Abstract
Large scale EST sequencing projects have been carried out for S. mansoni and S. japonicum. This update will briefly review the most recent accomplishments in the area and discuss the use of EST data for the purposes of gene discovery, gene model development, genome annotation and SNP analysis. In addition, the use of ESTs for studying other features of the transcriptome such as splice site and transcription initiation variants will be discussed as well as approaches to assigning function to unknown transcripts. Although EST sequencing has contributed much for schistosome research, other data mining possibilities exist, including the identification of putative drug and vaccine targets.
Keywords: Trematode, schistosomiasis, expressed sequence tags, transcriptome
The transcriptome can be defined as a collection of the genes transcribed in an organism, tissue, or cell. Transcriptomic information has been extremely useful for gene discovery, the elaboration of gene models, training of gene finding algorithms (once a genome is available) the design of microarrays and has tremendously facilitated the cloning of genes of interest by the availability of cDNA clones. However, the mRNA transcribed is not expressed in equal amounts even in a single cell and they also differ in length. Therefore, experimental efforts towards obtaining the transcriptome of an organism generally will provide partial mRNA sequences and will be enriched for genes that are more highly transcribed. The partial cDNA sequences generated are called expressed sequence tags, ESTs. Despite some technical difficulties for producing full length cDNAs and incomplete views of the transcriptome provided, the approach has proven to be a powerful tool for the study of genes and their expression pattern.
The original and main motivation for the study of transcriptomes was the possibility of discovering the gene content of an organism and generating STSs that pointed to a gene, without the need to obtain the complete genome sequence (Adams et al., 1991). Although the unfinished genome sequences of S. mansoni and S. japonicum have been produced (El-Sayed et al., 2004;McManus et al., 2004), and are currently in the annotation stage, the transcriptome of both species have been studied on a large scale and have contributed to the discovery of genes and genome annotation, among other uses that will be further discussed in this update (genome sequencing will be discussed in this issue). Currently, GenBank contains information for over 650 species with at least 1,000 ESTs (Table 1).
Table 1.
Online resources related to Schistosoma ESTs available.
There are two main approaches for the large scale production of cDNA sequences. The conventional method is based on reverse-transcribing mRNA, cloning and sequencing the extremities of the inserted cDNA (Fulton et al., 1995). The alternative approach named ORESTES is to sequence short randomly amplified and cloned cDNAs (Dias et al., 2000) (Figure 1). Both methods can potentially result in large numbers of redundant sequences of cDNAs. The sequences need to be assembled so that transcripts from one gene are grouped together. The assembly process yields, in many cases, full length virtual cDNA sequences. The methods are complimentary. The ORESTES tend to accumulate at the central region of the original mRNA and the conventional method at the 5' and 3' ends. Therefore, by combining both methods, the chance of assembling a full virtual cDNA sequence increases. The ORESTES method allows sampling of a larger number of small cDNA libraries, which is useful especially in cases of difficult to obtain mRNA. The method also normalizes the mini cDNA libraries, making the relative number of ESTs similar irrespective of the different gene transcription levels in the sampled tissue. Normalization permits the identification of rare transcripts. The conventional method, on the other hand, permits an in depth sampling of much larger cDNA libraries. In addition, the conventional method permits the estimation of the relative numbers of each transcript by not normalizing the libraries. Also importantly, the conventional cDNA sequencing method produces frozen stocks of clones, many of which are complete or near complete cDNAs. These cDNA clones are an important resource to the research community.
Figure 1.
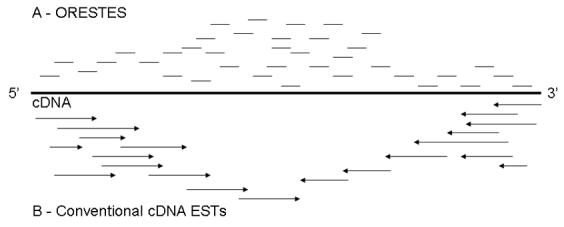
Methods for producing expressed sequence tags (ESTs). The complete cDNA clone is represented by the central line. A – ORESTES. B – Conventional cDNA sequencing. Conventional cDNAs are usually 100 to 500 bp sequences produced from cDNA cloned and sequenced directionally in respect to the 5' or 3' end, and tend to accumulate at the end of the representing mRNA. On the other hand, ORESTES are shorter sequences that tend to accumulate on the central region of the representing mRNA.
The study of the transcriptome can provide many different types of information about an organism. The main type of information generated is the discovery of new genes which can be accomplished efficiently and quickly with a transcriptomic approach, especially for organisms with large genomes, such as schistosomes. In the pre-genome era of schistosome research, this approach was used and yielded a large number of new genes (Franco et al., 1995;Oliveira, 2001) cDNA clones were also widely distributed to the research community. More recently, larger scale projects on S. mansoni have immensely expanded the knowledge and made mining the information much more effective (Oliveira and Johnston, 2001;Hu et al., 2003;Verjovski-Almeida et al., 2003;Verjovski-Almeida et al., 2004; Oliveira, unpublished effort of the RGMG). For S. japonicum one large project was published (Hu et al., 2003). The Wellcome Trust Sanger Institute also maintains at its ftp site EST sequences of S. mansoni and S. haematobium (Table 1).
The annotation, or attribution of possible function, of the sequences can be carried out with each individual read and with assembled sequences. Generally the methods will involve the search for similarities between the EST or contig and a database of DNA or protein sequence with functional annotation. This type of analysis will initially yield information about which genes are known and which are unknown. In order to annotate gene function using similarity search, curated databases of proteins are preferably used. Some of the interesting databases are: CDD (Marchler-Bauer et al., 2007) or Pfam (Finn et al., 2006) for conserved protein domains and families and PIR (Wu et al., 2003), SwissProt or UniProt (Wu et al., 2006a) for annotated proteins, among many others. InterPro contains several databases with information on protein families, domains and functional sites (Mulder et al., 2002), which can be queried with InterProScan (Quevillon et al., 2005). One of the problems with annotating a gene or a protein is the use of a structured vocabulary shared among different organisms. For this reason, it is also useful to annotate a sequence with a Gene Ontology term (Ashburner et al., 2000). One example of an annotated transcriptome database is the Gene Index (Merrick et al., 2003), but others are also available (Table 1).
The results obtained with schistosome EST sequences indicate that under the Molecular Function category, most of the ESTs belong to the binding (nucleic acid binding) and enzyme (mainly hydrolases and transferases) categories (Hu et al., 2003;Verjovski-Almeida et al., 2003). Under Biological Process the most prevalent categories were metabolism or transport. The finding of these categories as the most prevalent was expected, as metabolism is usually the major physiological activity of an organism. Some interesting features were observed, such as the presence of glucose importers, proteins involved in the uptake of amino acids and lipids and storage of lipids. The identification of the metabolic pathways in which parasite gene products participate will contribute to the understanding of their metabolic processes. This investigation may also contribute to the identification of possible drug targets (Fairlamb, 2002). One of the possibilities is to use KEGG that provides biochemical pathways, among other resources (Kanehisa et al., 2004). A visualization of the KEGG pathways obtained with the use of the transcriptome of S. mansoni is available (Oliveira, unpublished; Table 1).
After annotation, lists of genes possibly involved with a certain physiological function in the organism are usually produced. One example is genes involved in sex differentiation. This group of genes is of interest because interfering with sex differentiation or maturation may be one way of preventing the disease by interrupting egg production by the female parasite, as the eggs cause most of the pathology (LoVerde, 2002). In addition, sex differentiation in schistosomes should be an interesting model for study since most platyhelminths are hermaphrodites (Mone and Boissier, 2004). Several homologues of genes involved in sex differentiation, such as fox-1, mog-1, tra-2 and fem-1 were observed (Verjovski-Almeida et al., 2003). Hu and colleagues (2003), focused on genes differentially expressed between sexes and obtained results comparable to those of microarray experiments (Hoffmann et al., 2002). Among the differentially expressed genes are those coding for egg shell proteins, maleless, epididymal secretory protein E1 and transformer-2β.
One interesting approach for analyzing the EST content is the comparison of the frequency of ESTs of one organism with those of model organisms. This will provide information on conserved genes (Hu et al., 2003), but also can be informative in relation to the expected number of ESTs, indicating genes that follow unusual patterns of expression in comparison with other organisms (Mudado and Ortega, 2006).
Interestingly, from the first descriptions with fewer sequences (Franco et al., 1995), to the larger projects (Hu et al., 2003;Verjovski-Almeida et al., 2003), at least 50% of all transcripts identified yield no similar transcripts in GenBank. The number may decrease as the clusters grow in size and with the availability of better gene models with the genomic data. Nevertheless, clearly, schistosomes contain a large set of unique genes. One of the main challenges is the characterization of their function.
Functional genomics will contribute to describe gene function. In this issue recent developments in knocking down gene expression by RNA interference and over expression by the introduction of exogenous DNA are discussed. However, there is a need for observable phenotypes in order to see an effect resulting from knock-out, knock-down or transgene approaches. The function of a gene product may sometimes also be inferred by a guilt-by-association approach (although experimental evidence is necessary to corroborate the indications). Microarray experiments, for example, can reveal genes that are co-expressed in different situations and may, therefore, work in conjunction, or interact to generate a certain phenotype (Quackenbush, 2003;Voy et al., 2006). Proteomics results may also be analyzed in this fashion (Shin et al., 2005). Novel computational approaches may prove to be powerful tools in assigning function to an unknown gene product (Wolfe et al., 2005;Wu et al., 2006b). Microarray (Hoffmann et al., 2001;Hoffmann et al., 2002;Fitzpatrick et al., 2004;Fitzpatrick et al., 2005;Chai et al., 2006;Dillon et al., 2006;Gobert et al., 2006;Moertel et al., 2006;Vermeire et al., 2006) and proteomic (Curwen et al., 2004;Cheng et al., 2005;Knudsen et al., 2005;van Balkom et al., 2005;Braschi et al., 2006;Liu et al., 2006;Van Hellemond et al., 2006) data have been produced for schistosomes, but not yet fully explored with powerful computational methods.
The identification of genes in a genome is still an arduous task for large and complex genomes. ESTs have also been very useful, providing experimental evidence for the elaboration of gene models. EST evidence has been incorporated into various algorithms for gene identification (Stanke et al., 2006). New ESTs may also reveal previously unknown genes. It has been shown for the human genome, for example, that new EST information is useful for identifying transcribed regions in the genome even with the availability of an already large cDNA dataset (de Souza et al., 2000). In the case of schistosomes, for which a finished complete genome sequence is not available, ESTs will also provide evidence for genes that may not have been sequenced.
As previously stated, some effort has been dedicated to producing full length cDNA clones from existing libraries for S. mansoni and S. japonicum. The interests in obtaining full length cDNA clones are several. Full length clones can provide extremely useful information for the production of accurate gene models and provide better annotation (Castelli et al., 2004). Distinct intron/exon boundaries, transcription start sites, antisense RNA are some examples of the richness of the genome that can be further explored (Miura et al., 2006). The existing ESTs and those made available in the future will greatly contribute towards the full understanding of many features of the genome. Full length clones have also been used in novel global approaches towards the identification of candidate antigens for the development of malaria vaccines using a DNA vaccine approach (Shibui et al., 2005). Towards the goal of providing the research community with a set of full length cDNA clones, Faria-Campos and colleagues (2006) identified within the S. mansoni cDNAs clones from the RGMG work, a set of clones that potentially contained the initiating methionine. A similar approach was undertaken for S. japonicum (Hu et al., 2003). The nature of the selection process however yielded mostly short sequences. An effort toward the production of a large number of long full length cDNAs would benefit from the construction of capped cDNA libraries (Suzuki et al., 1997). As well as providing full length cDNAs, a finishing approach to the transcriptome should target ESTs pairs from longer transcripts or not fully covered gene models (Sogayar et al., 2004). Full length cDNAs are the gold standard for defining a transcript and will enhance genome characterization and gene annotation.
ESTs clones have been an invaluable resource for the research community. Obtaining cDNA clones for a gene of interest will not always be trivial. Several efforts towards providing the research community with already cloned cDNAs has made the cloning effort much easier and cheaper. Some of the efforts for human cDNA clones are for example I.M.A.G.E. clones from ATCC or the German Resource Center for Genome Research (Lennon et al., 1996), the Japanese NITE and Riken Resource Centers that distribute full length human cDNA clones, TAIR for Arabidopsis full length cDNA clones (Rhee et al., 2003) and several commercial suppliers. For parasite material there is the MR4 that provides a variety of types of clones (Wu et al., 2001). With the idea of making resources readily available and inexpensive for the research community, recently the SR3 was created and will need community support for its success (Table 1).
The study of the transcriptome using microarrays has been carried out in Schistosoma and is discussed in depth in this issue. In addition, SAGE analysis have also been conducted to some extent in Schistosoma (Verjovski-Almeida et al., 2003), Williams, see this issue of EP). Recently it was shown by long-SAGE that in vitro exposure to nitric oxide modulates the expression of several genes, among them the upregulation of superoxide dismutase (Masserli et al. 2006). EST data was been used for the design of microarrays, both of the cDNA and oligonucleotide types (Hoffmann et al., 2001; Hoffmann et al., 2002; Fitzpatrick et al., 2004; Fitzpatrick et al., 2005; Chai et al., 2006; Dillon et al., 2006; Gobert et al., 2006; Moertel et al., 2006; Vermeire et al., 2006). The identification of SAGE tags will enhance the knowledge of transcription patterns as well as corroborate experimental evidence for transcribed genomic regions.
In addition to finding and annotating genes, ESTs can be mined for other types of information. Simões at al. (2007) have used EST and base call quality information made available by one of the published projects (Verjovski-Almeida et al., 2003) to identify SNPs. The authors identified 15,615 putative SNPs, of which 1,832 resulted in non-synonymous amino acid substitutions. Many of the known antigens and vaccine candidates contained amino acid polymorphisms. This kind of approach will provide valuable information for researchers interested in vaccine development and the identification of drug targets.
In addition to SNPs, differences in splicing can be observed by investigating EST data. Differentially spliced genes, linked to development or sex, for example, are increasingly being observed in schistosomes (Shoemaker et al., 1992;Hamdan and Ribeiro, 1998;De Mendonca et al., 2002;Ram et al., 2004;DeMarco et al., 2006;Bahia et al., 2006). Alternative transcription initiation and polyadenylation sites could also be explored with the use of EST data (Kan et al., 2001;Zavolan et al., 2003;Le, V et al., 2006;Seoighe et al., 2006;Tian et al., 2007).
In conclusion, ESTs have been produced in large scale for S. mansoni and S. japonicum and contributed significantly to the discovery of new genes, annotation of the genome and production of microarrays. There are, however, several other uses that have been less explored, such as the identification of polymorphisms, alternative splice sites and transcription initiation sites, among others.
Acknowledgements
GO receives financial support from NIH grants 5D43TW007012-03 and 5R03TW007358-02; CNPq/FIOCRUZ 400315/2006-8; FAPEMIG REDE-2829/05 and EDT 17001/01.
abbreviations
- cDNA
complementary DNA
- mRNA
messenger RNA
- STS
sequence-tagged sites
- ORESTES
Open Reading Frame Expressed Sequence Tags
- RGMG
Minas Gerais Genome Network
- CDD
Conserved Domain Database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RNA
ribonucleic acid
- DNA
deoxyribonucleic acid
- SNP
single nucleotide polymorphism
- MR4
Malaria Research and Reference Reagent Resource
- SR3
Schistosome Related Reagent Repository
- SAGE
Serial Analysis of Gene Expression
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahia D, Avelar L, Mortara RA, Khayath N, Yan Y, Noel C, Capron M, Dissous C, Pierce RJ, Oliveira G. SmPKC1, a new protein kinase C identified in the platyhelminth parasite Schistosoma mansoni. Biochemical and Biophysical Research Communications. 2006;345:1138–1148. doi: 10.1016/j.bbrc.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Braschi S, Curwen RS, Ashton PD, Verjovski-Almeida S, Wilson A. The tegument surface membranes of the human blood parasite Schistosoma mansoni: a proteomic analysis after differential extraction. Proteomics. 2006;6:1471–1482. doi: 10.1002/pmic.200500368. [DOI] [PubMed] [Google Scholar]
- Castelli V, Aury JM, Jaillon O, Wincker P, Clepet C, Menard M, Cruaud C, Quetier F, Scarpelli C, Schachter V, Temple G, Caboche M, Weissenbach J, Salanoubat M. Whole genome sequence comparisons and “full-length” cDNA sequences: a combined approach to evaluate and improve Arabidopsis genome annotation. Genome Research. 2004;14:406–413. doi: 10.1101/gr.1515604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai M, McManus DP, McInnes R, Moertel L, Tran M, Loukas A, Jonesa MK, Gobert GN. Transcriptome profiling of lung schistosomula,in vitro cultured schistosomula and adult Schistosoma japonicum. Cellular and Molecular Life Sciences. 2006;63:919–929. doi: 10.1007/s00018-005-5578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng GF, Lin JJ, Feng XG, Fu ZQ, Jin YM, Yuan CX, Zhou YC, Cai YM. Proteomic analysis of differentially expressed proteins between the male and female worm of Schistosoma japonicum after pairing. Proteomics. 2005;5:511–521. doi: 10.1002/pmic.200400953. [DOI] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Johnston DA, Wilson RA. The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Molecular and Biochemical Parasitology. 2004;138:57–66. doi: 10.1016/j.molbiopara.2004.06.016. [DOI] [PubMed] [Google Scholar]
- De Mendonca RL, Bouton D, Bertin B, Escriva H, Noel C, Vanacker JM, Cornette J, Laudet V, Pierce RJ. A functionally conserved member of the FTZ-F1 nuclear receptor family from Schistosoma mansoni. European Journal of Biochemistry. 2002;269:5700–5711. doi: 10.1046/j.1432-1033.2002.03287.x. [DOI] [PubMed] [Google Scholar]
- de Souza SJ, Camargo AA, Briones MR, Costa FF, Nagai MA, Verjovski-Almeida S, Zago MA, Andrade LE, Carrer H, El-Dorry HF, Espreafico EM, Habr-Gama A, Giannella-Neto D, Goldman GH, Gruber A, Hackel C, Kimura ET, Maciel RM, Marie SK, Martins EA, Nobrega MP, Paco-Larson ML, Pardini MI, Pereira GG, Pesquero JB, Rodrigues V, Rogatto SR, da S, I, Sogayar MC, de Fatima SM, Tajara EH, Valentini SR, Acencio M, Alberto FL, Amaral ME, Aneas I, Bengtson MH, Carraro DM, Carvalho AF, Carvalho LH, Cerutti JM, Correa ML, Costa MC, Curcio C, Gushiken T, Ho PL, Kimura E, Leite LC, Maia G, Majumder P, Marins M, Matsukuma A, Melo AS, Mestriner CA, Miracca EC, Miranda DC, Nascimento AN, Nobrega FG, Ojopi EP, Pandolfi JR, Pessoa LG, Rahal P, Rainho CA, da RN, de Sa RG, Sales MM, da Silva NP, Silva TC, da SW, Jr., Simao DF, Sousa JF, Stecconi D, Tsukumo F, Valente V, Zalcbeg H, Brentani RR, Reis FL, as-Neto E, Simpson AJ. Identification of human chromosome 22 transcribed sequences with ORF expressed sequence tags. Proceedings of the National Academy of Sciences U.S.A. 2000;97:12690–12693. doi: 10.1073/pnas.97.23.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco R, Oliveira KC, Venancio TM, Verjovski-Almeida S. Gender biased differential alternative splicing patterns of the transcriptional cofactor CA150 gene in Schistosoma mansoni. Molecular and Biochemical Parasitology. 2006;150:123–131. doi: 10.1016/j.molbiopara.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Dias NE, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da SW, Jr., Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, de Oliveira PS, Bucher P, Jongeneel CV, O'Hare MJ, Soares F, Brentani RR, Reis LF, de Souza SJ, Simpson AJ. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proceedings of the National Academy of Sciences U.S.A. 2000;97:3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon GP, Feltwell T, Skelton JP, Ashton PD, Coulson PS, Quail MA, Nikolaidou-Katsaridou N, Wilson RA, Ivens AC. Microarray analysis identifies genes preferentially expressed in the lung schistosomulum of Schistosoma mansoni. International Journal for Parasitology. 2006;36:1–8. doi: 10.1016/j.ijpara.2005.10.008. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Bartholomeu D, Ivens A, Johnston DA, LoVerde PT. Advances in schistosome genomics. Trends in Parasitology. 2004;20:154–157. doi: 10.1016/j.pt.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Fairlamb AH. Metabolic pathway analysis in trypanosomes and malaria parasites. Philosophical.Transactions.: Biological.Sciences. 2002;357:101–107. doi: 10.1098/rstb.2001.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria-Campos AC, Moratelli FS, Mendes IK, Ortolani PL, Oliveira GC, Campos SVA, Ortega JM, Franco GR. Production of full-length cDNA sequences by sequencing and analysis of expressed sequence tags from Schistosoma mansoni. Memórias do Instituto Oswaldo Cruz. 2006;101:161–165. doi: 10.1590/s0074-02762006000900026. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Research. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick JM, Johansen MV, Johnston DA, Dunne DW, Hoffmann KF. Gender-associated gene expression in two related strains of Schistosoma japonicum. Molecular and Biochemical Parasitology. 2004;136:191–209. doi: 10.1016/j.molbiopara.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Johnston DA, Williams GW, Williams DJ, Freeman TC, Dunne DW, Hoffmann KF. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Molecular and Biochemical Parasitology. 2005;141:1–13. doi: 10.1016/j.molbiopara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Franco GR, Adams MD, Soares MB, Simpson AJ, Venter JC, Pena SD. Identification of new Schistosoma mansoni genes by the EST strategy using a directional cDNA library. Gene. 1995;152:141–147. doi: 10.1016/0378-1119(94)00747-g. [DOI] [PubMed] [Google Scholar]
- Fulton LL, Hillier LD, Wilson RK. Large-scale complementary DNA sequencing methods. Methods in Cell Biology. 1995;48:571–582. [PubMed] [Google Scholar]
- Gobert GN, McInnes R, Moertel L, Nelson C, Jones MK, Hu W, McManus DP. Transcriptomics tool for the human Schistosoma blood flukes using microarray gene expression profiling. Experimental Parasitology. 2006;114:160–172. doi: 10.1016/j.exppara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Ribeiro P. Cloning and sequence analysis of a lysophospholipase homologue from Schistosoma mansoni. Parasitology Research. 1998;84:839–842. doi: 10.1007/s004360050497. [DOI] [PubMed] [Google Scholar]
- Hoffmann KF, Johnston DA, Dunne DW. Identification of Schistosoma mansoni gender-associated gene transcripts by cDNA microarray profiling. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0041. RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann KF, McCarty TC, Segal DH, Chiaramonte M, Hesse M, Davis EM, Cheever AW, Meltzer PS, Morse HC, III, Wynn TA. Disease fingerprinting with cDNA microarrays reveals distinct gene expression profiles in lethal type 1 and type 2 cytokine-mediated inflammatory reactions. FASEB Journal. 2001;15:2545–2547. doi: 10.1096/fj.01-0306fje. [DOI] [PubMed] [Google Scholar]
- Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus DP, Xue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nature Genetics. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- Kan Z, Rouchka EC, Gish WR, States DJ. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Research. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Research. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Molecular & Cellular Proteomics. 2005;4:1862–1875. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- Le T, V, Riethoven JJ, Kumanduri V, Gopalakrishnan C, Lopez F, Gautheret D, Thanaraj TA. AltTrans: transcript pattern variants annotated for both alternative splicing and alternative polyadenylation. BioMed Central Bioinformatics. 2006;7:169. doi: 10.1186/1471-2105-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Liu F, Lu J, Hu W, Wang SY, Cui SJ, Chi M, Yan Q, Wang XR, Song HD, Xu XN, Wang JJ, Zhang XL, Zhang X, Wang ZQ, Xue CL, Brindley PJ, McManus DP, Yang PY, Feng Z, Chen Z, Han ZG. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. Public Library of Science Pathogens. 2006;2:e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVerde PT. Presidential address. Sex and schistosomes: an interesting biological interplay with control implications. Journal of Parasitology. 2002;88:3–13. doi: 10.1645/0022-3395(2002)088[0003:PASASA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, Weese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Research. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus DP, Le TH, Blair D. Genomics of parasitic flatworms. International Journal for Parasitology. 2004;34:153–158. doi: 10.1016/j.ijpara.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Merrick JM, Osman A, Tsai J, Quackenbush J, LoVerde PT, Lee NH. The Schistosoma mansoni Gene Index: gene discovery and biology by reconstruction and analysis of expressed gene sequences. Journal of Parasitology. 2003;89:261–269. doi: 10.1645/0022-3395(2003)089[0261:TSMGIG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Messerli SM, Morgan W, Birkeland SR, Bernier J, Cipriano MJ, McArthur AG, Greenberg RM. Nitric oxide-dependent changes in Schistosoma mansoni gene expression. Molecular and Biochemical Parasitology. 2006;150:367–370. doi: 10.1016/j.molbiopara.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proceedings of the National Academy of Sciences U.S.A. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moertel L, McManus DP, Piva TJ, Young L, McInnes RL, Gobert GN. Oligonucleotide microarray analysis of strain- and gender-associated gene expression in the human blood fluke, Schistosoma japonicum. Molecular and Cellular Probes. 2006;20:280–289. doi: 10.1016/j.mcp.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Mone H, Boissier J. Sexual biology of schistosomes. Advances in Parasitology. 2004;57:89–189. doi: 10.1016/S0065-308X(04)57002-1. [DOI] [PubMed] [Google Scholar]
- Mudado MA, Ortega JM. A picture of gene sampling/expression in model organisms using ESTs and KOG proteins. Genetics and Molecular Research. 2006:242–253. [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischmann W, Gouzy J, Griffith-Jones S, Haft D, Hermjakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJ. InterPro: an integrated documentation resource for protein families, domains and functional sites. Briefings in Bioinformatics. 2002;3:225–235. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- Oliveira G. Schistosoma gene discovery project update. Trends in Parasitology. 2001;17:108–109. [Google Scholar]
- Oliveira G, Johnston DA. Mining the schistosome DNA sequence database. Trends in Parasitology. 2001;17:501–503. doi: 10.1016/s1471-4922(01)02019-0. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Genomics. Microarrays-guilt by association. Science. 2003;302:240–241. doi: 10.1126/science.1090887. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Research. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram D, Ziv E, Lantner F, Lardans V, Schechter I. Stage-specific alternative splicing of the heat-shock transcription factor during the life-cycle of Schistosoma mansoni. Parasitology. 2004;129:587–596. doi: 10.1017/s003118200400602x. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, Mundodi S, Reiser L, Tacklind J, Weems DC, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Nembaware V, Scheffler K. Maximum likelihood inference of imprinting and allele-specific expression from EST data. Bioinformatics. 2006;22:3032–3039. doi: 10.1093/bioinformatics/btl521. [DOI] [PubMed] [Google Scholar]
- Shibui A, Shiibashi T, Nogami S, Sugano S, Watanabe J. A novel method for development of malaria vaccines using full-length cDNA libraries. Vaccine. 2005;23:4359–4366. doi: 10.1016/j.vaccine.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Shin H, Sheu B, Markey MK. Guilt-By-Association feature selection applied to simulated proteomic data. American Medical Informatics Association Annual Symposium Proceedings. 2005;1114 [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CB, Ramachandran H, Landa A, dos Reis MG, Stein LD. Alternative splicing of the Schistosoma mansoni gene encoding a homologue of epidermal growth factor receptor. Molecular and Biochemical Parasitology. 1992;53:17–32. doi: 10.1016/0166-6851(92)90003-3. [DOI] [PubMed] [Google Scholar]
- Simões M, Bahia D, Zerlotini A, Torres K, Artiguenave F, Neshich G, Kuser P, Oliveira G. Single nucleotide polymorphisms identification in expressed genes of Schistosoma mansoni. Molecular and Biochemical Parasitology. 2007 doi: 10.1016/j.molbiopara.2007.04.003. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogayar MC, Camargo AA, Bettoni F, Carraro DM, Pires LC, Parmigiani RB, Ferreira EN, de Sa ME, do Rosario D.d., Simpson AJ, Cruz LO, Degaki TL, Festa F, Massirer KB, Sogayar MC, Filho FC, Camargo LP, Cunha MA, de Souza SJ, Faria M, Jr., Giuliatti S, Kopp L, de Oliveira PS, Paiva PB, Pereira AA, Pinheiro DG, Puga RD, Souza JE S.d., Albuquerque DM, Andrade LE, Baia GS, Briones MR, Cavaleiro-Luna AM, Cerutti JM, Costa FF, Costanzi-Strauss E, Espreafico EM, Ferrasi AC, Ferro ES, Fortes MA, Furchi JR, Giannella-Neto D, Goldman GH, Goldman MH, Gruber A, Guimaraes GS, Hackel C, Henrique-Silva F, Kimura ET, Leoni SG, Macedo C, Malnic B, Manzini BC, Marie SK, Martinez-Rossi NM, Menossi M, Miracca EC, Nagai MA, Nobrega FG, Nobrega MP, Oba-Shinjo SM, Oliveira MK, Orabona GM, Otsuka AY, Paco-Larson ML, Paixao BM, Pandolfi JR, Pardini MI, Passos Bueno MR, Passos GA, Pesquero JB, Pessoa JG, Rahal P, Rainho CA, Reis CP, Ricca TI, Rodrigues V, Rogatto SR, Romano CM, Romeiro JG, Rossi A, Sa RG, Sales MM, Sant'Anna SC, Santarosa PL, Segato F, Silva WA, Jr., Silva ID, Silva NP, Soares-Costa A, Sonati MF, Strauss BE, Tajara EH, Valentini SR, Villanova FE, Ward LS, Zanette DL. A transcript finishing initiative for closing gaps in the human transcriptome. Genome Research. 2004;14:1413–1423. doi: 10.1101/gr.2111304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Schoffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BioMed Central Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Research. 2007 doi: 10.1101/gr.5532707. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Balkom BW, van Gestel RA, Brouwers JF, Krijgsveld J, Tielens AG, Heck AJ, Van Hellemond JJ. Mass spectrometric analysis of the Schistosoma mansoni tegumental sub-proteome. Journal of Proteome Research. 2005;4:958–966. doi: 10.1021/pr050036w. [DOI] [PubMed] [Google Scholar]
- Van Hellemond JJ, Retra K, Brouwers JF, van Balkom BW, Yazdanbakhsh M, Shoemaker CB, Tielens AG. Functions of the tegument of schistosomes: clues from the proteome and lipidome. International Journal for Parasitology. 2006;36:691–699. doi: 10.1016/j.ijpara.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Verjovski-Almeida S, DeMarco R, Martins EA, Guimaraes PE, Ojopi EP, Paquola AC, Piazza JP, Nishiyama MY, Jr., Kitajima JP, Adamson RE, Ashton PD, Bonaldo MF, Coulson PS, Dillon GP, Farias LP, Gregorio SP, Ho PL, Leite RA, Malaquias LC, Marques RC, Miyasato PA, Nascimento AL, Ohlweiler FP, Reis EM, Ribeiro MA, Sa RG, Stukart GC, Soares MB, Gargioni C, Kawano T, Rodrigues V, Madeira AM, Wilson RA, Menck CF, Setubal JC, Leite LC, as-Neto E. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nature Genetics. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- Verjovski-Almeida S, Leite LC, as-Neto E, Menck CF, Wilson RA. Schistosome transcriptome: insights and perspectives for functional genomics. Trends in Parasitology. 2004;20:304–308. doi: 10.1016/j.pt.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Vermeire JJ, Taft AS, Hoffmann KF, Fitzpatrick JM, Yoshino TP. Schistosoma mansoni: DNA microarray gene expression profiling during the miracidium-to-mother sporocyst transformation. Molecular and Biochemical Parasitology. 2006;147:39–47. doi: 10.1016/j.molbiopara.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Voy BH, Scharff JA, Perkins AD, Saxton AM, Borate B, Chesler EJ, Branstetter LK, Langston MA. Extracting gene networks for low-dose radiation using graph theoretical algorithms. Public Library of Science Computational Biology. 2006;2:e89. doi: 10.1371/journal.pcbi.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BioMed Central Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Research. 2006;34:D10–D15. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. The Protein Information Resource. Nucleic Acids Research. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hu Z, DeLisi C. Gene annotation and network inference by phylogenetic profiling. BioMed Central Bioinformatics. 2006;7:80. doi: 10.1186/1471-2105-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Fairfield AS, Oduola A, Cypess RH. The Malaria Research and Reference Reagent Resource (MR4) Center-creating African opportunities. African Journal of Mededicine and Medical Science. 2001;30(Suppl):52–54. [PubMed] [Google Scholar]
- Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y, Gaasterland T. Impact of Alternative Initiation, Splicing, and Termination on the Diversity of the mRNA Transcripts Encoded by the Mouse Transcriptome. Genome Research. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]


