Abstract
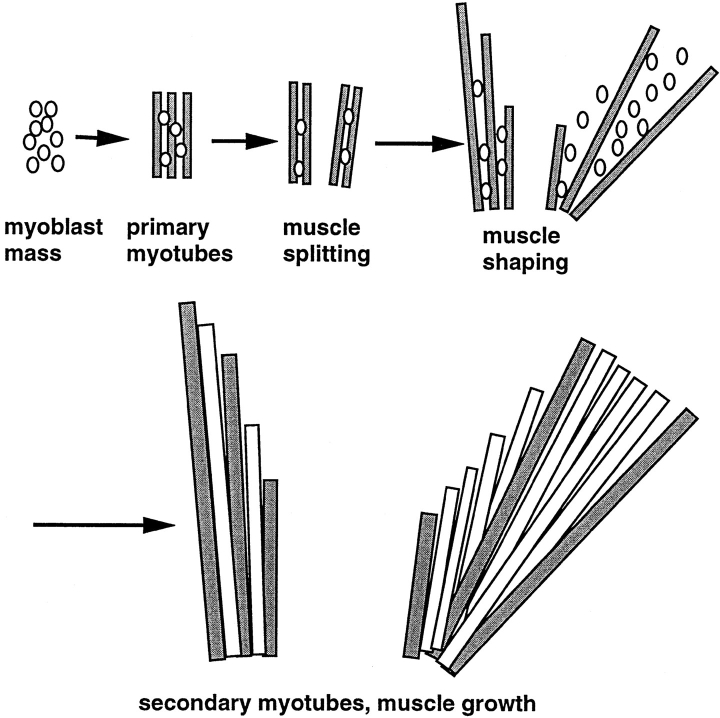
The characteristic shapes and positions of each individual body muscle are established during the process of muscle morphogenesis in response to patterning information from the surrounding mesenchyme. Throughout muscle morphogenesis, primary myotubes are arranged in small parallel bundles, each myotube spanning the forming muscles from end to end. This unique arrangement potentially assigns a crucial role to primary myotube end regions for muscle morphogenesis.
We have cloned muscle ankyrin repeat protein (MARP) as a gene induced in adult rat skeletal muscle by denervation. MARP is the rodent homologue of human C-193 (Chu, W., D.K. Burns, R.A. Swerick, and D.H. Presky. 1995. J. Biol. Chem. 270:10236–10245) and is identical to rat cardiac ankyrin repeat protein. (Zou, Y., S. Evans, J. Chen, H.-C. Kuo, R.P. Harvey, and K.R. Chien. 1997. Development. 124:793–804). In denervated muscle fibers, MARP transcript accumulated in a unique perisynaptic pattern. MARP was also expressed in large blood vessels and in cardiac muscle, where it was further induced by cardiac hypertrophy. During embryonic development, MARP was expressed in forming skeletal muscle. In situ hybridization analysis in mouse embryos revealed that MARP transcript exclusively accumulates at the end regions of primary myotubes during muscle morphogenesis. This closely coincided with the expression of thrombospondin-4 in adjacent prospective tendon mesenchyme, suggesting that these two compartments may constitute a functional unit involved in muscle morphogenesis. Transfection experiments established that MARP protein accumulates in the nucleus and that the levels of both MARP mRNA and protein are controlled by rapid degradation mechanisms characteristic of regulatory early response genes. The results establish the existence of novel regulatory muscle fiber subcompartments associated with muscle morphogenesis and denervation and suggest that MARP may be a crucial nuclear cofactor in local signaling pathways from prospective tendon mesenchyme to forming muscle and from activated muscle interstitial cells to denervated muscle fibers.
In their physiological context in vivo, cells must integrate multiple signals from their environment to fulfill their specific roles within the organism. This crucial role of integration is particularly evident during development, when cells display coordinate migration, differentiation, and morphogenesis in response to a variety of specific long-range and local signals. The formation of the skeletal musculature is an attractive model system in which to study how patterns of gene expression lead to the formation of a tissue. Due to major breakthroughs such as the establishment of differentiating myogenic cell lines, and the discovery of the converting activity of myogenic determination factors, the early events in myogenesis, from predetermined cells to terminally differentiated myotubes, are particularly well understood at the cellular level (Buckingham, 1992; Olson and Klein, 1994). In contrast, we know very little about the mechanisms that control muscle formation within the context of the developing organism. For example, the morphogenetic events that orchestrate the formation of the definitive muscle pattern are poorly understood. These include specific successive splitting of primordial muscle masses to generate all the different muscles, directed expansion of the newly formed muscles to generate their unique shapes, sizes, and positions, and attachment of the muscles to their specific tendon organs (Lance-Jones, 1979; Schroeter and Tosney, 1991a , b ; Hauschka, 1994). In the developing mouse limb, these events begin around embryonic day (E)1 12.5, and the definitive muscle pattern is established by E16 (Lance-Jones, 1979). Throughout muscle morphogenesis, the muscle pattern is reflected in the parallel arrangement of primary myotubes that span the forming muscles from end to end (Fig. 1) (Hilfer et al., 1973; Hauschka, 1974; Ontell and Kozeka, 1984; Duxson and Usson, 1989; Sweeney et al., 1989). After completion of muscle morphogenesis, primary myotubes then serve as scaffolds for secondary muscle fibers, which are generated through the proliferation and differentiation of intramuscular precursor cells (Ross et al., 1987; Condon et al., 1990; Ashby et al., 1993).
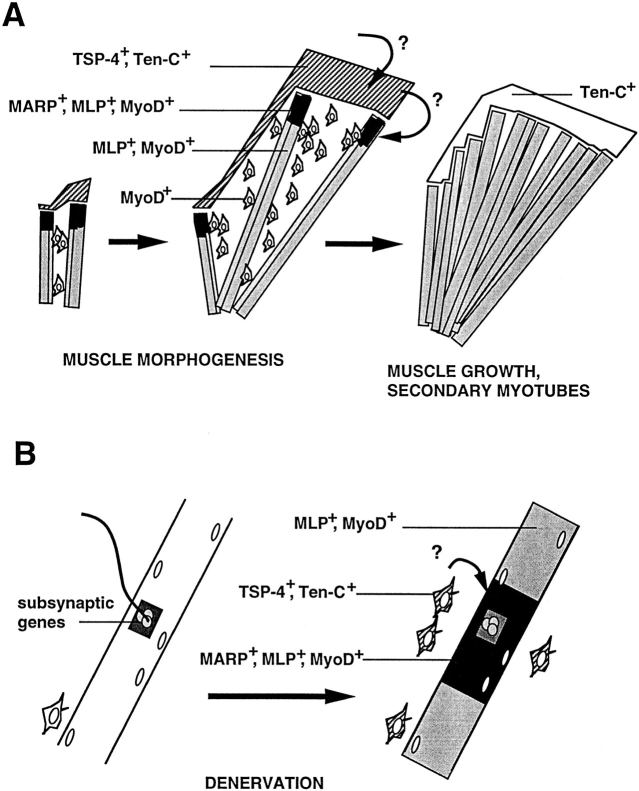
Figure 1.
Schematic representation of the spatial arrangement of myogenic cells during muscle formation. Muscle morphogenesis produces the definitive shape and arrangement of each individual muscle and precedes the formation of secondary myotubes. From their first appearance, primary myotubes (shaded rectangles) span forming muscles in a parallel end-to-end arrangement. During muscle morphogenesis, a substantial part of the muscle mass contains undifferentiated myoblasts (circles), and primary myotubes are arranged in parallel bundles of three to seven. When morphogenesis is completed, primary myotube bundles dissociate, and single primary myotubes serve as scaffolds along which secondary myotubes (white rectangles) fuse and elongate. Note that due to the particular arrangement of the primary myotubes, muscle splitting and shaping may in principle be regulated by controlling the position and local elongation of primary myotube end regions. The latter process may involve selective fusion of myoblasts to primary myotube end regions.
Classical embryological experiments have established that myogenic precursor cells are not predetermined with respect to the particular muscles that they will contribute to (Chevallier et al., 1977). Potential instructive roles by forming nerve or vasculature patterns have been essentially ruled out, and it is now clear that most morphogenetic information is derived from the surrounding mesenchyme (Chevallier et al., 1977; Lanser and Fallon, 1987; Grim, 1991; Ashby et al., 1993). How does patterned mesenchyme induce and control muscle morphogenesis? The fact that the spatial and temporal patterns of muscle splitting and shaping are unique for each muscle and do not reflect general gradients of limb differentiation (Grim, 1991; Schroeter and Tosney 1991a ,b) argues for specific local signaling processes. In principle, the end-to-end and parallel arrangement of primary myotubes within the forming muscles assigns a unique potential role to myotube end regions for muscle morphogenesis (Fig. 1). Accordingly, one possibility is that mesenchyme-to-muscle signaling may involve local regulation of myotube formation, orientation, and growth at the interface between myotube end regions and adjacent mesenchyme. Such local signaling should be reflected in the expression of corresponding molecular markers in the mesenchyme and in the responding muscle cells as well as in the existence of a spatially restricted subcompartment at primary myotube end regions.
In a search for genes involved in the regulation of muscle gene expression and the formation of neuromuscular synapses, we constructed and screened a cDNA library enriched in messages induced in adult skeletal muscle by denervation. As reported elsewhere, two clones coded for a striated muscle-specific positive regulator of myogenesis: muscle LIM protein (MLP; Arber et al., 1994) and thrombospondin-4 (TSP-4; Arber and Caroni, 1995), whose expression is induced in denervated muscle interstitial cells. We report here that a third clone codes for a nuclear protein with four ankyrin repeats that is expressed in striated muscle and vascular endothelial cells. Due to its predominant expression in striated muscle we have called it MARP (muscle ankyrin repeat protein). During the course of this work, isolation of the homologous human protein (C-139; Chu et al., 1995) and of the identical rat cDNA (cardiac ankyrin repeat protein [CARP]; Zou et al., 1997) were reported. MARP mRNA has degradation-promoting motifs characteristic of regulatory factors. Consistent with a possible role in signaling, MARP protein is rapidly degraded in the cell, and degradation can be substantially delayed by the addition of short carboxyl-terminal extentions. The distribution of MARP mRNA in developing mouse skeletal muscle revealed a unique and highly localized pattern associated with muscle morphogenesis. This was accompanied by a complementary pattern of TSP-4 expression in adjacent mesenchyme. The results establish the existence of novel muscle subcompartments associated with muscle morphogenesis and the perisynaptic region of denervated muscle and suggest that MARP may be a key nuclear cofactor in novel local signaling pathways to muscle.
Materials and Methods
Nucleic Acid Reagents
A partial clone coding for rat MARP was isolated from a subtracted plasmid cDNA library enriched in transcripts induced in adult diaphragm muscle 7 d after denervation. The construction of the library and the screening procedure were as described (Arber et al., 1994). Subsequently, several additional clones, including a 1.7-kb putative full length clone were isolated through rescreening of the same library. Mouse MARP cDNA clones were isolated from an adult mouse heart cDNA library, due to their hybridization to a rat MARP cDNA probe. A genomic clone containing MARP proximal promoter sequences was isolated with mouse MARP cDNA probes from a mouse (strain 129Sv) genomic library (Stratagene GmbH, Heidelberg, Germany). The promoter fragment shown in Fig. 2 C drives the expression of a reporter gene in transfected myogenic cell lines, indicating that it is a functional promoter (data not shown). cDNAs for Northern blot analysis and in situ hybridization were from the following sources: rat MLP (Arber et al., 1994), rat TSP-4 (Arber and Caroni, 1995), rat clone-16 (a cDNA isolated in our screen for denervation- induced genes; see Aigner et al., 1995), rat MyoD (kind gift from A. Buonanno, National Institutes of Health, Bethesda, MD), mouse tenascin-C (kind gift from R. Chiquet-Ehrismann, Friedrich Miescher Institute, Basel, Switzerland), mouse FGF6 (probe corresponding to 434 bp of 3′ untranslated sequences, as in deLapeyriére et al., 1993; kind gift of D. Birnbaum, U119 Institut National de la Sante et de la Recherche Medicale, Marseille, France), mouse protein kinase A subunits Riα and Cα (kind gift of M.C. Weiss, Institut Pasteur, Paris, France).
Figure 2.
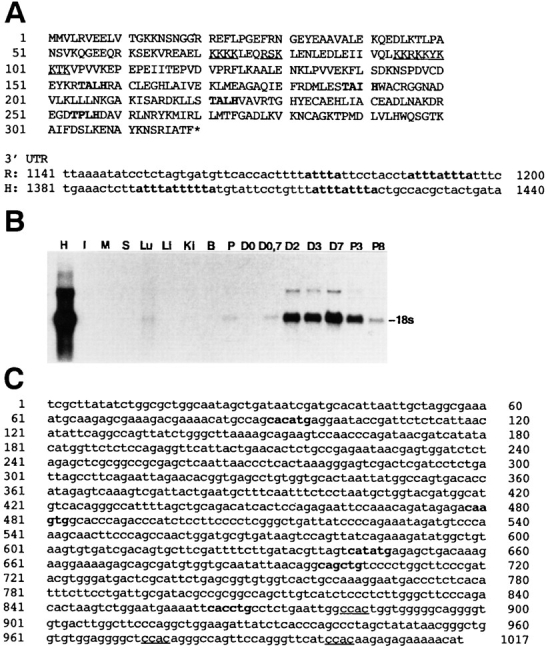
Sequence of rat MARP and of the mouse MARP promoter. Induction of MARP in denervated skeletal muscle. (A) Deduced amino acid sequence of rat MARP. The bipartite nuclear localization sequence is underlined, and the first residues of the four ankyrin repeats are in bold. (3′ UTR) Partial sequences of rat (R) and human (H) MARP mRNA with the ATTTA degradation sequences (bold). (B) MARP mRNA contents in adult rat tissues. Northern blot of comparable amounts of total RNA from heart (H), intestine (I), diaphragm muscle (M), skin (S), lung (Lu), liver (Li), kidney (Ki), brain (B), placenta (P), and gastrocnemius muscle before (D0) and 0.7, 2, 3, and 7 d after denervation, or 3 and 8 d after Botulinum toxin-A–induced paralysis (P). Note strong induction by denervation or paralysis of skeletal muscle. (C) Nucleotide sequence of the proximal promoter of mouse MARP. Muscle E-box sequences are in bold; muscle CCAC boxes are underlined.
Nucleic Acid Hybridization
RNA isolation and Northern blot analysis with digoxigenin-labeled riboprobes (reagents from Boehringer Mannheim Corp., Indianapolis, IN) were carried out as described in a previous study (Arber et al., 1994). For Northern blots, equal amounts of total RNA were loaded on a fomaldehyde gel, as verified by staining with methylene blue. In situ hybridization with digoxigenin-labeled cRNA probes was carried out according to a published protocol (Schaeren-Wiemers and Gerfin-Moser, 1993). For mouse embryos, the morning when a vaginal plug was detected was considered as E0.5. Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C. After extensive cryoprotection with 30% sucrose in PBS (4°C), fixed embryos were mounted in embedding medium and quickly frozen at −40°C in 2-methybutane. 12–14 μm cryosections were pretreated and hybridized overnight at 72°C, as described. The conditions for the proteinase K (Boehringer Mannheim Corp.) pretreatments of the sections (at room temperature and in 50 mM Tris, pH 7.5, 5 mM EDTA) were as follows: E11.5–13.5 embryos: 5 min, 1 μg/ml; E14.5–17.5 embryos: 8 min, 1 μg/ml; P1 tissue: 5 min, 5 μg/ml; adult muscle: 6 min, 20 μg/ml. Complete sets of consecutive sections for the comparative analysis of expression patterns were processed in parallel, and the experiments were carried out with several independent groups of embryos, with comparable results. In control experiments, the specificity of the in situ hybridization signals was verified by the complete absence of any detectable signal with corresponding sense probes. Visualization of histological features on adjacent sections with hematoxylin–eosin stains or with an acetylcholine esterase reaction was according to standard procedures.
Transfection and Coculture Experiments
All cell lines (mouse 3T3 fibroblasts; mouse C2C12 myoblasts; rat L6 myoblasts; rat 10T1/2 fibroblasts; monkey kidney epithelial cells COS-1) were from American Type Cell Culture Collection (Rockville, MD). Cells were cultured, transfected with the lipofectamine reagent (GIBCO BRL, Gaithersburg, MD), and differentiated as described (Arber et al., 1994). When indicated, cultures were treated for 6 to 8 h with 10–50 μg/ml cycloheximide (Fluka, Buchs, Switzerland). For transfections, a CMV promoter-based eukaryotic expression vector (pcDNA3; Invitrogen, San Diego, CA) was used. To obtain a MARP construct devoid of 3′ ATTTA motifs, cDNA sequences downstream of nucleotide 999 (i.e., 37 bp 3′ from the MARP stop codon) were deleted, and the deletion construct was subcloned in pcDNA3. Epitope tags were added to MARP cDNA using the PCR, with appropriate primers. The corresponding MARP constructs had the following amino acid sequences: MEQKLISEEDLN-MM . . . (Myc-MARP); ..ATFAAAPMEQKLISEEDLN (MARP-Myc); MEQKLISEEDLNI-MMV. . . .ATFAAAPGLVVMNIT (Myc-MARP-GLVVMNIT). The rabbit antibody against the synthetic carboxyl-terminal peptide VLRVEELVTGKKC of MARP was produced by coupling the peptide to keyhole lympet hemocyanin (Sigma Chemical Co., St. Louis, MO) via a carboxyl-terminal Cys. Further antibodies were from the following sources: rabbit antiserum to carboxyl-terminal peptide GLVVMNIT (kind gift of R. Chiquet-Ehrismann, Friedrich Miescher Institute), monoclonal antibody to Myc epitope (kind gift of A. Matus, Friedrich Miescher Institute), monoclonal antibody to sarcomeric α-actinin (Sigma Chemical Co.). For immunoblots, tissues or cells were homogenized in SDS-PAGE sample buffer, homogenates were boiled, and solubilized proteins were fractionated. For immunocytochemistry, cells (usually 24 h after transfection) were fixed for 10 min at room temperature with 4% paraformaldehyde in PBS and then permeabilized for 7 min at room temperature with 0.2% Triton X-100 in PBS. Antibody incubations were carried out in PBS with 5% BSA, and all washes were in PBS. Double-labeling immunocytochemistry was carried out with biotinylated secondary antibodies, followed by lucifer yellow–conjugated streptavidin for the first channel and rhodamine-coupled second antibodies for the second epifluorescence channel. All secondary antibodies were from Molecular Probes Inc. (Eugene, OR).
4–5 h before the addition of muscle-derived cells, COS or COS-TSP-4 cells (Arber and Caroni, 1997) were plated at densities of 40,000–80,000 cells on laminin-coated 35-mm culture dishes in the presence of DME with 10% horse serum and 5% chick embryo extract. Muscle-derived cells consisted mainly of myoblasts and fibroblasts and were obtained and cultured according to standard procedures. Briefly, muscle from the hind limbs of P0-P1 mice was minced, trypsinised for 45 min at 37°C, and triturated with a pasteur pipette in the presence of DNaseI. Cells were then filtered through a cell strainer (70 μm) and preplated for 1.5 h in DME with 10% horse serum and 5% chick embryo extract. Nonadherent cells were counted, and 500,000 cells were plated onto the laminin/COS or laminin/ COS-TSP-4 dishes. After 2 d of coculture the medium was changed to DME with 2% horse serum (differentiation medium), and myotube formation was scored 1 and 2 d later.
Results
MARP Is an Inducible Gene of Adult Skeletal and Heart Muscle
We isolated MARP cDNA as a clone that was strongly induced by denervation in adult skeletal muscle (see Arber et al., 1994, where MARP was designated as clone 4). The deduced amino acid sequence codes for a protein of 319 amino acids, with four ankyrin repeats. The sequence of rat MARP is shown in Fig. 2 A. It is identical to that of rat CARP (Zou et al., 1997) and 90% homologous to the human cDNA C-193, which was identified as an immediate early gene of vascular endothelial cells (Chu et al., 1995). Shared elements include a bipartite nuclear localization motif (residues 71–80 and 94–103), a PEST-like sequence characteristic of rapidly degraded proteins (residues 108– 126; Rechsteiner and Rogers, 1996), and the four ankyrin repeats (Blank et al., 1992; residues 152–184, 185–217, 218– 250, and 251–283) that are flanked by a carboxyl-terminal extension of 36 residues. In addition, the MARP (rat, mouse) and C-193 (human) transcripts contain multiple ATTTA degradation motifs (Shaw and Kamen, 1986) in their 3′ untranslated sequences (Fig. 2 A). Based on the high degree of sequence homology and the conserved regulatory properties of the transcripts and proteins (see below), we tentatively conclude that MARP is the rodent homologue of C-193. As shown in Fig. 2 B, adult rat heart contained high levels of MARP mRNA, whereas very low levels were detected in lung and placenta, and either no or extremely low signals were detected in intestine, skeletal muscle, skin, liver, kidney, and brain. Denervation led to massive induction of MARP mRNA in skeletal muscle (Fig. 2 B). Comparison with other denervation-sensitive transcripts showed that MARP induction was particularly rapid (significantly faster than MLP and clone-16 and at least as fast as myogenin; not shown). Finally, Fig. 2 C shows the sequence of the proximal promoter of mouse MARP. Consistent with the expression of MARP in skeletal and heart muscle, this sequence includes five muscle E-boxes and three CCAC-boxes that regulate transcription in skeletal and cardiac muscle cells (Fig. 2 C; Buckingham, 1992; Grayson et al., 1995).
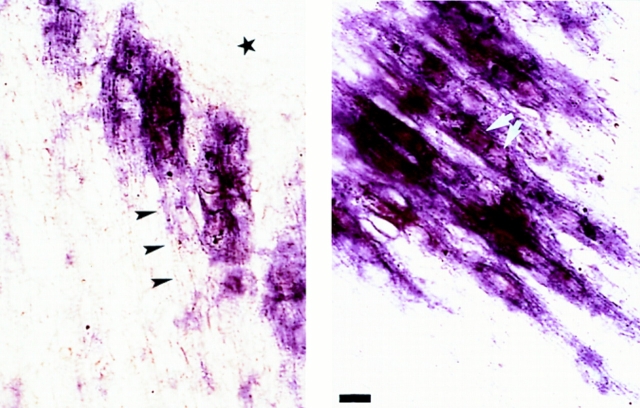
Fig. 3 A shows that no MARP transcript was detectable in adult medial gastrocnemius muscle, where strong expression in the vicinity of acetylcholine esterase–positive neuromuscular synapses was induced by denervation. At closer inspection, the MARP signal frequently highlighted striations and accumulated in a longitudinal pattern along the inner face of muscle fibers, indicating that it was localized inside skeletal muscle fibers (Fig. 3 B). This dramatic accumulation pattern in denervated muscle had features unique to MARP. Thus: (a) MyoD and MLP transcripts accumulated throughout denervated muscle fibers, and the signal for clone-16 mRNA only showed partial synaptic enrichment (data not shown); (b) although the α subunit of the acetylcholine receptor displays some synaptic enrichment, it is largely due to its synaptic expression in innervated muscle, and synaptic accumulation is in fact reduced in denervated muscle (Fontaine and Changeux, 1989; Witzemann et al., 1991); (c) while typical synapse- associated transcripts such as the α subunit of the acetylcholine receptor or the regulatory subunit RIα of protein kinase A display local accumulation restricted to the subsynaptic region (i.e., coincident with acetylcholine esterase reaction product; data not shown; see Witzemann et al., 1991; Imaizumi-Scherrer et al., 1996), MARP transcript accumulated in a much broader perisynaptic pattern, suggesting local regulation by signals in the vicinity of denervated synapses. Unlike these muscle fiber transcripts, and as reported previously, TSP-4 mRNA accumulated in muscle interstitial cells (Fig. 3 B; see also Arber and Caroni, 1995). In innervated muscle it was restricted to epimysium, whereas denervation induced expression in the endomysium (Arber and Caroni, 1995). Interestingly, induction was distinctly more pronounced in perisynaptic regions, providing a first example of spatial correlation between the accumulation of MARP mRNA in muscle fibers and that of TSP-4 in adjacent cells (see below). The dramatic perisynaptic accumulation of MARP transcript in denervated muscle is highly unusual. It suggests that this transcript is subject to local regulatory mechanisms and that even several days after removal of the motor nerve the perisynaptic region of muscle is under the influence of distinct local signals.
Figure 3.
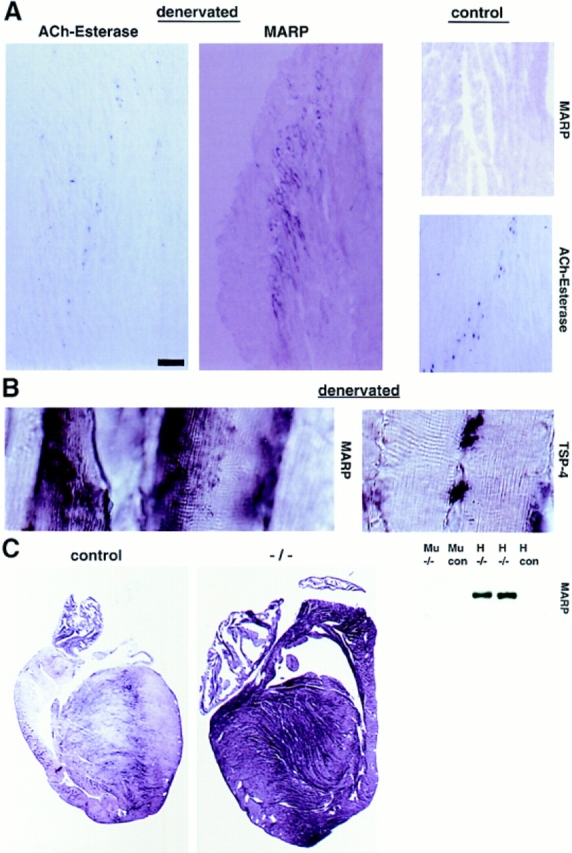
Local patterns of MARP transcript induction in adult denervated muscle and hypertrophic heart. (A) Perisynaptic accumulation of MARP transcript in denervated muscle. Gastrocnemius muscle of adult mice was collected 7 d after resection of the sciatic nerve. Note prominent MARP in situ hybridization signal at a broad region near to acetylcholine esterase reaction product (adjacent section on the right). As expected from the absence of the corresponding mRNA on Northern blots, no MARP signal was detected in control muscle (right). (B) High-magnification photographs of MARP (left) and TSP-4 (right) transcript localization in the perisynaptic region of 7-d denervated gastrocnemius muscle. Note MARP signal along the inner face of striated muscle fibers. Distribution of the TSP-4 transcript, which accumulates in a muscle interstitial cell pattern clearly distinct from that of MARP is shown for comparison. (C) Induction of MARP transcript in the hypertrophic heart. (Left) In situ hybridization for MARP mRNA in adult mouse heart. In control heart MARP mRNA is expressed at high levels in atria and at low levels in ventricles. In the hearts of MLP-deficient mice (−/−) with dilated cardiomyopathy and hypertrophy (Arber et al., 1997), MARP transcript is strongly upregulated in ventricles. (Right) Selective upregulation of MARP mRNA in hypertrophic ventricles. The Northern blot was hybridized with a MARP probe and comparable amounts of total RNA from adult ventricle (H) or gastrocnemius muscle (Mu) from control or MLP-deficient (−/−) mice were applied to the gel. Bar: (A) 70 μm, (B, MARP) 8 μm, (B, TSP-4) 16 μm.
Fig. 3 C shows the localization of MARP transcript in a control and an MLP-deficient mouse heart with dilated cardiomyopathy and hypertrophy (Arber et al., 1997). In control hearts, MARP mRNA levels were very high in atria, whereas ventricles displayed a characteristic inhomogeneous distribution. Interestingly, and similar to atrial natriuretic factor (Chien et al., 1992), cardiac hypertrophy led to massive induction of MARP message in ventricular cardiomyocytes, whereas atrial levels were much less affected. Fig. 3 C also shows that although by Northern blot MARP was expressed at substantial levels in the heart of normal adult mice (Fig. 2 B), hypertrophic hearts contained markedly elevated levels of MARP transcript. Unlike MARP, the denervation-sensitive transcripts MLP and clone-16 were not elevated in hypertrophic heart (data not shown). MARP is therefore an inducible gene in both adult skeletal and heart muscle.
MARP Is a Nuclear and Cytosolic Protein Subject to Tight Posttranscriptional Regulation
To investigate the cellular regulation of MARP mRNA we analyzed its levels in non- and myogenic cell lines. As shown in Fig. 4 A, the transcript could be detected in the myogenic cell line C2C12, in 10T1/2 fibroblasts, but not in 3T3 fibroblasts. Significantly, when C2C12 cells were transferred to differentiation medium, MARP transcript levels were selectively upregulated in myotubes (Fig. 4 B). Myogenic cell lines exhibited varying degrees of coupling between MARP transcript levels and differentiation. Thus, while one batch of C2C12 cells displayed detectable expression only upon differentiation (see Arber et al., 1994), a second C2C12 batch (Fig. 4 A) and L6 cells (data not shown) already expressed significant levels under nondifferentiating conditions. In 10T1/2 cells, but much less so in C2C12 cells MARP mRNA levels were elevated in the presence of the protein synthesis inhibitor cycloheximide (Fig. 4 A). This is reminiscent of human vascular endothelial cells, where C-193 transcript has early response properties in small vessel cells and is expressed at much higher levels in large vessel cells (Chu et al., 1995).
Figure 4.
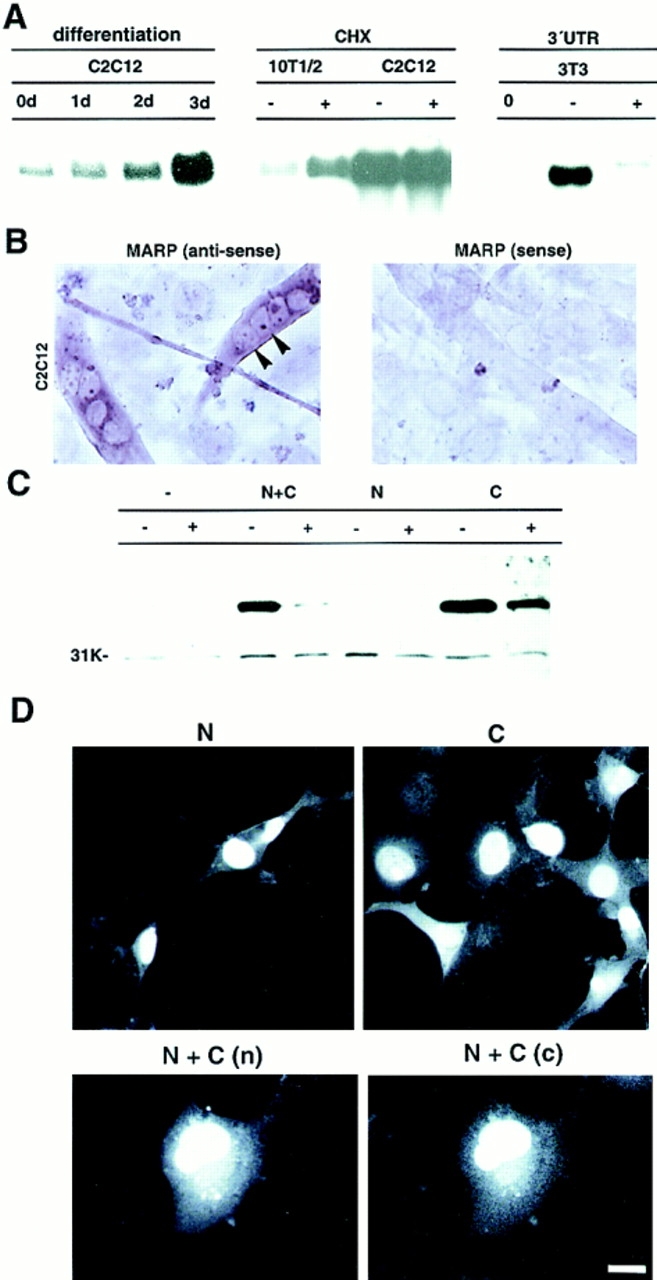
MARP has properties of a nuclear early response gene. (A) Regulation of MARP mRNA contents by myogenic differentiation, cycloheximide, and 3′ untranslated sequences (UTR). The RNA blots (equivalent amounts of total RNA for each blot) were hybridized with a rat MARP probe. The experiment on the left was carried out with a C2C12 subclone exhibiting MARP induction during differentiation (days). The C2C12 subclone used for the cycloheximide (CHX) induction experiment already expressed high levels of MARP in high-serum medium, whereas CHX induced MARP transcript in 10T1/2 fibroblasts. (− and +) RNA collected after 6 h without or with CHX. For the experiments on the right, Swiss 3T3 fibroblasts were transfected with MARP expression constructs containing (+) or devoid (−) of 3′ untranslated region sequences, and RNA was collected 2 d after transfection. The presence of MARP 3′ untranslated region sequences led to greatly reduced transcript levels. (0) Nontransfected cells. (B) In differentiating C2C12 cells MARP transcript selectively accumulates in multinucleated myotubes (arrows). In situ hybridization of cultures 3 d after switch to differentiation medium. (Right) MARP sense probe as a negative control. The C2C12 subclone was as in (A) (Left) Note homogeneous pattern of MARP transcript accumulation in myotubes in vitro. (C) MARP protein is rapidly degraded in transfected cells, and degradation can be delayed by the fusion of short peptide sequences to the carboxyl-terminal end of the protein. COS cells were transfected with MARP expression constructs, and MARP contents were determined on the immunoblot (anti-MARP antiserum). Equal amounts of cell homogenate protein were applied for each sample (see background band at ∼31 kD). (N + C) myc-tag at the amino terminus and GLVVMNIT-tag at the carboxy terminus; (N) myc-tag at the amino terminus; (C) myc-tag at the carboxyl terminus; (−) no epitope tags. For each MARP construct, cells were incubated with (+) or without (−) CHX during the last 8 h before homogenization. (D) Nuclear and cytosolic accumulation of MARP in transfected COS cells. The epitope-tagged constructs are defined as described in (C). (Top) Anti-myc labeling of amino- (left) and carboxyl-terminal–tagged (right) MARP. Note higher frequency of labeled cells and of cytosolic signal in the presence of carboxyl-terminal tag sequence. Double-tagged (N + C) MARP was visualized by double labeling for the amino terminus (n) and carboxyl terminus (c) epitope tag. Note nuclear accumulation of undegraded MARP. Bar: (B) 15 μm; (D, single labelings) 9 μm; (D, double labeling) 6 μm.
The rat MARP transcript has five closely spaced ATTTA motifs in its 3′ untranslated sequence (Fig. 2 A). Deletion of 3′ untranslated sequences, including the ATTTA motifs, resulted in dramatically higher transcript levels in transfected 3T3 cells, confirming that MARP mRNA carries functional degradation signals (Fig. 4 A). Similar results were obtained with transfected C2C12 and 10T1/2 cells (data not shown). To analyze the regulation and subcellular localization of MARP we generated an antiserum against the amino-terminal end of the protein. Surprisingly, in spite of the substantial corresponding Northern blot signals, MARP protein signals on immunoblots from heart homogenates were very low, and no signals could be detected on immunoblots of differentiated L6 and C2C12 cells (data not shown). A weak signal could be detected on immunoblots of transfected COS (Fig. 4 C) and C2C12 cells. To exclude effects due to the instability of MARP transcripts, all transfection experiments were carried out with 3′ untranslated region–deleted constructs (Fig. 4 A), and the presence of comparable transcript levels was verified on corresponding Northern blots. To determine whether detection difficulties were due to poor antisera we analyzed immunoblots of cells transfected with epitope-tagged MARP constructs. These experiments led to the surprising finding that the addition of short amino acid extensions to the carboxyl-terminal end of MARP greatly protected this protein against degradation. The analysis revealed that addition of any of two different epitope tags to the carboxyl-terminal end of MARP resulted in greatly elevated levels of the corresponding protein. In contrast, amino-terminal epitope tags were ineffective (Fig. 4 C; note that all constructs were detected with antiserum against a sequence from the amino-terminal end of MARP). The fact that the carboxyl-terminal tags also reduced the decline of MARP contents in the presence of protein synthesis inhibitor (Fig. 4 C) further argues for protection against protein degradation. The data of Fig. 4 C were obtained with COS cells, but similar results were obtained in transfected C2C12 cells (data not shown).
MARP contains a classical nuclear localization sequence, suggesting that it may be a nuclear protein. Detection with MARP-specific antiserum revealed accumulation in the nucleus and in the cytosol of transfected cells (Chu et al., 1995; Zou et al., 1997). About two thirds of the labeled cells showed an exclusively nuclear pattern, whereas the rest of the cells also displayed cytosolic signal (Fig. 4 D). Cytosolic MARP frequently accumulated in clump-like structures, suggesting aggregation and/or degradation. To determine whether the entire MARP protein translocates to the nucleus and whether stabilization against proteolytic degradation may reveal selective accumulation in a cellular compartment, we analyzed the subcellular localization of epitope-tagged MARP constructs. When amino and carboxyl-terminus-tagged protein was double labeled for both epitopes, a majority of the labeled cells displayed comparable distribution of the two epitopes (Fig. 4 D). These results indicate that a substantial proportion of apparently intact MARP accumulates in the nucleus (Chu et al., 1995; Zou et al., 1997). It is therefore likely that this highly regulated early response protein exerts its function in this compartment. In addition, the experiments revealed that besides greatly elevating the number of cells with detectable MARP signals (∼20-fold increase), carboxyl-terminal tags also led to a substantially higher proportion of cytosolic MARP in the transfected cells (Fig. 4 D). A possible interpretation of these findings is that carboxyl-terminal epitope tags protect MARP from degradation in the cytosol.
The Spatial and Temporal Distribution of MARP mRNA in Embryonic Mouse Skeletal Muscle Coincides with Muscle Morphogenesis
In mouse embryos before E11.5, MARP mRNA was only detectable in the heart and in large blood vessels. Both tissues expressed high levels of MARP transcript at all later stages of embryogenesis. Prominent expression in the developing and adult heart is consistent with the findings of Zou et al. (1997), and selective accumulation of MARP mRNA in large blood vessels is consistent with the constitutive expression of the human homologue of MARP (C-193) in endothelial cell lines derived from large, but not small blood vessels (Chu et al., 1995). Based on the tissue culture results with myogenic cell lines, the absence of MARP mRNA in myogenic regions of E10.5 embryos was unexpected, since in these embryos, determined myogenic cells already express MyoD, which in mouse skeletal muscle cells is the latest of the myogenic determination factors (Buckingham, 1992). At E11.5, MARP mRNA was detectable in the myotome region (Fig. 5 A). Comparison with the somitic expression patterns of MyoD, and of MLP, which is only expressed in terminally differentiated skeletal muscle cells (Arber et al., 1994), revealed the presence of MARP transcript in a subset of cells within MLP-positive territories (Fig. 5 A). In contrast, no MARP expression was detected in E11.5 forelimb, which contained MyoD-positive, but MLP-negative cells (Fig. 5 A). This coincidence between MLP- and MARP-positive areas was consistently observed between E11.5 and 16.5. The observation that within MLP-expressing areas MARP-positive cells were consistently less numerous than those expressing MLP prompted us to analyze these cells in E14.5 embryos, where myotubes are easier to identify. As shown in Fig. 5 B, MLP-positive myotubes were arranged in small clusters of primary myotubes. These clusters were oriented parallel to each other and spanned the muscle from end to end (Fig. 5 B; see Fig. 7, left, for E12.5 data and Fig. 1 for a schematic representation of myotube arrangement during muscle morphogenesis). In contrast, MyoD-positive cells, which included myotubes and muscle precursor cells, displayed a more homogeneous distribution. Hybridization for MARP mRNA revealed a striking pattern specifically associated with the end regions of primary myotube clusters (Figs. 5 B, 6, and 7). The higher magnification photographs show that MARP mRNA accumulated inside multinucleated striated muscle fibers (Fig. 6). Systematic analysis revealed that all MLP-positive muscles in these embryos contained MARP-positive muscle fibers.
Figure 5.
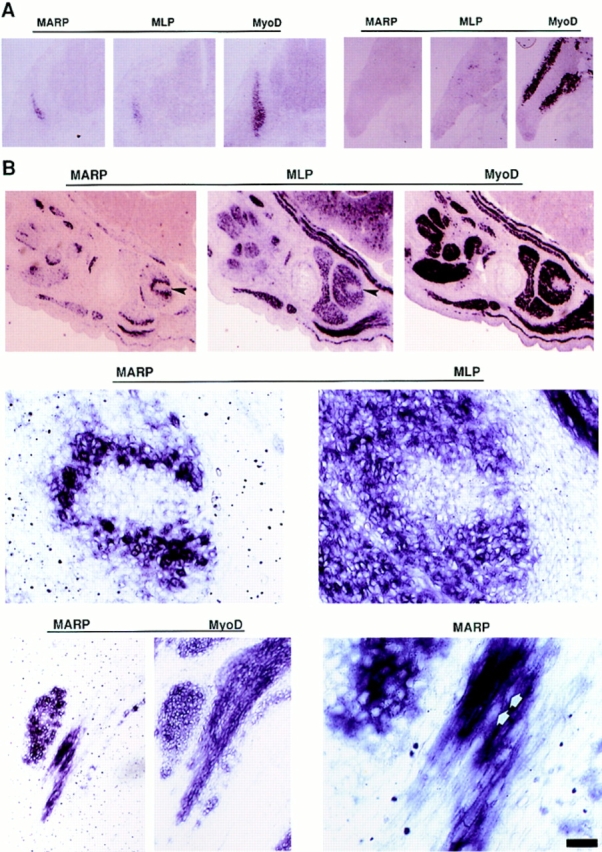
During muscle formation in vivo, appearance of MARP transcript coincides with terminal differentiation, and the transcript selectively accumulates at muscle end regions. In situ hybridization of mouse embryo sections. MyoD is expressed in myogenic cells before and after terminal differentiation; MLP is a marker for terminally differentiated striated muscle cells. The horizontal lines indicate consecutive sections (14 μm). (A) At E11.5, MARP mRNA is detectable within MLP-positive myotome (left), whereas in the forelimb, MyoD-positive myogenic progenitor cells were negative for MLP and MARP transcripts. (B) At E14.5, MARP transcript accumulated at end regions of MLP- and MyoD-positive muscles (top; the example is from foot muscles). A detail from the top panels (arrows) is shown in the middle panels. Note restriction of MARP transcript to edge of the indentation in the MLP-positive muscle. The bottom panels show an example of MARP transcript accumulation at the end of a MyoD-positive muscle (between ribs). In the corresponding higher magnification photograph on the right, MARP transcript can be detected inside striated cells, i.e., primary myotubes (arrows point to spared nuclei). Bar: (A) 480 μm; (B, top) 240 μm; (B, middle) 30 μm; (B, bottom, left) 60 μm; (B, bottom, right) 15 μm.
Figure 7.
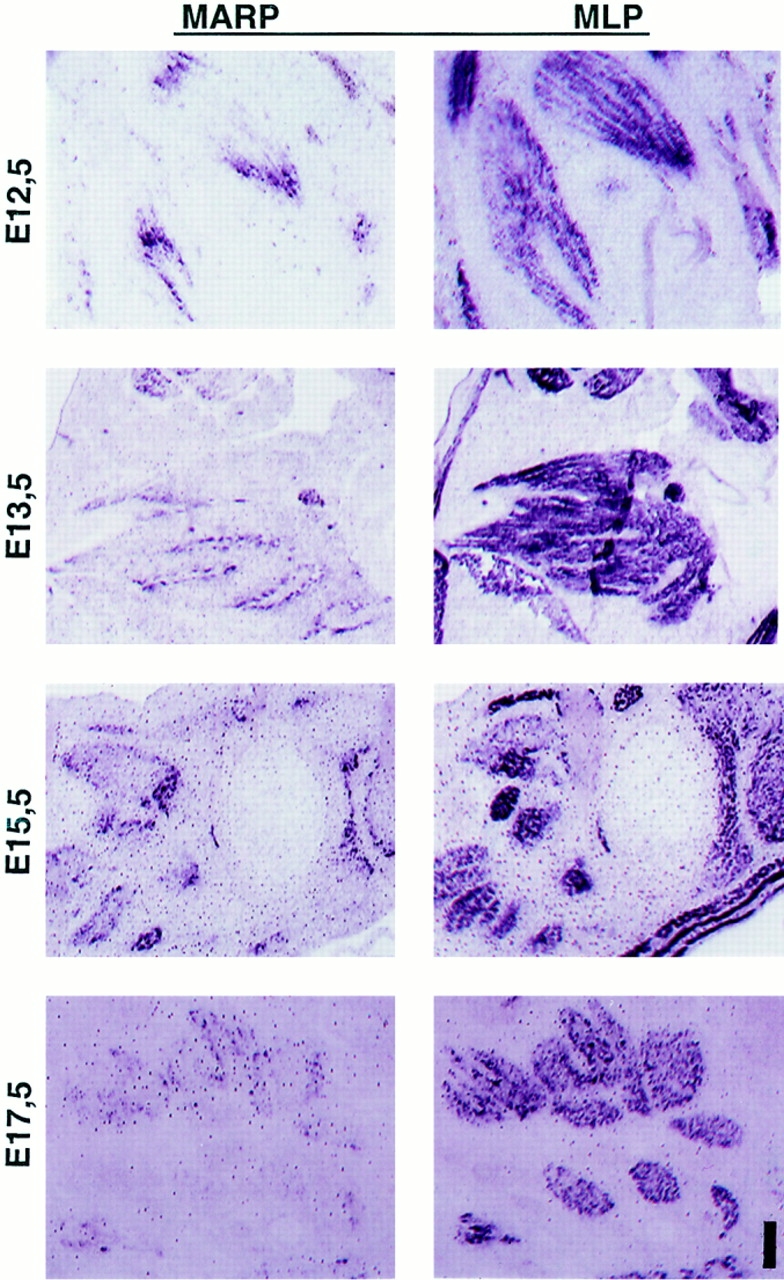
Accumulation of MARP transcript at end regions of skeletal muscle during muscle morphogenesis. In situ hybridization of consecutive sections labeled for MARP (left) and MLP (right) transcript. The sections display different groups of muscles between E12.5 and 17.5 (E12.5, face; E13.5 and 15.5, hindpaw; E17.5, front paw). Note consistent restriction of MARP transcript to myotube end regions. MARP expression was strongly downregulated at E17.5. Bar: (E12.5) 180 μm; (E13.5, 15.5, and 17.5) 360 μm.
Figure 6.
Accumulation of MARP transcript at end regions of primary myotubes. Two high-magnification examples of MARP transcript localization at end regions of facial muscles in an E14.5 mouse embryo are shown. (Left) Nonmuscle region (top right) is marked with an asterisk, and the three arrows delineate the nonlabeled part of a primary myotube bundle (striations) that extends towards the opposite end of the muscle; MARP signal is restricted to the end regions of the primary myotube bundles. (Right) In this example the muscle end is at the top left corner; multinucleated (arrows; nuclei are spared in the in situ hybridization labeling) primary myotube bundles are labeled from the muscle end, and signal intensity decreases towards the lower right corner of the figure, i.e., towards the opposite end of the muscle. Bar, 11 μm.
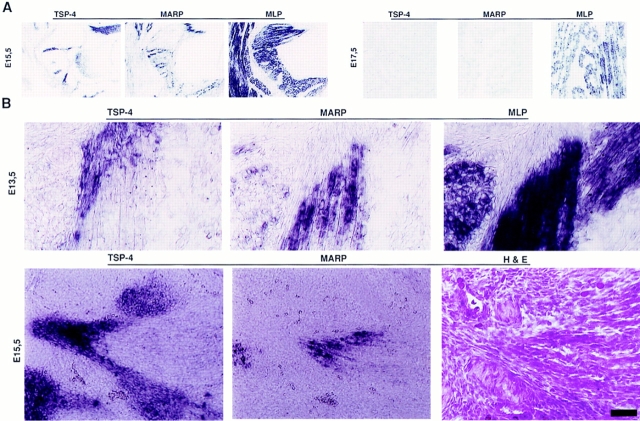
The expression of MARP in embryonic skeletal muscle was restricted to early stages of muscle formation. As shown in Fig. 7, MARP transcript (left) was detectable in all MLP-positive muscles (right) between E12.5 and 15.5, with highest signal levels around E14.5. Throughout this period, MARP mRNA was consistently restricted to muscle end regions. MARP mRNA levels started to decline after E15.5, and by E17.5 signals were absent from most muscles (Fig. 7). Therefore, the temporal and spatial pattern of MARP expression coincides with splitting, shaping, and tendon attachment of developing muscles, i.e., with muscle morphogenesis.
During Muscle Morphogenesis Expression of TSP-4 in Prospective Tendon Mesenchyme Is Adjacent to that of MARP-positive Skeletal Muscle Fibers
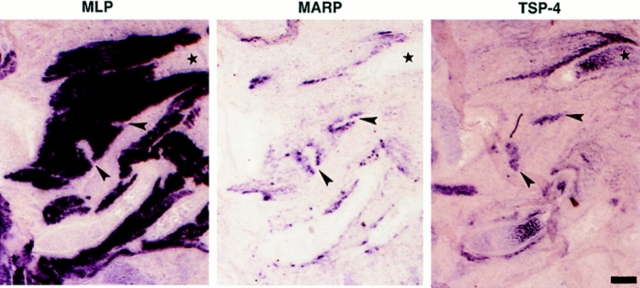
What signals are responsible for the striking accumulation of MARP transcript at the end regions of primary myotubes in forming muscles? Because of the role of mesenchyme in muscle morphogenesis, we searched for transcripts expressed in regions adjacent to MARP-positive muscle. Due to its association with tendon regions (Tucker et al., 1994) and its upregulation in interstitial cells of denervated muscle (Arber and Caroni, 1995), TSP-4 was one potential candidate. Before E12.5, TSP-4 mRNA was detected in the developing heart (data not shown). In addition, like in the chick (Tucker et al., 1995), TSP-4 mRNA accumulated in chondrogenic regions of the mouse embryo. Close examination of the TSP-4 signal in the vicinity of developing cartilage revealed a striking complementarity to the localization of MARP mRNA (Fig. 8). The high-magnification photographs (Fig. 9 B) show that the MARP and TSP-4 signals precisely apposed each other, and that TSP-4 expressing tissue was coextensive to muscle tissue. Exclusion of muscle transcripts from TSP-4–expressing end regions was also observed for MLP (Fig. 9) and MyoD mRNA. Comparable relative localizations of MARP and TSP-4 transcripts were detected from E13.5 to 17.5 (Figs. 8 and 9). As shown in Fig. 9 B, spatial coincidence was not restricted to regions adjacent to developing bone. The characteristic arrangement of the TSP-4 signal with respect to forming muscle suggested that it was associated with prospective tendon mesenchyme. This conclusion was supported by the fact that TSP-4–expressing tissue also accumulated tenascin-C transcript (data not shown; Chiquet and Fambrough, 1984).
Figure 8.
At muscle end regions, MARP and TSP-4 transcripts accumulate at adjacent locations. Consecutive sections (chest muscles, above heart) from an E14.5 embryo were hybridized for MLP, MARP, and TSP-4 transcript, as indicated. Note similar distribution and complementarity of MARP and TSP-4 labeling patterns (two examples are indicated by the arrowheads). In addition to regions adjacent to muscle ends, TSP-4 transcript also accumulated in chondrogenic areas (one example indicated by the asterisk). Bar, 500 μm.
Figure 9.
TSP-4 transcript accumulates at prospective tendon mesenchyme regions adjacent to MARP-positive primary myotube ends. In situ hybridization experiments; horizontal lines indicate adjacent sections. (A) Spatial and temporal correlation of MARP and TSP-4 transcript accumulation. Trunk muscles, cervical region. Note TSP-4 signal adjacent to MARP (e.g., at muscle indentations). Also note that at E17.5 both MARP and TSP-4 signal were downregulated. (B) MARP transcript accumulates at the ends of primary myotubes, adjacent to TSP-4–expressing prospective tendon mesenchyme. Consecutive sections from E13.5 (top) and E15.5 (bottom) intercostal muscle. Note coextensive arrangement of MARP- and TSP-4–expressing structures. H&E, Haematoxilin and eosin stain. Bar: (A) 1.1 mm, (B, top) 35 μm, (B, bottom) 140 μm.
In addition to a close spatial correlation, MARP and TSP-4 transcripts were also correlated temporally: they appeared together around E12.5, declined after E15.5, and were nearly undetectable at most muscle end regions at E17.5 (Fig. 9 A). This was in contrast to tenascin-C mRNA, whose expression at the forming tendon persisted beyond the period of muscle morphogenesis (data not shown). The tongue provided a striking example of the spatial and temporal correlation of TSP-4 and MARP transcript accumulation. In this muscle-rich organ with no cartilage, muscle groups are linked by tendinous scaffold. Particularly high levels of TSP-4 and MARP mRNA were detected in the developing tongue, where the two transcripts accumulated at complementary locations. Unlike in other muscles, expression of both transcripts was maintained throughout late embryogenesis and was still detectable at postnatal day 1 (not shown). These observations demonstrate that prospective tendon mesenchyme adajacent to sites of MARP mRNA accumulation in myotubes can be visualized by the expression of TSP-4. The close temporal and spatial relation between the two transcripts and the likely role of muscle attachment regions in muscle morphogenesis (Kardon, G. 1996. Dev. Biol. 175:393a) suggest that accumulation of MARP transcript may be a local response of primary myotubes to TSP-4–expressing mesenchyme.
What types of functional responses may be induced at muscle end regions by prospective tendon mesenchyme? As mentioned above, one possibility is local induction of muscle growth, including proliferation and fusion of myoblasts to primary myotubes, myogenic differentiation, and myotube extension. A previous in vitro study demonstrated that myoblasts adhere to a peptide corresponding to the cell-binding carboxyl-terminal end of TSP-4 (Adams and Lawler, 1994). Accordingly, to determine whether TSP-4 can promote myogenic differentiation we carried out coculture experiments with primary myoblasts from newborn mice and TSP-4–expressing COS cells (COS-TSP-4; Arber and Caroni, 1995). The latter secrete substantial amounts of TSP-4, which was shown in a previous study to efficiently promote neurite outgrowth (Arber and Caroni, 1995). COS-TSP-4 or naive COS cells were plated at low density on laminin-coated culture dishes, and hind limb muscle-derived cells were added 4–5 h later. After 2 d in growth-promoting medium the cultures were switched to differentiation medium, and myotube formation was scored 1 and 2 d later. As shown in Fig. 10, under these experimental conditions, myogenic differentiation and myotube formation were markedly potentiated in the presence of the COS-TSP-4 cells. Muscle-derived cells, which were noticably smaller and thus easily distinguishable from cocultured COS cells, could be detected in comparable amounts in the two types of cultures. However, in the absence of TSP-4 these cells only expressed low levels of muscle-specific α actinin and only rarely fused to myotubes (Fig. 10). The finding that TSP-4 can promote myogenic differentiation under these in vitro conditions is consistent with the possibility that secretion of this extracellular matrix protein by prospective tendon mesenchyme cells is part of a local signaling process to couple muscle formation to tendon morphogenesis.
Figure 10.
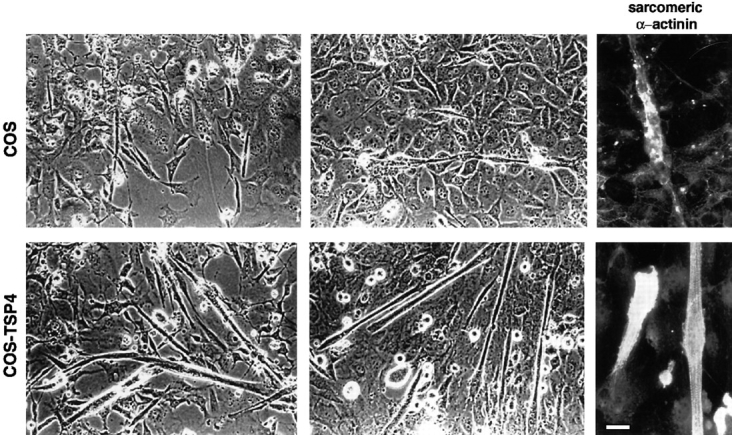
Myogenic differentiation in the presence of COS-TSP-4 cells. Cocultures of control (top; COS) or TSP-4–expressing (bottom; COS-TSP-4) COS cells with newborn hind limb muscle- derived cells. Culture dishes were precoated with laminin, and experimental details were as described in Materials and Methods. The phase contrast photographs of living cells were taken 1 (left) and 2 (right) d after switch to differentiation medium. The photographs on the right show immunocytochemistry of muscle-specific α actinin 2 d after induction of differentiation. Note that in the presence of naive COS cells, muscle-derived cells frequently formed chains (top, center) of cells that expressed low levels of sarcomeric α actinin (top, right) but in most cases failed to fuse to myotubes. In contrast, efficient formation of myotubes that expressed high levels of sarcomeric α actinin was detected in the presence of COS-TSP-4 cells (bottom). Bar: (Left and middle; phase-contrast) 24 μm; (right; immunofluorescence) 12 μm.
Discussion
MARP Is a Nuclear Protein with Regulatory Properties of a Specialized Early Response Gene
The tight regulation of MARP at the posttranscriptional and posttranslational levels is characteristic of a regulatory early response gene (McMahon and Monroe, 1992) and suggests that MARP may be a control component in signaling to the nucleus (Chu et al., 1995). Thus the rat, mouse, and human mRNAs contain multiple ATTTA degradation motifs in their 3′ untranslated sequence; these motifs affect mRNA levels in transfected cells, and MARP mRNA contents are elevated in cycloheximide-treated cells. The latter property is a hallmark of early response genes whose transcripts are subject to rapid degradation involving short-lived cellular proteins (McMahon and Monroe, 1992). In addition, possibly due to the presence of a PEST protein degradation motif (Rechsteiner and Rogers, 1996), MARP was difficult to detect in homogenates from tissues and cell lines with substantial MARP mRNA contents. Transfection of cDNA constructs devoid of ATTTA motifs led to transient accumulation of MARP protein with half-lives <30 min. Addition of short carboxyl-terminal peptide extensions specifically retarded MARP degradation, suggesting the existence of regulatory mechanisms affecting MARP levels in the cell. Potential phosphorylation sites of MARP have been noted adjacent to the PEST-like motif and near the carboxyl-terminal end of the protein (Chu et al., 1995). Accordingly, and in analogy to the regulated degradation of other short-lived proteins, MARP stabilization may be regulated by phosphorylation.
Regulation of MARP levels has cell type- and signal-specific features. Thus a human cell line with properties of large vessel vascular endothelial cells expresses comparatively high levels of MARP mRNA, whereas corresponding small vessel cells require activation with IL1, lysophosphatidic acid, or cycloheximide for detectable MARP expression (Chu et al., 1995). Regulation of MARP mRNA levels in vascular endothelial cells may have similar properties in vivo, since in mouse embryos, corresponding in situ hybridization signals were only detected along subdomains of large blood vessels (not shown). Similar cell-type specific regulation was detected in myogenic and fibroblastic cell lines. In addition, cell type- and signal-specific regulation was detected in developing and adult mouse muscle in vivo, where MARP expression was restricted to subtypes and subdomains of striated muscle cells under defined reactive conditions. The unique temporal and spatial patterns of MARP transcript accumulation suggest that MARP levels may control specific signaling pathways to the nucleus. The observation that certain cell lines can contain unexpectedly high levels of MARP mRNA may reflect the absence or attenuation of physiological inhibitory mechanisms under tissue culture conditions. Similar considerations probably apply to primary myotubes from newborn hindlimb myoblasts, which expressed substantial levels of MARP mRNA, although the corresponding muscle cells in vivo contained no detectable MARP transcript (not shown).
What cellular processes may be affected by MARP? Although it accumulates in the nucleus, MARP does not appear to bind DNA with high affinity (Chou et al., 1995). A recent independent study focusing on the possible role of CARP/MARP in heart-specific gene expression provided evidence that this protein is a nuclear cofactor that promotes the expression of certain muscle-specific genes in cardiomyocytes (Zou et al., 1997). In that study CARP/ MARP was isolated by a two-hybrid approach as a nuclear cofactor that interacts with the ubiquitous transcription factor YB-1. Negative regulation of a YB-1–responsive minimal promoter construct suggested that CARP/MARP may specifically regulate HF-1–dependent pathways for ventricular muscle gene expression (Zou et al., 1997). Possibly due to technical reasons, although a weak CARP/ MARP signal was detected in that study on a Northern blot of adult skeletal muscle, no in situ hybridization signal was detected in skeletal muscle at any stage of mouse embryogenesis (hence the name CARP; Zou et al., 1997). Assuming that the findings on YB-1 regulation can be extended to skeletal muscle genes, then MARP may be a nuclear cofactor in a signal transduction pathway for local cell activation in forming and denervated muscle.
Besides a classical bipartite nuclear localization sequence and the PEST motif mentioned above, MARP has a cluster of four ankyrin repeats near its carboxyl-terminal end. Clusters containing different numbers of this 30-amino acid motif have been found in a variety of apparently unrelated proteins, where they are thought to mediate protein–protein interactions (Blank et al., 1992). The molecular size of MARP, the arrangement and number of its ankyrin repeats, and the fact that it is subject to degradative regulation in the cell are most reminiscent of IκB proteins. These are negative regulators of NF-κB proteins, a family of transcription factors mediating acute cellular responses. To explore the possibility that MARP may function as an IκB we carried out cotransfection experiments with various NF-κB constructs, assaying for protein stability in the presence and absence of phorbol ester and for transcriptional activation. So far, however, these experiments provided no positive indications (data not shown). Therefore, while it is tempting to speculate that MARP may be involved in novel signal transduction pathways mediating local alterations in gene expression, elucidation of the actual nature of the processes affected by MARP will require further experimentation.
Primary Myotube Endcompartments and Adjacent Prospective Tendon Mesenchyme May Constitute a Functional Unit Involved in Muscle Morphogenesis
A main finding of this study is that the accumulation of MARP transcript visualizes a novel muscle subcompartment associated with muscle morphogenesis. MARP mRNA accumulated at the ends of MLP-positive primary myotubes from about E12 to 16. Later in development, MARP skeletal muscle signal was restricted to the tongue, where it could still be detected during the first postnatal week. In addition, denervation rapidly induced skeletal muscle MARP mRNA in the vicinity of vacated neuromuscular synapses. Therefore, in developing and adult skeletal muscle, MARP transcript is under tight spatial and temporal regulation. Due to the unique size and shape of myotubes and skeletal muscle fibers, this narrow regulation of MARP mRNA reveals regulatory subcompartments within muscle fibers. The perisynaptic compartment of functionally denervated skeletal muscle fibers may reflect signaling from activated muscle interstitial cells (Fig. 11; Connor and McMahan, 1987; Gatchalian et al., 1989). The unique properties of primary myotube end regions during muscle morphogenesis have not been detected before, and the possible implications of these observations are discussed below.
Figure 11.
The local accumulation of MARP mRNA visualizes novel regulatory muscle subcompartments during muscle morphogenesis and in denervated muscle. (A) During muscle morphogenesis, MARP mRNA accumulates at primary myotube end regions, adjacent to TSP-4–expressing prospective tendon mesenchyme. In contrast, MLP transcript accumulates throughout primary myotubes, and that for MyoD is also detected in myoblasts. The arrows with question marks indicate the possible direction of signaling, from surrounding mesenchyme to tendon mesenchyme to primary myotube end regions. During muscle morphogenesis, extensive myoblast proliferation, fusion to preexisting primary myotubes, and myotube elongation may be restricted to muscle end regions under the control of prospective tendon mesenchyme. (B) In denervated adult skeletal muscle, MARP mRNA accumulates in the perisynaptic region of muscle fibers, in the vicinity of activated muscle interstitial cells. The arrow with question mark indicates the possible direction of signaling from activated muscle interstitial cells to the perisynaptic region of denervated muscle. Unlike MARP mRNA, transcripts for classical denervation- induced genes such as MyoD and MLP accumulate throughout denervated muscle fibers. In addition, subsynaptic muscle nuclei express distinct genes in innervated and denervated muscle.
Muscles form from initial masses of terminally differentiated primary myotubes through a series of stereotyped splitting processes (e.g., Schroeter and Tosney, 1991a , b ). These are followed by directed lateral extension to yield the final configurations of shapes, attachment sites, and positions characteristic for each individual muscle. In the mouse, this process of muscle morphogenesis is completed by E15–16 and precedes a second wave of myoblast proliferation and formation of secondary myotubes (Lance-Jones, 1979; Ross et al., 1987). Primary myotubes then serve as scaffold for muscle growth (Ashby et al., 1993; Jellies, 1990; Hauschka, 1994). From the earliest time when primary myotubes could be detected, they were found to span muscles from end to end in small parallel bundles (Hilfer et al., 1973; Hauschka, 1974; Duxson and Usson, 1989). This configuration potentially assigns unique properties to end regions of muscle during morphogenesis (Fig. 11). Such unique properties may be crucial to the morphogenesis process. Thus, (a) muscle splitting always proceeds in one defined direction, starting from one muscle end (either from the origin or the attachment site, depending on the particular muscle; Schroeter and Tosney, 1991b ), and (b) instead of involving cell migration, the morphogenesis of individual muscles appears to involve local orientation and growth through myoblast fusion at primary myotube end regions (e.g., Hauschka, 1994). These regions are now shown to accumulate MARP transcript in primary myotubes and TSP-4 mRNA in adjacent prospective tendon mesenchyme. The close temporal and spatial correlation in the expression of these two transcripts during muscle morphogenesis suggests that these domains at the ends of forming muscles constitute a functional unit involved in the process of morphogenesis (Kardon, G. 1996. Dev. Biol. 175:393a). This interpretation is consistent with the observation that no obvious physical boundaries could be detected between adjacent TSP-4- and MARP-expressing tissue and that the two transcripts appeared to be expressed in coextensive tissue (Fig. 9). Due to the fact that it is the surrounding mesenchyme that provides the information for the formation and shaping of individual muscles, it seems likely that TSP-4- and MARP-expressing muscle end regions respond to local patterning cues during the shaping process. One possibility is that local signals induce and guide the formation and shaping of TSP-4–expressing prospective tendon mesenchyme (Kardon, G. 1996. Dev. Biol. 175:393a). This in turn would signal to adjacent muscle, as visualized by the accumulation of MARP transcript and locally promote proliferation, fusion, and elongation of primary myoblasts and myotubes (Fig. 11). Within this context, extracellular matrix TSP-4 could contribute to a local environment promoting myogenesis. In addition, the muscle end properties of the MARP-expressing subdomain of activated primary myotubes may also lead to feedback signaling from muscle ends to muscle- associated and surrounding mesenchyme. One such signal may be mediated by FGF-6. Thus this transcript preferentially accumulates at the ends of forming muscles between E12 and 16 (deLapeyriére et al., 1993). Although local accumulation of FGF-6 mRNA was much less pronounced than that of MARP transcript, a spatial and temporal correlation between the two transcripts could be detected (data not shown).
In conclusion, this study has revealed the existence of a novel muscle subcompartment at the ends of primary myotubes and has provided molecular markers for that subcompartment and prospective tendon mesenchyme during muscle morphogenesis. The spatial and temporal relation between the compartments defined by the expression of MARP and TSP-4 suggests that they may define a functional unit involved in muscle morphogenesis. These findings should provide a molecular basis to elucidate novel local signaling pathways leading to the specification and shaping of defined skeletal muscles. Likewise, the visualization of a perisynaptic subcompartment in denervated skeletal muscle may provide a starting point to define novel signaling pathways at denervated neuromuscular junctions.
Acknowledgments
We are grateful to T. Jessell (Columbia University, New York, NY), R. Chiquet-Ehrismann, C. Hagios (Friedrich Miescher Institute) and M. Rüegg (Biocenter, Basel, Switzerland) for critically reading the manuscript. We thank M. Adam for substantial help during the initial phases of this project and M.-P. Chevron for help with TSP-4 experiments. We thank K. Scheidereit (Max-Delbrück Center, Berlin, Germany) for generously providing us with reagents and advice on I-κB proteins, M.C. Weiss for specific protein kinase A cDNA probes (RIα and Cα), and D. Birnbaum for a specific FGF6 cDNA probe. We are particularly grateful to G. Kardon (Duke University, Durham, NC) and C. Lance-Jones (University of Pittsburgh, Pittsburgh, PA) for precious information and insight about muscle morphogenesis.
Abbreviations used in this paper
- CARP
cardiac ankyrin repeat protein
- E
embryonic day
- MARP
muscle ankyrin repeat domain
- MLP
muscle LIM protein
- TSP-4
thrombospondin-4
Footnotes
Address all correspondence to Pico Caroni, Friederich Miescher Institute, P.O. Box 2543, CH-4002 Basel, Switzerland. Tel.: (41) 61-6973727. Fax: (41) 61-6973976. E-mail: caroni@fmi.ch
Silvia Arber's present address is Howard Hughes Medical Institute, Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY.
References
- Adams JC, Lawler J. Cell-type specific adhesive interactions of skeletal myoblasts with Thrombospondin-1. Mol Biol Cell. 1994;5:423–437. doi: 10.1091/mbc.5.4.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner L, Arber S, Kapfhammer JP, Laux T, Schneider C, Botteri F, Brenner H-R, Caroni P. Overexpression of the neural growth-associated protein GAP-43 induces nerve sprouting in the adult nervous system of transgenic mice. Cell. 1995;83:269–278. doi: 10.1016/0092-8674(95)90168-x. [DOI] [PubMed] [Google Scholar]
- Arber S, Caroni P. Thrombospondin-4, an extracellular matrix protein expressed in the developing and adult nervous system promotes neurite outgrowth. J Cell Biol. 1995;131:1083–1094. doi: 10.1083/jcb.131.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel positive regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard J-C, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Ashby PR, Wilson SJ, Harris AJ. Formation of primary and secondary myotubes in aneural muscles in the mouse mutant peroneal muscular atrophy. Dev Biol. 1993;156:519–528. doi: 10.1006/dbio.1993.1098. [DOI] [PubMed] [Google Scholar]
- Blank V, Kourislky P, Israel A. NF-κB and related proteins: Rel/ dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–149. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- Chevallier A, Kieny M, Mauger A. Limb-somite relationship: origin of the limb musculature. J Embryol Exp Morphol. 1977;41:245–258. [PubMed] [Google Scholar]
- Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1992;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- Chiquet M, Fambrough DM. Chick myotendinous antigen. I. A monoclonal antibody as a marker for tendon and muscle morphogenesis. J Cell Biol. 1984;98:1926–1936. doi: 10.1083/jcb.98.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W, Burns DK, Swerick RA, Presky DH. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J Biol Chem. 1995;270:10236–10245. doi: 10.1074/jbc.270.17.10236. [DOI] [PubMed] [Google Scholar]
- Condon K, Silberstein L, Blau HM, Thompson WJ. Development of muscle fiber types in the prenatal rat hindlimb. Dev Biol. 1990;138:256–274. doi: 10.1016/0012-1606(90)90196-p. [DOI] [PubMed] [Google Scholar]
- Connor EA, McMahan UJ. Cell accumulation in the junctional region of denervated muscle. JCell Biol. 1987;104:109–120. doi: 10.1083/jcb.104.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLapeyriére O, Ollendorf V, Planche J, Ott MO, Pizette S, Coulier F, Birnbaum D. Expression of the Fgf6 gene is restricted to developing skeletal muscle in the mouse embryo. Development. 1993;118:601–611. doi: 10.1242/dev.118.2.601. [DOI] [PubMed] [Google Scholar]
- Duxson MJ, Usson Y. Cellular insertion of primary and secondary myotubes in embryonic rat muscle. Development. 1989;107:243–252. doi: 10.1242/dev.107.2.243. [DOI] [PubMed] [Google Scholar]
- Fontaine B, Changeux J-P. Localization of nicotinic acetylcholine receptor α-subunit transcripts during myogenesis and motor endplate development in the chick. J Cell Biol. 1989;108:1025–1037. doi: 10.1083/jcb.108.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian CL, Schachner M, Sanes JR. Fibroblasts that proliferate near denervated synaptic sites in skeletal muscle synthesize the adhesive molecules tenascin(J1), N-CAM, fibronectin, and a heparan sulfate proteoglycan. J Cell Biol. 1989;108:1873–1890. doi: 10.1083/jcb.108.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson J, Sanders R, Williams, Yu Y-T, Bassel-Duby R. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol Cell Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim, M. 1991. Control of muscle morphogenesis and endplate pattern in limb muscles of avian chimeras. In Developmental Patterning of the Vertebrate Limb. J.R. Hinchliffe, J.M. Hurle, and D. Summerbell, editors. Plenum Press, New York. 293–297.
- Hauschka SD. Clonal analysis of vertebrate myogenesis. 3. Developmental changes in the muscle-colony-forming cells of the human fetal limb. Dev Biol. 1974;37:345–368. doi: 10.1016/0012-1606(74)90154-7. [DOI] [PubMed] [Google Scholar]
- Hauschka, S.D. 1994. The embryonic origin of muscle. In Myology. Second Edition. A.G. Engel and C. Franzini-Armstrong, editors. McGraw Hill, New York. 3–73.
- Hilfer SR, Searls RL, Fonte VG. An ultrastructural study of early myogenesis in the chick wing bud. Dev Biol. 1973;30:374–391. doi: 10.1016/0012-1606(73)90095-x. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Scherrer T, Faust DM, Benichon J-C, Hellio R, Weiss MC. Accumulation in fetal muscle and localization to the neuromuscular junction of cAMP-dependent protein kinase A regulatory and catalytic subunits RIα and Cα. JCell Biol. 1996;134:1241–1254. doi: 10.1083/jcb.134.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellies J. Muscle assembly in simple systems. Trends Neurosci. 1990;13:126–131. doi: 10.1016/0166-2236(90)90003-s. [DOI] [PubMed] [Google Scholar]
- Lance-Jones C. The morphogenesis of the thigh of the mouse with special reference to tetrapod muscle homologies. JMorphol. 1979;162:275–309. doi: 10.1002/jmor.1051620207. [DOI] [PubMed] [Google Scholar]
- Lanser HE, Fallon JF. Development of wing bud-derived muscles in normal and wingless chick embryos: a computer assisted three-dimensional reconstruction study of muscle pattern formation in the absence of skeletal elements. Anat Rec. 1987;217:61–85. doi: 10.1002/ar.1092170110. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Monroe JG. Role of primary response genes in generating cellular responses to growth factors. FASEB J. 1992;6:2707–2715. doi: 10.1096/fasebj.6.9.1612295. [DOI] [PubMed] [Google Scholar]
- Olson EN, Klein WH. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- Ontell M, Kozeka K. The organogenesis of murine striated muscle: a cytoarchitectural study. Am J Anat. 1984;171:133–148. doi: 10.1002/aja.1001710202. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Ross JJ, Duxson MJ, Harris AJ. Formation of primary and secondary myotubes in rat lumbrical muscles. Development. 1987;100:383–394. doi: 10.1242/dev.100.3.383. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types of expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labeled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Schroeter S, Tosney KW. Spatial and temporal patterns of muscle cleavage in the chick thigh and their value as criteria for homology. Am J Anat. 1991a;191:325–350. doi: 10.1002/aja.1001910402. [DOI] [PubMed] [Google Scholar]
- Schroeter S, Tosney KW. Ultrastructural and morphometric analysis of the separation of two thigh muscles in the chick. Am J Anat. 1991b;191:351–368. doi: 10.1002/aja.1001910403. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Sweeney LJ, Kennedy JM, Zak R, Kokjohn K, Kelley SW. Evidence for expression of a common myosin heavy chain phenotype in future fast and slow skeletal muscle during initial stages of avian embryogenesis. Dev Biol. 1989;133:361–374. doi: 10.1016/0012-1606(89)90040-7. [DOI] [PubMed] [Google Scholar]
- Tucker RP, Adams JC, Lawler J. Thrombospondin-4 is expressed by early osteogenic tissues in the chick embryo. Dev Dyn. 1995;203:477–490. doi: 10.1002/aja.1002030410. [DOI] [PubMed] [Google Scholar]
- Witzemann V, Brenner H-R, Sakmann B. Neural factors regulate AChR subunit mRNAs at rat neuromuscular synapses. J Cell Biol. 1991;114:125–141. doi: 10.1083/jcb.114.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Evans S, Chen J, Kuo H-C, Harvey RP, Chien KR. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]