Abstract
A hairpin polyamide-chlorambucil conjugate linked by α-diaminobutyric acid (α-DABA) has been shown to have interesting biological properties in cellular and small animal models. Remarkably, this new class of hairpin polyamides has not been previously characterized with regard to energetics and sequence specificity. Herein we present a series of pyrrole-imidazole hairpin polyamides linked by α-DABA and compare them to polyamides containing the standard γ-DABA turn unit. The α-DABA hairpins have overall decreased binding affinities. However, α-DABA polyamide-chlorambucil conjugates are sequence-specific DNA alkylators with increased specificities. Affinity cleavage studies of α-DABA polyamide-EDTA conjugates confirmed their preference for binding DNA in a forward hairpin conformation. In contrast, an unsubstituted glycine-linked polyamide prefers to bind in an extended binding mode. Thus, substitution on the turn unit locks the α-DABA polyamide into the forward hairpin binding motif.
1. Introduction
Pyrrole-imidazole polyamides are a class of synthetic ligands that can be programmed to bind a broad repertoire of DNA sequences with affinities and specificities similar to DNA-binding proteins.1, 2 Eight-ring hairpin polyamides have been shown to localize to the nuclei of living cells3, 4 and regulate transcription by interfering with transcription factor-DNA interfaces in the promoters of endogenous genes.5–9 Conjugation of polyamides to the DNA alkylating agent chlorambucil produces molecules capable of sequence-specific covalent reaction with DNA,10 previously demonstrated for targeted sequences in the HIV-1 promoter, simian virus 40, and the histone H4c gene.11–13
DNA recognition by polyamides depends on side-by-side, aromatic amino acid pairings in the minor groove. Pairing rules have been established for DNA base pair recognition: an N-methylpyrrole/N-methylimidazole (Py/Im) pair recognizes C•G, the reverse (Im/Py) specifies G•C, and a Py/Py pair targets both A•T and T•A.2 For some sequences, replacement of Py with a flexible β-alanine (β) residue can enhance polyamide binding affinity by relaxing ligand curvature, affording a more optimal fit in the minor groove of DNA.14
Although Py-Im polyamides will bind in the minor groove as anti-parallel dimers,15 polyamides bind DNA optimally as a single oligomer in a hairpin structure.16 Early studies demonstrated the optimal length for the turn element is three methylene units, via the use of γ-aminobutyric acid as the turn residue.16 The polyamide hairpin binds in the ‘forward’ direction N→C with respect to the 5′→3′ direction of the DNA strand.17 Utilization of the substituted turn (R)-2,4-diaminobutyric acid (γ-DABA) further enhances DNA binding affinity.18 Notably, employment of a linkage containing one methylene unit – provided by the use of glycine as the turn amino acid – resulted in polyamides that bound DNA in an extended 2:1 binding mode, rather than as a 1:1 hairpin complex.16, 19–21 In contrast, the γ-aminobutyric acid linkage appears to prefer the U-shaped conformation rather than an extended dimeric conformation.20
We recently reported a new turn unit with remarkable properties in small animal models: (R)-α-diaminobutyric acid (α-DABA), which is formally a substituted glycine linkage since it also includes a single methylene unit within the hairpin turn.22 This initial study characterized an α-DABA polyamide-chlorambucil conjugate and compared its biological activity to its γ-DABA analog. A dramatic difference in toxicity was observed for the two conjugates: normal mice treated with the α-DABA polyamide-chlorambucil conjugate did not differ significantly from control, while the analogous γ-DABA conjugate was lethal.22 Notably, only the α-DABA conjugate has been demonstrated to arrest cancer growth in a SW620 xenograft nude mouse tumor model without apparent toxicity.13
Given the striking in vivo effects demonstrated by the α-DABA polyamide conjugate, this new turn unit merits further study. In particular, α-DABA polyamides appear to bind in a hairpin conformation and not as an extended dimer, as would be expected based on studies of glycine-linked polyamides.
We report herein the characterization of α-DABA polyamides. A series of α-DABA polyamides and their chlorambucil conjugates were synthesized, along with their γ-DABA analogs. DNA binding properties of the parent molecules were established by DNase I footprinting titration experiments, and DNA alkylation properties of the chlorambucil conjugates were determined by thermal cleavage assay experiments. Importantly, we study the binding orientation of α-DABA polyamides and address their ability to bind as hairpins by comparing them to their glycine-linked analogs.
2. Results and discussion
2.1 Synthesis of the polyamide series
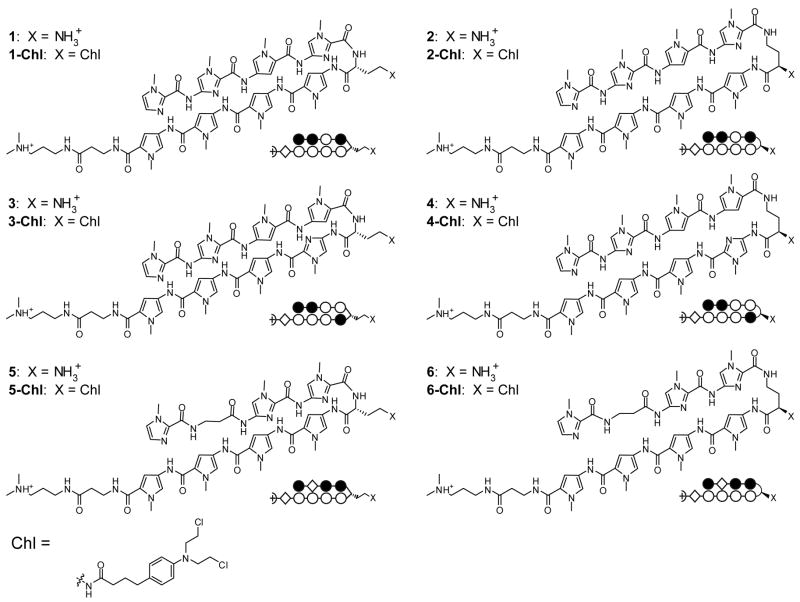
Three core ring sequences were selected for characterization (Fig. 1). Two standard eight-ring hairpin sequences, as well as a hairpin containing a β/Py pair, were chosen. Six parent hairpin polyamides were thus prepared: three molecule pairs which code for different sequences according to the pairing rules. For each pair, one molecule was linked via the α-DABA turn unit (polyamides 1, 3, and 5), and the other was connected with the γ-DABA turn (polyamides 2, 4, and 6). Polyamides were synthesized on Pam resin using standard solid phase methods.22, 23 The chlorambucil conjugates of the six parent hairpin polyamides were also prepared based on previously published methods.22
Figure 1.
Chemical and ball-and-stick structures of polyamides 1-6 and their Chl conjugates. (Left) α-diaminobutyric acid-linked hairpin polyamides. (Right) γ-diaminobutyric acid-linked hairpin polyamides. The ball and stick symbols are defined as follows: an open circle denotes a pyrrole ring, a filled circle denotes an imidazole, and a diamond shape denotes β-alanine.
2.2 Plasmid design
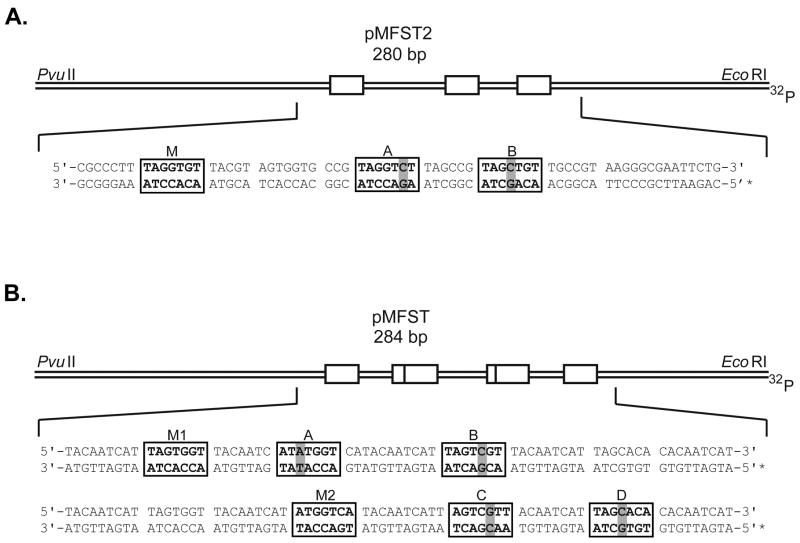
Polyamides 1 and 2 were designed to bind the sequence 5′-WWGGWGW-3′ (where W = A or T). The plasmid pMFST2 was designed with an insert containing a single match site (‘M’ = 5′-TAGGTGT-3′) and two single base pair mismatch sites (‘A’ = 5′-TAGGTCT-3′ and ‘B’ = 5′-TAGCTGT-3′) for polyamides 1 and 2 (Fig. 2A).22
Figure 2.
Illustration of the EcoRI/PvuII restriction fragments derived from plasmids (A) pMFST2 and (B) pMFST. Polyamides 1 and 2 were characterized on pMFST2. Polyamides 3-6 were characterized on pMFST. The designed polyamide binding sites are indicated by boxes. Single base pair mismatches are indicated by shaded regions. For pMFST, the designed insert is shown twice with binding sites for (B, bottom) polyamides 3 and 4 and (B, top) polyamides 5 and 6.
A single plasmid, pMFST, was prepared to characterize polyamides 3-6 (Fig. 2B). Polyamides 3 and 4 were targeted to the sequence 5′-WWGGWCW-3′, and polyamides 5 and 6 were designed to bind 5′-WWGWGGW-3′. The designed insert for pMFST contains match sites for each set of polyamides (‘M1’ = 5′-TAGTGGT-3′, targeted to 5 and 6, and ‘M2’ = 5′-ATGGTCA-3′, targeted to 3 and 4), as well as two single base pair mismatch sites for each (‘A’ = 5′-ATATGGT-3′ and ‘B’ = 5′-TAGTCGT-3′ for 5 and 6, and ‘C’ = 5′-AACGACT-3′ and ‘D’ = 5′-TAGCACA-3′ for 3 and 4). The chlorambucil moiety is expected to alkylate at adenines proximal to the polyamide binding site.11, 22 Thus, for the purposes of the thermal cleavage assays, adenines were placed next to the 3′ end of the match sites and at least one of the mismatch sites.
2.3 DNA binding affinity and sequence specificity
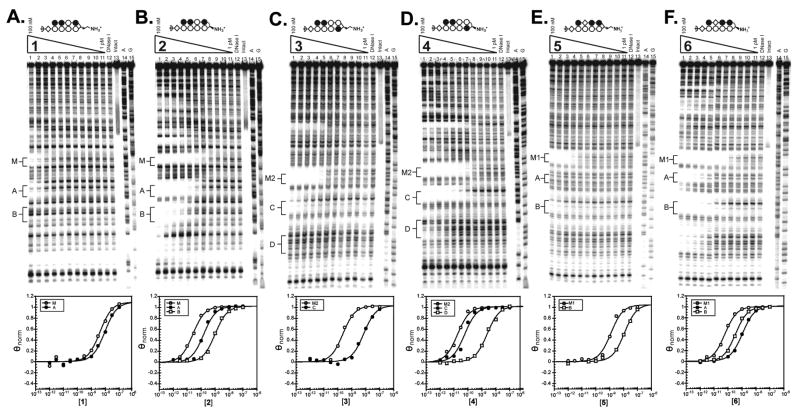
Quantitative DNase I footprinting titrations were performed with the parent polyamides to measure their binding site affinities and specificities (Fig. 3 and Table 1). The α-DABA polyamide 1 and its γ-DABA analog 2 were footprinted on the 5′ 32P-labeled 280 base pair PCR product of plasmid pMFST2 (Fig. 3A–3B). α-DABA polyamide 1 bound the match site M with Ka = 3.0 × 108 M−1 and the mismatch site A with Ka = 8.5 × 107 M−1 (Table 1). Binding of 1 at the mismatch site B was not detected. In contrast, the γ-DABA polyamide 2 bound the match site M with Ka = 4.4 × 1010 M−1 and the mismatch sites A and B with Ka = 1.1 × 1010 M−1 and Ka = 1.1 × 109 M−1, respectively. The γ-DABA polyamide 2 displays a 147-fold increased binding affinity over the α-DABA polyamide 1 for their match site M.
Figure 3.
Quantitative DNase I footprinting titration experiments for polyamides (A) 1 and (B) 2 on the 280 base pair, 5′ end-labeled PCR product of plasmid pMFST2; and for polyamides (C) 3, (D) 4, (E) 5, and (F) 6 on the 284 base pair, 5′ end-labeled PCR product of plasmid pMFST: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. (Below) Each footprinting gel is accompanied by its respective binding isotherms.
Table 1.
Binding affinities (M−1) for parent polyamides on plasmid pMFST2 (1–2) and plasmid pMFST (3–6). Equilibrium association constants reported are mean values from three DNase I footprinting titration experiments. Standard deviations are shown in parentheses. Bolded numbers in brackets indicate fold decrease in match site binding affinity for α-DABA-linked versus γ-DABA-linked polyamides. Mismatch base pairs are indicated in bold.
| Polyamide | Match Site | Ka (M−1) | Mismatch Site | Ka (M−1) | Mismatch Site | Ka(M−1) | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5′-TAGGTGT-3′ | 3.0(±1.3)x108 | [147] | 5′-TAGGTCT-3′ | 8.5(±3.1)x107 | [129] | 5′-TAGCTGT-3′ | – | |
| 2 | 5′-TAGGTGT-3′ | 4.4(±0.4)x1010 | 5′-TAGGTCT-3′ | 1.1 (±0.8)x1010 | 5′-TAGCTGT-3′ | 1.1 (±0.3)x109 | |||
|
| |||||||||
| 3 | 5′-ATGGTCA-3′ | 4.4(±0.6)x109 | [11] | 5′-AACGACT-3′ | 1.5(±0.3)x108 | [120] | 5′-TAGCACA-3′ | – | |
| 4 | 5′-ATGGTCA-3′ | 4.9(±0.7)x1010 | 5′-AACGACT-3′ | 1.8(±0.3)x1010 | 5′-TAGCACA-3′ | 3.9(±1.0)x108 | |||
|
| |||||||||
| 5 | 5′-TAGTGGT-3′ | 8.0(±0.6)x108 | [54] | 5′-ATATGGT-3′ | – | 5′-TAGTCGT-3′ | 6.2(±3.0)x107 | [100] | |
| 6 | 5′-TAGTGGT-3′ | 4.3(±0.3)x1010 | 5′-ATATGGT-3′ | 9.9(±0.3)x108 | 5′-TAGTCGT-3′ | 6.2(±0.2)x109 | |||
The α-DABA polyamides 3 and 5 and their γ-DABA analogs 4 and 6, respectively, were footprinted on the 5′ 32P-labeled 284 base pair PCR product of plasmid pMFST (Fig. 3C–3F). α-DABA polyamide 3 bound its match site M2 with Ka = 4.4 × 109 M−1 and the mismatch site C with Ka = 1.5 × 108 M−1. There was no detectable binding at mismatch site D. However, the analogous γ-DABA polyamide 4 bound the match site M2 with Ka = 4.9 × 1010 M−1 and the mismatch sites C and D with Ka = 1.8 × 1010 M−1 and Ka = 3.9 × 108 M−1, respectively. The α-DABA polyamide 3 binds its match site with 11-fold decreased affinity relative to the analogous γ-DABA polyamide 4.
α-DABA polyamide 5 bound its match site M1 with Ka = 8.0 × 108 M−1 and its mismatch site B with Ka = 6.2 × 107 M−1. Binding was not observed at mismatch site A. In comparison, the γ-DABA polyamide 6 binds the match site M1 with Ka = 4.3 × 1010 M−1 and the mismatch sites A and B with Ka = 9.9 × 108 M−1 and Ka = 6.2 × 109 M−1, respectively. The α-DABA polyamide 5 shows a 54-fold decreased affinity for its match site compared to the analogous γ-DABA polyamide 6.
These results indicate that α-DABA polyamides are proficient DNA binders, with decreased affinity and comparable or improved specificity relative to γ-DABA molecules. The decrease in affinity is dependent on the polyamide core ring sequence. Additional comparison of molecules corroborates these conclusions (Supplemental Fig. 1). Varying the heterocyclic order of polyamides has also been shown to affect binding affinities in γ-DABA molecules.24
We previously reported the α-DABA polyamide of sequence ImImβIm-(R)H2Nα-PyPyPyPy-βDp (where Dp = 3-(dimethylamino)-propylamine) binds the match site M of pMFST2 with Ka = 6.7 × 109 M−1.22 Polyamide 1 has an identical structure except for the replacement of a β residue in the core sequence with a Py, and it binds the same match site M of pMFST2 with 22-fold decreased affinity. Polyamide 5 also has the same structure except for the movement of the β residue from the third position in the sequence to the second, and it binds its designed match site with 8-fold decreased affinity. These results suggest the presence and position of the β residue is important in determining the DNA binding affinities of α-DABA polyamides. We hypothesize that the shortened α-DABA turn unit requires the flexibility provided by the β to optimize binding.
2.4 DNA alkylation properties
Thermal cleavage assays were performed with the polyamide-chlorambucil conjugates to determine their DNA alkylation properties (Fig. 4). Previous studies have shown conjugates to alkylate at N3 of adenines proximal to DNA binding sites.11, 13 Attachment of the chlorambucil moiety to the parent polyamide does not appear to affect DNA binding properties.11
Figure 4.
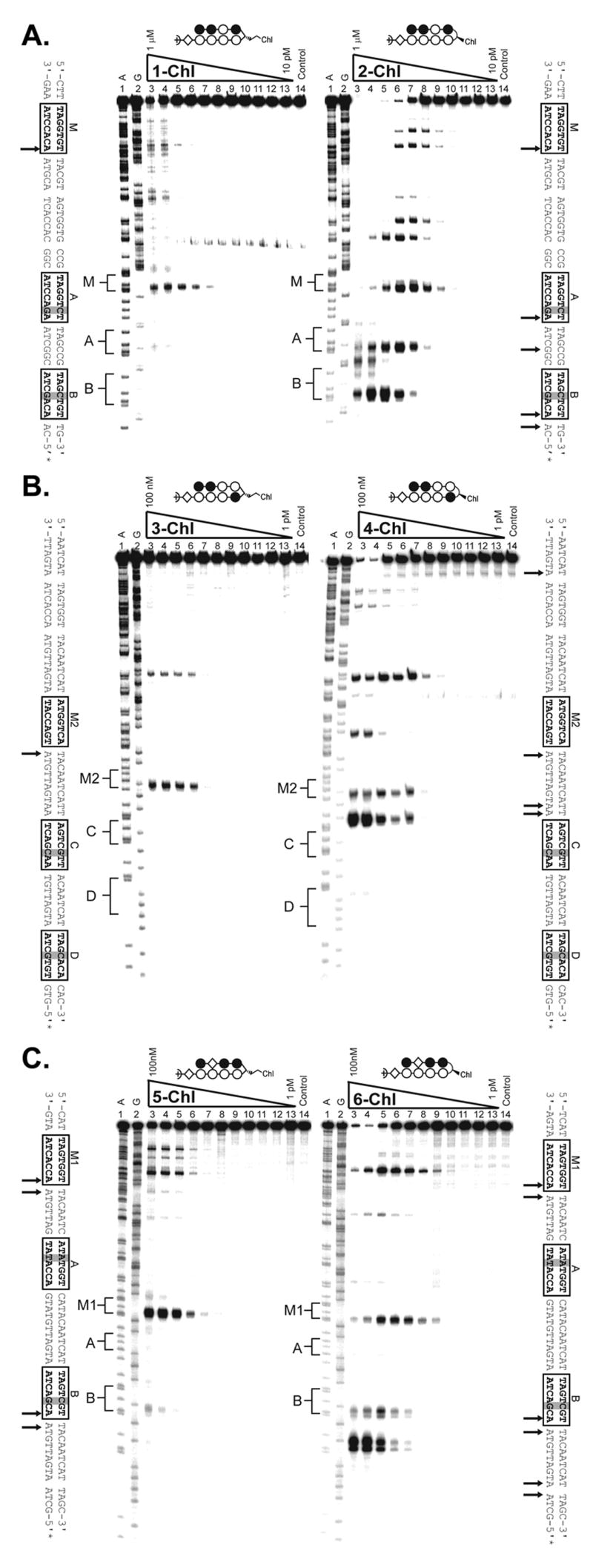
(A) Thermal cleavage assay experiments with polyamide-chlorambucil conjugates (left) 1-Chl and (right) 2-Chl on the on the 280 base pair, 5′ end-labeled PCR product of plasmid pMFST2: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (C-D) Thermal cleavage assay experiments with polyamide-chlorambucil conjugates (C, left) 3-Chl and (C, right) 4-Chl and (D, left) 5-Chl and (D, right) 6-Chl on the on the 284 base pair, 5′ end-labeled PCR product of plasmid pMFST: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 14, intact DNA. Putative major sites of alkylation on the DNA fragments are indicated by arrows on the sequences adjacent to each gel.
The α-DABA conjugate 1-Chl and its γ-DABA analog 2-Chl were assayed on the 5′ 32P-labeled 280 base pair PCR product of plasmid pMFST2 (Fig. 4A). Alkylation was only observed in the designed insert of pMFST2 at the match site M for the α-DABA conjugate 1-Chl, whereas the γ-DABA conjugate 2-Chl is more promiscuous, alkylating at the match site M and both of the mismatch sites A and B, as well as at additional, unexpected sites. γ-DABA conjugate 2-Chl also alkylates at lower concentrations than conjugate 1-Chl (Fig 4A, lanes 9).
Conjugates 3-Chl and 4-Chl were assayed on the 5′ 32P-labeled 284 base pair PCR product of plasmid pMFST (Fig. 4B). While parent polyamide 3 binds at site M2 and site C, α-DABA conjugate 3-Chl only alkylates at match site M2 in the designed insert. Alkylation is observed at both the match site M2 and the mismatch site C for the γ-DABA conjugate 4-Chl. Although the parent polyamide 4 binds mismatch site D, alkylation would not be expected to occur there because an A•T base pair is not located adjacent to the turn unit upon conjugate binding. Again, alkylation is observed at lower concentrations for the γ-DABA conjugate 4-Chl than the α-DABA conjugate 3-Chl (Fig. 4B, lanes 7).
Conjugates 5-Chl and 6-Chl were also assayed on the PCR product of plasmid pMFST (Fig. 4C). For the α-DABA conjugate 5-Chl, strong alkylation is only observed at match site M1 in the designed insert, with minor alkylation occurring at mismatch site B. In contrast, the γ-DABA conjugate 6-Chl strongly alkylates at both match site M1 and mismatch site B, as well as at other unexpected sites. As previously, alkylation is not expected at mismatch site A since an A•T base pair is not located next to the turn unit upon conjugate binding. In addition, γ-DABA conjugate 6-Chl is able to alkylate at a lower concentration than α-DABA conjugate 5-Chl (Fig. 4C, lanes 9).
These results indicate α-DABA conjugates to be alkylators with increased specificity and decreased reactivity relative to γ-DABA conjugates. Thermal cleavage data from additional conjugates further support these conclusions (Supplemental Fig. 1).
2.5 Polyamide binding orientation
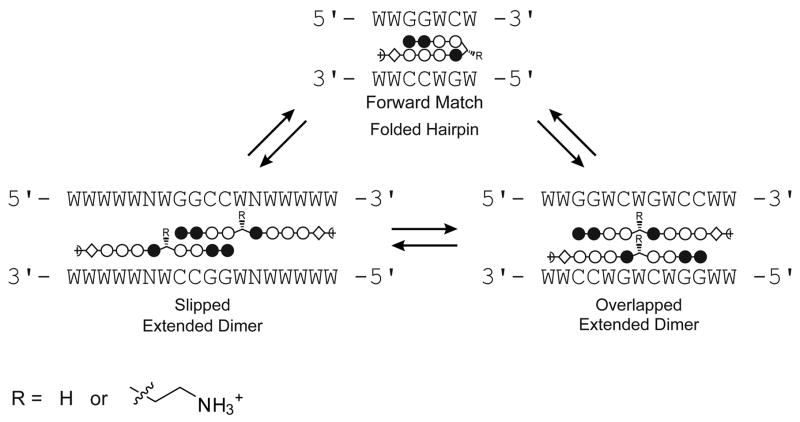
Previous footprinting studies indicated that glycine-linked polyamides bind DNA in extended, dimeric 2:1 complexes,16, 20 and the structures were confirmed by NMR studies.19, 21 Two possible binding modes were established (Fig. 5). In the “overlapped” binding mode, the two polyamides bind directly opposite one another with each heterocycle paired to another on the other strand. In the “slipped” binding mode, the polyamides integrate the 2:1 and 1:1 polyamide-DNA binding motifs at a single site.
Figure 5.
Illustration of the binding modes for polyamides 3 and 7. W signifies A or T; N represents any nucleotide. Sequence specificity for the slipped binding mode has been presumed based on the polyamide literature.
Formally, the α-DABA moiety is a substituted glycine turn, as glycine-linked polyamides also contain a single methylene group linking the heterocyclic subunits. Yet, based on their footprinting data (i.e. binding site sequence and size, and cooperativity as measured in the binding isotherms), α-DABA polyamides bind in a 1:1 hairpin conformation.22 In addition, although it is well-established that γ-DABA polyamides bind in the forward direction,17 there is still ambiguity as to the preferred binding orientation of α-DABA polyamides.
We hypothesized that a polyamide with the unsubstituted glycine linkage would favor the extended conformation, whereas substitution of the turn using the α-DABA moiety would force the polyamide to bind in a forward hairpin conformation. A similar phenomenon has been observed by Boger and coworkers, who studied a series of α-substituted β-linked polyamides and found that a polyamide linked by (R)-α-methoxy-β preferentially adopts a hairpin conformation, versus the unsubstituted molecule which binds in an extended conformation.25 Interestingly, other substitutions disrupted binding affinity.
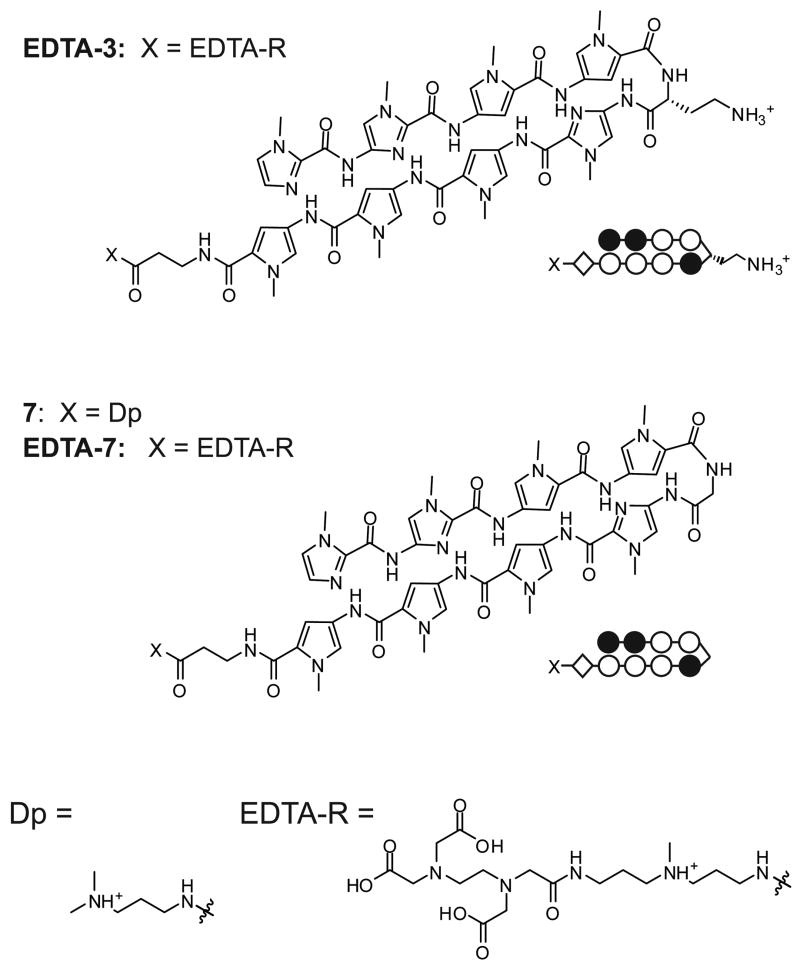
α-DABA polyamide 3 was chosen as the parent molecule for study. To verify its binding orientation, the EDTA conjugate EDTA-3 was synthesized for affinity cleavage studies (Fig. 6). Polyamide 7 – the glycine-linked analog of 3 – was prepared, as well as its EDTA conjugate, EDTA-7.
Figure 6.
Chemical and ball-and-stick structures of EDTA-3, and of the glycine-linked polyamide 7 and its EDTA conjugate EDTA-7.
Four binding orientations are possible for polyamides 3 and 7. The expected match site for the molecules binding as forward hairpins is 5′-WWGGWCW-3′, whereas for reverse-binding hairpins the expected match site is 5′-WCWGGWW-3′. In the overlapped binding mode, the expected target sequence is 5′-WWGGWCWGWCCWW-3′. It should be noted that the overlapped match site is inherently ambiguous because it contains two side-by-side copies of the forward hairpin match site. Binding affinity in the slipped mode is sensitive to the particular sequence of the 1:1 binding portion, and there are multiple slipped binding modes for a given polyamide core ring sequence.20 Since an Im/Im pair is disfavored,26 we studied the slipped orientation for 3 and 7 believed to be preferred for binding. We expect this binding model to give an 18 base pair match site with sequence 5′-WWWWWNWGGCCWNWWWWW-3′ where N = A, T, G, or C.20, 27
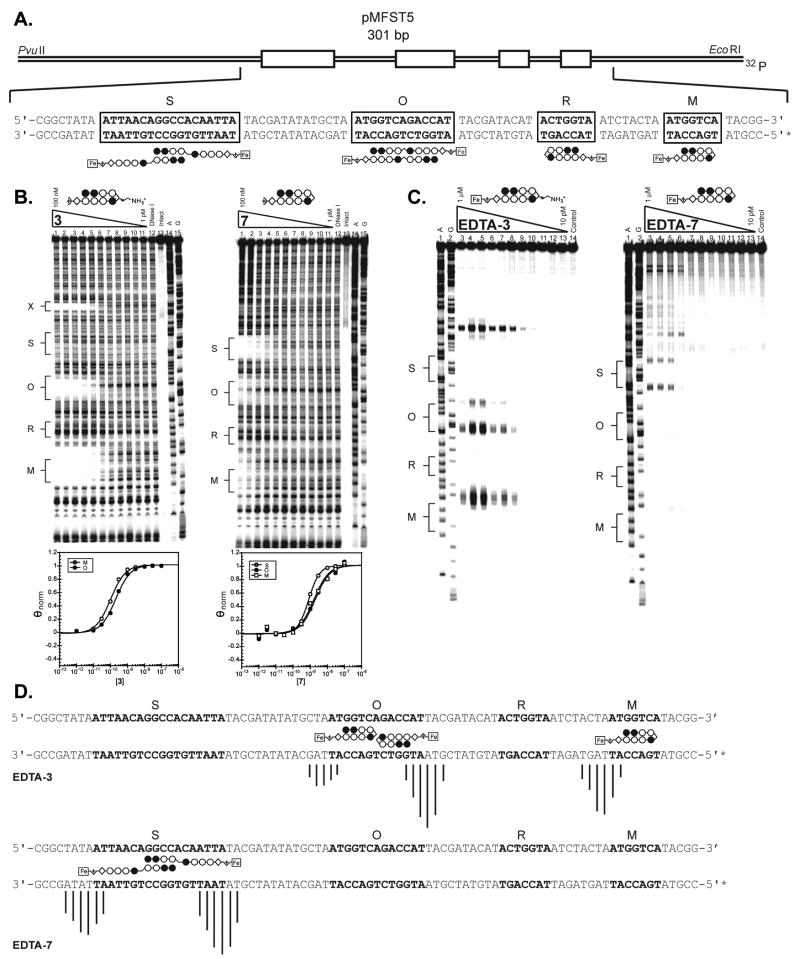
Plasmid pMFST5 was prepared to study the binding preferences of the selected molecules (Fig. 7A). The designed insert contains four match sites for each of the possible binding modes for polyamides 3 and 7: slipped extended binding (‘S’ = 5′-ATTAACAGGCCACAATTA-3′), overlapped extended binding (‘O’ = 5′-ATGGTCAGACCAT-3′), reverse hairpin binding (‘R’ = 5′-ACTGGTA-3′), and forward hairpin binding (‘M’ = 5′-ATGGTCA-3′).
Figure 7.
(A) Illustration of the EcoRI/PvuII restriction fragment derived from plasmid pMFST5. The designed polyamide binding sites are indicated by boxes. Binding models are illustrated below the DNA sequence by ball-and-stick structures of EDTA-7. (B) Quantitative DNase I footprinting titration experiments for polyamides (left) 3 and (right) 7 on the 301 base pair, 5′ end-labeled PCR product of plasmid pMFST5: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. Site X indicates an undesigned match site inherent in the plasmid sequence. (Below) Each footprinting gel is accompanied by its respective binding isotherms. (C) Affinity cleavage experiments with polyamide-EDTA conjugates (left) EDTA-3 and (right) EDTA-7 on the 301 base pair, 5′ end-labeled PCR product of plasmid pMFST5: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (D) Summary of affinity cleavage patterns for (top) EDTA-3 and (bottom) EDTA-7. Bar heights are proportional to the relative cleavage intensities at each base pair, normalized to each gel. Ball-and-stick structures illustrating their respective proposed modes of binding are shown using EDTA-7 for clarity. Designed binding sites are indicated in bold in the DNA sequence.
Quantitative DNase I footprinting titrations were performed with α-DABA polyamide 3 and glycine polyamide 7 (Fig. 7B and Table 2), and affinity cleavage studies were performed with α-DABA conjugate EDTA-3 and glycine conjugate EDTA-7 (Fig. 7C–7D) on the 5′ 32P-labeled 301 base pair PCR product of plasmid pMFST5. A dramatic difference between the two molecules was observed. The α-DABA polyamide 3 binds the forward hairpin binding site M with Ka = 9.8 × 109 M−1, while no binding was detected at the slipped binding site S or reverse hairpin site R. Cleavage by EDTA-3 was observed at match site M at the locus in agreement with forward binding and is 3′-shifted, consistent with minor groove occupancy.28
Table 2.
Binding affinities (M−1) for parent polyamides 3 and 7 on plasmid pMFST5. Equilibrium association constants reported are mean values from three DNase I footprinting titration experiments. Standard deviations are shown in parentheses.
| Polyamide | M | O | S |
|---|---|---|---|
| 3 | 9.8(± 1.3) × 109 | 4.7(± 1.3) × 109 | -- |
| 7 | 7.0(± 1.0) × 108 | 5.9(± 2.1) × 108 | 1.3(± 0.2) × 109 |
Additional binding of polyamide 3 was observed at the overlapped site O with Ka = 4.7 × 109 M−1. Unequal cleavage intensities by EDTA-3 were observed at the 3′ and 5′ ends of the match site. A fully anti-parallel overlapped motif would be expected to afford equal intensities. Therefore it is likely that polyamide 3 is binding as a hairpin in either of the adjacent hairpin match sites embedded in the 13 base-pair sequence O (Fig. 7D, top).
In contrast, the glycine polyamide 7 binds the slipped extended site S with Ka = 1.3 × 109 M−1, and it binds the forward hairpin site M and overlapped site O with ~ 2-fold decreased affinities (Ka = 7.0 × 108 M−1 and Ka = 5.9 × 108 M−1, respectively). No binding was observed at the reverse hairpin site R. Strong cleavage by EDTA-7 was only observed at site S, probably due to the weaker binding affinities at the other sites. The cleavage pattern was 3′ shifted, verifying minor groove occupancy of the ligand, and the observed pattern was consistent with dimeric, antiparallel binding in the slipped mode. There is again ambiguity regarding the binding mode at the overlapped site O.
It is interesting to note that though polyamide 7 is a relatively small ligand (MW = 1196), it displays specificity for an 18 base pair sequence. It is likely that the binding affinity of 7 in the slipped binding mode could be enhanced by modifying the sequence of the W tracts in the 1:1 binding portions of the molecules to a DNA sequence that favors a narrower minor groove.20
These data show a clear distinction in preferred binding modes between α-DABA polyamide 3 and glycine polyamide 7. The α-DABA polyamide appears to bind exclusively in a forward hairpin conformation, while the glycine polyamide seems to exhibit mixed binding motifs, preferring the extended binding mode but also appearing to be capable of binding as a forward hairpin. Thus, substitution of the glycine turn ‘locks’ the polyamide into a hairpin conformation. Affinity cleavage studies performed on additional α-DABA polyamides confirm their general binding in the forward hairpin mode (Supplemental Fig. 2 and 3).
3. Conclusion
Herein we present a characterization of α-DABA polyamides. We have confirmed their ability to bind DNA and established their binding affinities to be decreased 2-fold to 147-fold relative to γ-DABA polyamides. The magnitude of this reduced affinity appears to be sensitive to the presence and positioning of an internal β residue in the polyamide structure. Remarkably, despite the reduced affinity, α-DABA polyamide-chlorambucil conjugates alkylate DNA with greater specificity and lower reactivity than γ-DABA conjugates. Importantly, we have demonstrated that polyamides linked by the (R)-α-diaminobutyric acid moiety bind DNA in a forward hairpin binding mode. Polyamides linked by a glycine amino acid prefer to bind DNA in an extended binding mode, indicating that it is the substitution of the glycine turn unit that locks the α-DABA polyamide into the hairpin conformation. While it has been previously shown that α-DABA polyamide-chlorambucil conjugates are efficacious in cellular and small animal models,13, 22, 29 cellular uptake experiments of polyamide-fluorescein conjugates displayed limited nuclear uptake properties (Supplemental Fig. 4, Table 1), possibly limiting the general use of α-DABA polyamides in vivo.
4. Experimental
4.1 General
Boc-β-Ala-Pam resin was from Peptides International. Trifluoroacetic acid (TFA) was purchased from Halocarbon. Dichloromethane (DCM) was from Fisher Scientific. (Benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBOP) was from Novabiochem. Chlorambucil, N,N-diisopropylethylamine, 3-(dimethylamino)-propylamine, 3,3′-diamino-N-methyl-dipropylamine, N,N-dimethylformamide, and 1-methyl-2-pyrrolidone (NMP) were purchased from Aldrich. Oligonucleotides were synthesized by Integrated DNA Technologies. Enzymes were purchased from Roche. RNase free water was from USB and used for all footprinting and cleavage reactions.
4.2 Polyamide synthesis
Polyamides were synthesized on Boc-β-Ala-Pam resin following manual solid-phase methods using Boc-protected amino acids as previously reported.23 Boc-Py-OBt (OBt = benzotriazol-1-yloxy), Boc-β-Im-OH, Im-CCl3, Boc-Py-Py-OBt, Boc-Py-Im-OH, Boc-Im-OH, and Im-Im-OH were used as building blocks. The α-DABA polyamides 1, 3, 5, 1-Chl, 3-Chl, 5-Chl, and EDTA-3 were synthesized using Boc-D-Dab(Fmoc)-OH as the turn unit. The γ-DABA polyamides 2, 4, 6, 2-Chl, 4-Chl, and 6-Chl were synthesized using Fmoc-D-Dab(Boc)-OH as the turn unit. The glycine-linked polyamides 7 and EDTA-7 were synthesized using Boc-Gly-OH as the turn unit.
Boc deprotections were conducted at room temperature for 25 min using 80% TFA/DCM. Carboxylic acids (3 eq) were activated with DIEA (5 eq) and PyBOP (2.5 eq) for 30–45 min at 37 °C prior to coupling. Couplings were allowed to proceed in NMP for 2 h at room temperature. Polyamides were cleaved from resin with either 3-(dimethylamino)-propylamine (Dp) or 3,3′-diamino-N-methyl-dipropylamine (Da) neat at 55 °C overnight. Products were purified by preparatory reverse-phase high performance liquid chromatography (prep HPLC) and characterized by analytical HPLC, UV-visible spectroscopy, and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry.
Polyamide-chlorambucil conjugates were generated by activating the chlorambucil carboxylic acid (4 eq) with PyBOP (4 eq) and DIEA (12 eq) for 30 min at room temperature followed by addition of the polyamide. The reaction was allowed to proceed at room temperature for 1–2 h. Products were purified by reverse-phase prep HPLC, immediately lyophilized, and characterized as above.
Polyamide-EDTA conjugates were synthesized by slow addition of a 50% DMF/DIEA polyamide solution to a solution of EDTA dianhydride (20 eq) dissolved in a 1 DMSO:1 DMF:2 DIEA mixture, vigorously shaking at 55 °C. The reaction was allowed to proceed an additional 30 min, followed by hydrolysis of the remaining EDTA dianhydride with 0.1 N NaOH for 10 min. The reaction was diluted and purified by reverse-phase prep HPLC. Subsequent Boc deprotection was accomplished by dissolving the dried EDTA conjugate in a minimum volume of neat TFA doped with triethylsilane. The reaction was allowed to proceed for 10–30 min at room temperature, then immediately diluted with water and purified by reverse-phase prep HPLC.
1: ImImPyIm-(R)H2Nα-PyPyPyPy-βDp MALDI-TOF MS calcd for C57H72N23O10+ [M+H]+: 1238.58; found: 1238.82.
2: ImImPyIm-(R)H2Nγ-PyPyPyPy-βDp MALDI-TOF MS calcd for C57H72N23O10+ [M+H]+: 1238.58; found: 1238.59.
3: ImImPyPy-(R)H2Nα-ImPyPyPy-βDp MALDI-TOF MS calcd for C57H72N23O10+ [M+H]+: 1238.58; found: 1238.90.
4: ImImPyPy-(R)H2Nγ-ImPyPyPy-βDp MALDI-TOF MS calcd for C57H72N23O10+ [M+H]+: 1238.58; found: 1238.65.
5: ImβImIm-(R)H2Nα-PyPyPyPy-βDp MALDI-TOF MS calcd for C54H71N22O10+ [M+H]+: 1188.28; found: 1188.5.
6: ImβImIm-(R)H2Nγ-PyPyPyPy-βDp MALDI-TOF MS calcd for C54H71N22O10+ [M+H]+: 1188.28; found: 1188.86.
7: ImImPyPy-Gly-ImPyPyPy-βDp MALDI-TOF MS calcd for C55H67N22O10+ [M+H]+: 1195.54; found: 1195.43.
1-Chl: ImImPyIm-(R)Chlα-PyPyPyPy-βDp MALDI-TOF MS calcd for C71H89Cl2N24O11+ [M+H]+: 1523.65; found: 1523.66.
2-Chl: ImImPyIm-(R)Chlγ-PyPyPyPy-βDp MALDI-TOF MS calcd for C71H89Cl2N24O11+ [M+H]+: 1523.65; found: 1523.70.
3-Chl: ImImPyPy-(R)Chlα-ImPyPyPy-βDp MALDI-TOF MS calcd for C71H89Cl2N24O11+ [M+H]+: 1523.65; found: 1523.88.
4-Chl: ImImPyPy-(R)Chlγ-ImPyPyPy-βDp MALDI-TOF MS calcd for C71H89Cl2N24O11+ [M+H]+: 1523.65; found: 1523.57.
5-Chl: ImβImIm-(R)Chlα-PyPyPyPy-βDp MALDI-TOF MS calcd forC68H88Cl2N23O10+ [M+H]+: 1474.48, found: 1474.95.
6-Chl: ImβImIm-(R)Chlγ-PyPyPyPy-βDp MALDI-TOF MS calcd for C68H88Cl2N23O10+ [M+H]+: 1474.48, found: 1474.26.
EDTA-3: ImImPyPy-(R)H2Nα-ImPyPyPy-βDa-EDTA MALDI-TOF MS calcd for C69H91N26O17+ [M+H]+: 1555.71; found: 1555.73.
EDTA-7: ImImPyPy-Gly-ImPyPyPy-βDa-EDTA MALDI-TOF MS calcd for C67H86N25O17+ [M+H]+: 1512.66; found: 1512.54.
4.2 Construction of plasmids
Plasmids were constructed by annealing oligonucleotide pairs as follows: for pMFST – 5′-AGCTGTACAATCATTAGTGGTTACAATCATATGGTCATACAATC-ATTAGTCGTTACAATCATTAGCACACACAATCATC-3′ and 5′-GATCGATGATT-GTGTGTGCTAATGATTGTAACGACTAATGATTGTATGACCATATGATTGTAA-CCACTAATGATTGTAC-3′; for pMFST5 – 5′-AGCTGCGGCTATAATTAACAGGC-CACAATTATACGATATATGCTAATGGTCAGACCATTACGATACATACTGGTA-ATCTACTAATGGTCATACGGC-3′ and 5′-GATCGCCGTATGACCATTAGTAGAT-TACCAGTATGTATCGTAATGGTCTGACCATTAGCATATATCGTATAATTGTG-GCCTGTTAATTATAGCCGC-3′.
Annealed nucleotides were ligated into the BamHI/HindIII restriction fragment of pUC19 using T4 DNA ligase, and the plasmid was transformed into Escherichia coli JM109 competent cells. Ampicillin-resistant white colonies were selected from 25 mL Luria-Bertani agar plates containing 50 mg/mL ampicillin treated with XGAL and isopropyl-β-D-thiogalactopyranoside (IPTG) solutions and grown overnight at 37 °C. Cells were harvested the following day, and purification of the plasmids were performed with a Wizard Plus Midiprep DNA purification kit (Promega). DNA sequencing of the plasmid insert was performed by the sequence analysis facility at the California Institute of Technology.
4.3 Preparation of 5′ 32P-end-labeled DNA
The primer 5′-GAATTCGAGCTCGGTACCCGGG-3′ was 32P-labeled at the 5′ end and used with the primer 3′-CAGCCCTTTGGACAGCACGGTC-5′ to PCR amplify plasmids as described previously.28
4.4 DNase footprinting titrations
Polyamide equilibrations and DNase I footprinting titrations were conducted on the 5′ end-labeled PCR product of the appropriate plasmid according to standard protocols.28 DNA was incubated with polyamide or water (controls) for 12–16 h at room temperature prior to reaction. Determination of equilibrium association constants were performed as previously described.28 Data points were fitted to a Langmuir binding isotherm by using the modified Hill equation with n = 1, except for the quantitation of glycine polyamide 7, where n was optimized to the generated curve for the slipped site S and the overlapped site O.
4.5 Thermal cleavage assays
Thermal cleavage assay experiments were conducted on the 5′ end-labeled PCR products of the designated plasmids as described previously.11 DNA was incubated with polyamide conjugates or water (control) for 24 h at 37 °C prior to work-up.
4.6 Affinity cleavage assays
Affinity cleavage experiments were conducted on the 5′ end-labeled PCR product of plasmid pMFST5 as previously described.28 DNA was incubated with polyamide conjugates or water (control) for 13 h at room temperature prior to addition of ferrous ammonium sulfate.
Supplementary Material
Supplemental Table 1. Cellular uptake properties of α-DABA polyamide-fluorescein conjugates in 3 cell lines as determined by confocal microscopy. ++, Nuclear staining exceeds that of the medium; +, nuclear staining is less than or equal to that of the medium, but still prominent; −, very little nuclear staining, with the most fluorescence seen in the cytoplasm and/or medium; − −, no nuclear staining. Polyamides were assayed at 5 μM concentration.
Supplemental Figure 1. (A) Chemical and ball-and-stick structures for polyamides 8 and 9 and their Chl conjugates. (B) Illustration of the EcoRI/PvuII restriction fragments derived from plasmid pMFST3. The designed polyamide binding sites are indicated by boxes. Single base pair mismatches are indicated by shaded regions. (C) Quantitative DNase I footprinting titration experiments for polyamides (left) 8 and (right) 9 on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. (Below) Each footprinting gel is accompanied by its respective binding isotherms. (D) Thermal cleavage assay experiments with polyamide-chlorambucil conjugates (left) 8-Chl and (right) 9-Chl on the on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 14, intact DNA. Putative major sites of alkylation on the DNA fragments are indicated by arrows on the sequences adjacent to each gel. (E) Table of binding affinities (M−1) for parent polyamides 8 and 9 on plasmid pMFST3. Equilibrium association constants reported are mean values from at last three DNase I footprinting titration experiments. Standard deviations are shown in parentheses. Bolded numbers in brackets indicate fold decrease in match site binding affinity for α-DABA-linked versus γ-DABA-linked polyamides.
Supplemental Figure 2. (A) Chemical and ball-and-stick structures for polyamide 10 and its EDTA conjugate EDTA-10. (B) Illustration of the EcoRI/PvuII restriction fragment derived from plasmid pMFST4. The designed polyamide binding sites are indicated by boxes. (C) Quantitative DNase I footprinting titration experiments for polyamide 10 on the 283 base pair, 5′ end-labeled PCR product of plasmid pMFST4: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. (Below) Binding isotherms. Binding affinities for sites M and O are 9.8 (±0.5) × 108 M−1 and 1.1 (±0.6) × 109 M−1, respectively. (D) Affinity cleavage experiment with EDTA-10 on the 283 base pair, 5′ end-labeled PCR product of plasmid pMFST4: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (E) Summary of affinity cleavage patterns for EDTA-10. Bar heights are proportional to the relative cleavage intensities at each base pair. Ball-and-stick structures illustrating the proposed modes of binding are shown. Designed binding sites are indicated in bold in the DNA sequence.
Supplemental Figure 3. (A) Chemical and ball-and-stick structures for polyamide-EDTA conjugates EDTA-8 and EDTA-9. (B) Affinity cleavage experiments with polyamide-EDTA conjugates (left) EDTA-8 and (right) EDTA-9 on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (C) Summary of affinity cleavage patterns for (top) EDTA-8 and (bottom) EDTA-9. Bar heights are proportional to the relative cleavage intensities at each base pair. The designed polyamide binding sites are indicated by boxes.
Supplemental Figure 4. Chemical and ball-and-stick structures for polyamide-fluorescein conjugates prepared for studying the cellular uptake properties of α-DABA polyamides.
Acknowledgments
The authors thank the National Institutes of Health for research support and for a predoctoral NRSA training grant to M.E.F. Mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering at the California Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wemmer DE, Dervan PB. Curr Opin Struct Biol. 1997;7:355–361. doi: 10.1016/s0959-440x(97)80051-6. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Poulin-Kerstien AT, Fechter EJ, Edelson BS. Top Curr Chem. 2005;253:1–31. [Google Scholar]
- 3.Best TP, Edelson BS, Nickols NG, Dervan PB. Proc Natl Acad Sci U S A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc Natl Acad Sci U S A. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB, Gottesfeld JM. Proc Natl Acad Sci U S A. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kageyama Y, Sugiyama H, Ayame H, Iwai A, Fujii Y, Huang LE, Kizaka-Kondoh S, Hiraoka M, Kihara K. Acta Oncol. 2006;45:317–324. doi: 10.1080/02841860500486648. [DOI] [PubMed] [Google Scholar]
- 8.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickols NG, Dervan PB. Proc Natl Acad Sci U S A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bando T, Sugiyama H. Accounts Chem Res. 2006;39:935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- 11.Wurtz NR, Dervan PB. Chem Biol. 2000;7:153–161. doi: 10.1016/s1074-5521(00)00085-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang YD, Dziegielewski J, Wurtz NR, Dziegielewska B, Dervan PB, Beerman TA. Nucleic Acids Res. 2003;31:1208–1215. doi: 10.1093/nar/gkg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickinson LA, Burnett R, Melander C, Edelson BS, Arora PS, Dervan PB, Gottesfeld JM. Chem Biol. 2004;11:1583–1594. doi: 10.1016/j.chembiol.2004.09.004. Chem. Biol. 2006. 13, 339. [DOI] [PubMed] [Google Scholar]
- 14.Turner JM, Swalley SE, Baird EE, Dervan PB. J Am Chem Soc. 1998;120:6219–6226. [Google Scholar]
- 15.Mrksich M, Wade WS, Dwyer TJ, Geierstanger BH, Wemmer DE, Dervan PB. Proc Natl Acad Sci U S A. 1992;89:7586–7590. doi: 10.1073/pnas.89.16.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mrksich M, Parks ME, Dervan PB. J Am Chem Soc. 1994;116:7983–7988. [Google Scholar]
- 17.White S, Baird EE, Dervan PB. J Am Chem Soc. 1997;119:8756–8765. [Google Scholar]
- 18.Herman DM, Baird EE, Dervan PB. J Am Chem Soc. 1998;120:1382–1391. [Google Scholar]
- 19.Geierstanger BH, Mrksich M, Dervan PB, Wemmer DE. Nat Struct Biol. 1996;3:321–324. doi: 10.1038/nsb0496-321. [DOI] [PubMed] [Google Scholar]
- 20.Trauger JW, Baird EE, Mrksich M, Dervan PB. J Am Chem Soc. 1996;118:6160–6166. [Google Scholar]
- 21.de Clairac RPL, Seel CJ, Geierstanger BH, Mrksich M, Baird EE, Dervan PB, Wemmer DE. J Am Chem Soc. 1999;121:2956–2964. [Google Scholar]
- 22.Tsai SM, Farkas ME, Chou CJ, Gottesfeld JM, Dervan PB. Nucleic Acids Res. 2007;35:307–316. doi: 10.1093/nar/gkl1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baird EE, Dervan PB. J Am Chem Soc. 1996;118:6141–6146. [Google Scholar]
- 24.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk B, Puckett JW, Wang CC, Dervan PB. Tetrahedron. 2007;63:6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods CR, Ishii T, Wu B, Bair KW, Boger DL. J Am Chem Soc. 2002;124:2148–2152. doi: 10.1021/ja0122039. [DOI] [PubMed] [Google Scholar]
- 26.White S, Baird EE, Dervan PB. Chem Biol. 1997;4:569–578. doi: 10.1016/s1074-5521(97)90243-x. [DOI] [PubMed] [Google Scholar]
- 27.Urbach AR, Dervan PB. Proc Natl Acad Sci U S A. 2001;98:4343–4348. doi: 10.1073/pnas.081070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trauger JW, Dervan PB. Meth Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez D, Chou CJ, Latella L, Zeitlin SG, Ku S, Puri PL, Dervan PB, Gottesfeld JM. Cell Cycle. 2006;5:1537–1548. doi: 10.4161/cc.5.14.2913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Cellular uptake properties of α-DABA polyamide-fluorescein conjugates in 3 cell lines as determined by confocal microscopy. ++, Nuclear staining exceeds that of the medium; +, nuclear staining is less than or equal to that of the medium, but still prominent; −, very little nuclear staining, with the most fluorescence seen in the cytoplasm and/or medium; − −, no nuclear staining. Polyamides were assayed at 5 μM concentration.
Supplemental Figure 1. (A) Chemical and ball-and-stick structures for polyamides 8 and 9 and their Chl conjugates. (B) Illustration of the EcoRI/PvuII restriction fragments derived from plasmid pMFST3. The designed polyamide binding sites are indicated by boxes. Single base pair mismatches are indicated by shaded regions. (C) Quantitative DNase I footprinting titration experiments for polyamides (left) 8 and (right) 9 on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. (Below) Each footprinting gel is accompanied by its respective binding isotherms. (D) Thermal cleavage assay experiments with polyamide-chlorambucil conjugates (left) 8-Chl and (right) 9-Chl on the on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 14, intact DNA. Putative major sites of alkylation on the DNA fragments are indicated by arrows on the sequences adjacent to each gel. (E) Table of binding affinities (M−1) for parent polyamides 8 and 9 on plasmid pMFST3. Equilibrium association constants reported are mean values from at last three DNase I footprinting titration experiments. Standard deviations are shown in parentheses. Bolded numbers in brackets indicate fold decrease in match site binding affinity for α-DABA-linked versus γ-DABA-linked polyamides.
Supplemental Figure 2. (A) Chemical and ball-and-stick structures for polyamide 10 and its EDTA conjugate EDTA-10. (B) Illustration of the EcoRI/PvuII restriction fragment derived from plasmid pMFST4. The designed polyamide binding sites are indicated by boxes. (C) Quantitative DNase I footprinting titration experiments for polyamide 10 on the 283 base pair, 5′ end-labeled PCR product of plasmid pMFST4: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM, and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. (Below) Binding isotherms. Binding affinities for sites M and O are 9.8 (±0.5) × 108 M−1 and 1.1 (±0.6) × 109 M−1, respectively. (D) Affinity cleavage experiment with EDTA-10 on the 283 base pair, 5′ end-labeled PCR product of plasmid pMFST4: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (E) Summary of affinity cleavage patterns for EDTA-10. Bar heights are proportional to the relative cleavage intensities at each base pair. Ball-and-stick structures illustrating the proposed modes of binding are shown. Designed binding sites are indicated in bold in the DNA sequence.
Supplemental Figure 3. (A) Chemical and ball-and-stick structures for polyamide-EDTA conjugates EDTA-8 and EDTA-9. (B) Affinity cleavage experiments with polyamide-EDTA conjugates (left) EDTA-8 and (right) EDTA-9 on the 288 base pair, 5′ end-labeled PCR product of plasmid pMFST3: lane 1, A reaction; lane 2, G reaction; lanes 3–13, 1 μM, 300 nM, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, and 10 pM polyamide, respectively; lane 14, intact DNA. (C) Summary of affinity cleavage patterns for (top) EDTA-8 and (bottom) EDTA-9. Bar heights are proportional to the relative cleavage intensities at each base pair. The designed polyamide binding sites are indicated by boxes.
Supplemental Figure 4. Chemical and ball-and-stick structures for polyamide-fluorescein conjugates prepared for studying the cellular uptake properties of α-DABA polyamides.