microtubule-binding domain of Stu2p. The identical residues between these two repeats are indicated by vertical lines. (B) The Kd 's of Stu2p polypeptides were determined as in Fig. 6 and presented as the average of the measured values. The full-length Stu2p was assayed three times and standard error of the mean is shown. All the truncated polypeptides were assayed twice and the range is included. The top line represents the full-length Stu2p. The numbers adjacent to the endpoints represent the positions of the corresponding amino acids.
The 558–657 binding domain is highly basic with a predicted isoelectric point of 10.7 and is rich in serine (18%), threonine (8%), proline (7%), and basic amino acids (16%). In addition, this region consists of two imperfect repeats that we have named R1 (558–607) and R2 (612–657) (Fig. 9 A). The first 20 amino acids of R1 and R2 are 40% identical (regions R1a and R2a), and the last 20 amino acids of R1 and R2 are 40% identical (regions R1b and R2b). The sequences between the a and b regions differ in length and do not display any amino acid similarity. Interestingly, there is one putative Cdc28 phosphorylation site in each of the b repeats.
Contribution of the Individual Repeats to the Binding Affinity of Stu2p
We next determined the microtubule-binding affinities of Stu2p polypeptides with carboxy- and amino-terminal endpoints located between the repeat domains (Fig. 9 B). A carboxy-terminal truncation that removes R2 (C611) has a nearly sixfold higher Kd than the full length protein. An amino-terminal truncation that removes the R1 element (N612) has a 3.5-fold higher Kd. Thus, both the R1 and R2 regions contribute to microtubule binding and to similar extents.
We also generated truncations that removed half of each repeat. In three of the four cases, removing half of a repeat has nearly the same effect as removing the whole repeat. For example, C637 which lacks only element R2b has nearly the same binding affinity as the C611, which lacks both elements R2a and R2b. Similarly, C587, which contains only element R1a, and N638, which contains only element R2b, do not bind microtubules any better than the C557 and N658 constructs that lack the entire binding region. The one exception is the construct N583 which lacks only the element R1a and has a binding affinity that is equivalent to that for N558, which contains the complete R1 and R2 elements. We have not yet determined whether R1a is required for microtubule binding in the absence of R2.
Discussion
We report the identification of Stu2p, a yeast microtubule-binding protein that is located at the SPB. Both Stu2p-HA3 and -GFP fusion proteins localize primarily to the SPB in vegetative cells at all stages of the cell cycle and to a lesser extent along the length of microtubules. We believe these fusion proteins mimic the authentic Stu2p, because both rescue the stu2 null mutation. SPB localization is not dependent on the presence of detectable microtubules, indicating that Stu2p is an integral component of the SPB and must bind at least one other SPB component. This experiment has the caveat that a small amount of polymer that is undetectable by immunofluorescence may remain near the SPB after treatment of cells with nocodazole.
Both genetic and biochemical evidence indicate that Stu2p interacts directly with microtubules. Alleles of STU2 were identified as suppressors of tub2-423, a cold-sensitive mutation in the yeast β-tubulin gene. Thus, the stu2sup alleles compensate for the microtubule defect caused by the tub2-423 mutation. The simplest explanation for this result is that the tub2-423 mutation compromises the ability of Stu2p to associate with microtubules, and this situation is remedied by the corresponding alterations in Stu2p. This hypothesis is supported by the observation that suppression is allele specific; only one of 16 other tub2 mutations tested could be suppressed by one of the stu2sup alleles. In addition, stu2sup alleles are dominant suppressors as expected for such a reciprocal interaction. Finally, the synthetic lethality observed between stu2-1 and tub2-404 also indicates that the products of these genes interact.
Consistent with our genetic data, we have shown that Stu2p binds microtubules in vitro. We performed these assays using bovine brain microtubules, because yeast microtubules are difficult to obtain in large quantities and can not be stabilized by taxol (Barnes et al., 1992). Yeast and bovine tubulins share >70% identity and have been shown to bind the same profile of proteins from yeast extracts (Barnes et al., 1992). Therefore, we assume that the binding properties of Stu2p to bovine brain microtubules reflect the binding properties of Stu2p to yeast microtubules. Stu2p binds to microtubules with an apparent Kd of 0.54 μM tubulin. This value is about three times greater than that obtained for the neuronal microtubule-associated protein, tau, using the same assay (Goode and Feinstein, 1994). Thus, the affinity of Stu2p for microtubules is within the range expected for a bona fide microtubule-associated protein.
In vitro binding assays using truncations of Stu2p have defined a ∼100 amino acid region that is necessary for microtubule binding. This region does not have any significant sequence similarity to the related microtubule-binding domains of tau, MAP2, and MAP4 (Lee et al., 1988; Lewis et al., 1988; Chapin and Bulinski, 1991) or the unrelated microtubule-binding domain of MAP1B (Noble et al., 1989). However, like these domains, it is positively charged and contains repeated elements. The sequence alignment of Stu2p and its S. pombe homologue p93dis1 shows that the microtubule-binding region of Stu2p corresponds to a region of p93dis1 that falls well within its reported microtubule-binding domain (Nakaseko et al., 1996). However, the microtubule-binding regions of these proteins do not show any more sequence similarity than the remainder of the proteins, and the p93dis1 microtubule-binding domain does not contain repeated elements. The fact that the Stu2p microtubule-binding region is highly basic and the carboxyl-terminal portion of tubulins is acidic suggests that Stu2p could interact with microtubules through electrostatic interactions. This model is supported by the finding that Stu2p binding to microtubules, like that of other microtubule-associated proteins (Vallee, 1986), is sensitive to salt. The amino acid substitutions in stu2-1 and stu2-2 do not lie within the microtubule-binding domain of Stu2p as might be expected. However, these residues are very close to the microtubule-binding domain and may play a role in altering the Stu2p conformation to enable it to interact more effectively with tub2-423 microtubules.
The microtubule-binding region of Stu2p has two imperfect bipartite repeats. Truncations of Stu2p lacking either repeat bind microtubules with about a fivefold decrease in affinity, indicating that each repeat is capable of binding microtubules independently. This indicates that Stu2p may bind to two adjacent tubulin subunits within microtubules. There is one potential phosphorylation site for Cdc28 kinase in each repeat, suggesting that Stu2p might be phosphorylated in a cell cycle-dependent manner. It is known that the phosphorylation of tau reduces its in vitro microtubule-binding activity (Biernat et al., 1993; Bramblett et al., 1993). By analogy, phosphorylation of Stu2p might regulate its ability to bind microtubules.
In addition to Stu2p, three other proteins with the ability to bind microtubules have been shown to be located at the SPB. Two of these, the kinesin-related protein Kar3p (Meluh and Rose, 1990; Page et al., 1994; Saunders et al., 1997) and the dynein heavy chain protein Dhc1p (Yeh et al., 1995), are microtubule motors. The association Dhc1p with the SPB is dependent on the presence of microtubules; whether Kar3p localization to the SPB depends on microtubules is not known. The third is the S. cerevisiae γ-tubulin Tub4p (Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996). Like Stu2p, the localization of Tub4p to the SPB is not dependent on the presence of microtubules. However, unlike Stu2p, γ-tubulin from human and Xenopus has been shown to bind specifically to the minus ends of microtubules (Li and Joshi, 1995; Zheng et al., 1995). Thus, Stu2p may be unique as an integral component of the SPB that associates laterally with microtubules.
One potential role for Stu2p is in tethering microtubules to the SPB. The minus ends of microtubules in animal cells have been shown to exchange tubulin subunits during the process of poleward microtubule flux (Mitchison, 1989; Sawin and Mitchison, 1991). Because Stu2p binds laterally to microtubules, it could maintain the attachment of microtubules to the pole even during subunit exchange at the ends. In addition, Stu2p may be involved in the organization of microtubule ends at the SPB. We have evidence via the two-hybrid system that Stu2p can dimerize (Chen, P.X., and T.C. Huffaker, unpublished observations) which may allow it to crosslink microtubules. Thus, Stu2p might bundle microtubules near the SPB to help generate the parallel array of microtubules observed in yeast spindles. Finally, Stu2p could influence microtubule dynamics through its binding near the minus ends. These roles for Stu2p are not mutually exclusive, and Stu2p could act as both a structural and regulatory component of the yeast mitotic spindle.
Homologues of Stu2p are found in a variety of organisms, indicating that Stu2p has evolutionarily conserved functions. In addition to the S. pombe p93dis1 (Nabeshima et al., 1995), the C. elegans ZYG-9 (Matthews, 1997), and human ch-TOG (Charrasse et al., 1995), the Xenopus protein XMAP215 (Gard and Kirschner, 1987) also appears to be a member of this family. Partial sequencing of XMAP215 shows that it is similar to ch-TOG (Charrasse, S., M. Schroeder, C. Gauthier-Rouviere, L. Cassimeris, D.L. Garrd, and C. Larroque. 1996. Mol. Biol. Cell. 7:222a). Like Stu2p, p93dis1 (Nabeshima et al., 1995), ZYG-9 (Matthews, 1997), ch-TOG (Charrasse, S., M. Schroeder, C. Gauthier-Rouviere, L. Cassimeris, D.L. Garrd, and C. Larroque. 1996. Mol. Biol. Cell. 7:222a), and XMAP215 (Gard et al., 1995) localize at the spindle poles and along spindle microtubules during mitosis. However during interphase, p93dis1 localizes along cytoplasmic microtubules and ch-TOG colocalizes with endoplasmic reticulum markers. p93dis1 (Nakaseko et al., 1996), ch-TOG (Charrasse, S., M. Schroeder, C. Gauthier-Rouviere, L. Cassimeris, D.L. Garrd, and C. Larroque. 1996. Mol. Biol. Cell. 7: 222a), and XMAP215 (Gard and Kirschner, 1987) are all capable of binding microtubules in vitro, and XMAP215 has been shown to promote microtubule assembly and turnover (Vasquez et al., 1994). Therefore, it seems reasonable to conclude that the proteins in this family perform some overlapping functions. Further study of these homologues should provide insights into the specific roles they play in spindle function.
Acknowledgments
We thank (from Cornell University) William Brown and Ken Kemphues for comments on the manuscript, Daniel Starr and David Wolfgang for performing some preliminary experiments that led to this work, Xiaoyue Peter Chen for providing CUY1068, and Roger Y. Tsien (University of California, San Diego) for permission to use the mutant GFP.
This work was supported by a grant from the National Institutes of Health (GM40479).
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- HA
hemagglutinin, MTOC, microtubule-organizing center
- ORF
open reading frame
- SPB
spindle pole body
Footnotes
Address all correspondence to Tim Huffaker, Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, NY 14853. Tel.: (607) 255-9947. Fax: (607) 255-2428. E-mail:tch4@cornell.edu
References
- Barnes G, Louie KA, Botstein D. Yeast proteins associated with microtubules in vitro and in vivo. Mol Biol Cell. 1992;3:29–47. doi: 10.1091/mbc.3.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- Blose SH, Meltzer DI, Feramisco JR. 10-nm filaments are induced to collapse in living cells microinjected with monoclonal and polyclonal antibodies against tubulin. J Cell Biol. 1984;98:847–858. doi: 10.1083/jcb.98.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett GT, Goedert M, Jakes R, Merrick SE, Trojanowski JQ, Lee VM. Abnormal tau phosphorylation at Ser396 in Alzheimer's disease recapitulates development and contributes to reduced microtubule binding. Neuron. 1993;10:1089–1099. doi: 10.1016/0896-6273(93)90057-x. [DOI] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. 59–96.
- Chapin SJ, Bulinski JC. Non-neuronal 210 × 103Mr microtubule-associated protein (MAP4) contains a domain homologous to the microtubule-binding domains of neuronal MAP2 and tau. J Cell Sci. 1991;98:27–36. doi: 10.1242/jcs.98.1.27. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Mazel M, Taviaux S, Berta P, Chow T, Larroque C. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur J Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopussperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopuseggs that specifically promotes assembly at the plus end. J Cell Biol. 1987;105:2203–2216. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Cha BJ, Schroeder MM. Confocal immunofluorescence microscopy of microtubules, microtubule-associated proteins, and microtubule-organizing centers during amphibian oogenesis and early development. Curr Top Dev Biol. 1995;31:383–431. doi: 10.1016/s0070-2153(08)60234-3. [DOI] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Heinrich, P. 1991. Constructing nested deletions for use in DNA sequencing. In Current Protocols in Molecular Biology. Vol. 1. R.B.F.M. Ausubel, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, editors. John Wiley & Sons, New York. 7.2.1–7.2.8. [DOI] [PubMed]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Hoyt MA, Botstein D. Genetic analysis of the yeast cytoskeleton. Annu Rev Genet. 1987;21:259–284. doi: 10.1146/annurev.ge.21.120187.001355. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. γ-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee G, Cowan N, Kirschner M. The primary structure and heterogeneity of tau protein from mouse brain. Science. 1988;239:285–287. doi: 10.1126/science.3122323. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Wang D, Cowan NJ. Microtubule-associated protein MAP2 shares a microtubule binding motif with tau protein. Science. 1988;242:936–939. doi: 10.1126/science.3142041. [DOI] [PubMed] [Google Scholar]
- Li Q, Joshi HC. γ-Tubulin is a minus end–specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, VanDyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin–like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L. 1997. A molecular analysis of ZYG-9, a component of the Caenorhabditis elegans meiotic and mitotic spindle poles. PhD Thesis. Cornell University.
- Meluh P, Rose M. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin–containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Nabeshima K, Kinoshita K, Yanagida M. Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells. 1996;1:633–644. doi: 10.1046/j.1365-2443.1996.00253.x. [DOI] [PubMed] [Google Scholar]
- Noble M, Lewis SA, Cowan NJ. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J Cell Biol. 1989;109:3367–3376. doi: 10.1083/jcb.109.6.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. γ-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. . Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of Kar3 kinesin heavy chain–related protein requires the Cik1 interating protein. J Cell Biol. 1994;124:507–520. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L, Dutchik JE, Baktha A, McCauley BK, Thayer EC, Leckie MP, Braden VV, Depke JE, Olson MV. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiaeat a resolution of 2.6 kilobase pairs. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Broach JR. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–229. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rose M, Novick P, Thomas J, Botstein D, Fink G. A Saccharomyces cerevisiaegenomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiaekinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Poleward microtubule flux in mitotic spindles assembled in vitro. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-tubulin-like Tub4p of Saccharomyces cerevisiaeis associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Vallee R. Purification of brain microtubules and microtubule-associated protein 1 using taxol. Methods Enzymol. 1986;134:104–115. doi: 10.1016/0076-6879(86)34079-5. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopuseggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. γ-Tubulin is present in Drosophila melanogaster and Homo sapiensand is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]