Summary
PDZ protein interaction domains are typically selective for C-terminal ligands, however, non-C-terminal, “internal” ligands have also been identified. The PDZ domain from the cell polarity protein Par-6 binds C-terminal ligands and an internal sequence from the protein Pals1/Stardust. The structure of the Pals1-Par-6 PDZ complex reveals that the PDZ ligand-binding site is deformed to allow for internal binding. While binding of the Rho GTPase Cdc42 to a CRIB domain adjacent to the Par-6 PDZ regulates binding of C-terminal ligands, the conformational change that occurs upon binding of Pals1 renders its binding independent of Cdc42. These results suggest a mechanism by which the requirement for a C-terminus can be readily bypassed by PDZ ligands and reveal a complex set of cooperative and competitive interactions in Par-6 that are likely to be important for cell polarity regulation.
Protein interaction domains form the backbone of cellular information flow1. The PDZ protein interaction domain participates in a wide variety of signaling pathways and is one of the most common in metazoan genomes2. Because they often occur in multiple instances in the same polypeptide, PDZ domains are thought to serve an organizational role in signal transduction pathways3. Given the large number of PDZ domains, several modes of ligand recognition exist whose mechanisms are still being elucidated.
PDZ domains bind to short sequences of five to seven residues in their target proteins4,5. Although these recognition sequences have a low information content, specificity is typically enhanced by the requirement that the sequence occurs at the C-terminus. C-terminal recognition is found to fall into several different classes, depending on the identity of critical binding residues3. For example, class I PDZ ligands have a consensus sequence of -S-X-V-COOH.
The requirement for a C-terminus results from a steric rather than electrostatic mechanism. The peptide-binding pocket is constructed such that residues that extend past the C-terminus clash with a conserved PDZ segment known as the carboxylate-binding loop3,4. Although C-terminal ligands possess a negatively charged carboxylate, studies of salt effects on the binding reaction suggest that electrostatic contributions are negligible6. Because of the additional specificity provided by the C-terminus, enforcement of C-terminal binding is an important component of PDZ - ligand recognition.
While recognition of C-terminal motifs appears to be the dominant mode of PDZ-ligand interaction, non-C-terminal (also known as “internal”), PDZ ligands also exist. Compared to our understanding of C-terminal PDZ ligands, however, the mechanism of internal ligand recognition is much less clear. The best-characterized internal PDZ interactions involve ligands that adopt a specific conformation that adheres to the steric requirements of the PDZ binding pocket. For example, in the hetero-oligomerization of the nNOS and syntrophin PDZ domains, an extension of the nNOS PDZ domain, termed the β-finger, forms a sharp β-turn where the C-terminus would occur in a C-terminal PDZ ligand7. Likewise, an internal PDZ ligand identified in a phage display library contains a disulfide that presumably allows the chain to avoid the steric block as reduction of the disulfide abrogates binding8. The specialized sequences of these internal ligands suggest that bypassing the requirement for a C-terminus requires the ligand to adopt a specific, pre-formed conformation. For example, the nNOS PDZ domain is required for high-affinity binding to the syntrophin PDZ as the β-finger is stabilized through interactions with the domain9.
Several questions about recognition of internal sequences by PDZ domains remain. First, can PDZ domains bind to internal sequences that don’t have specialized structures such as the nNOS β-finger? PDZ’s have been shown to bind internal sequences that don’t themselves contain PDZ’s10 indicating that PDZ oligomerization is not the only mechanism for internal binding. Are these alternative internal ligands as dependent on conformation as the β-finger of nNOS-syntrophin? Answering these questions will allow us to better understand the diversity of ligand interactions in this important protein interaction domain family.
The cell polarity protein Par-6 is an excellent model system for studying internal PDZ recognition. Par-6 contains a single PDZ domain that has been shown to bind to both internal and C-terminal ligands. Par-6 binds to the C-terminus of the transmembrane receptor Crumbs (Crb)11 and a similar C-terminal sequence identified in a peptide library screen12. Par-6 binding to C-terminal ligands is regulated by binding of the Rho GTPase Cdc42 to a CRIB domain adjacent to the Par-6 PDZ. An allosteric transition in the CRIB-PDZ of Par-6 induced by Cdc42 binding leads to conversion from a low affinity to a high affinity PDZ conformation12. In addition, Par-6 has recently been shown to bind to Partner of Lin-7 (Pals1) and its Drosophila homologue Stardust (Sdt) through an internal interaction that is important for formation of tight junctions in epithelial cells10.
In order to further understand PDZ internal recognition, we have examined the interaction of the Par-6 PDZ with Pals1. Binding studies reveal that a remarkably small sequence from Pals1 is sufficient for binding to the Par-6 PDZ domain, which is inconsistent with the internal binding mode in nNOS-Syntrophin. In addition, Cdc42 binding to Par-6 has no effect on its affinity for Pals1, in contrast to C-terminal ligands. Comparison of the Par-6 - Pals1 internal complex with a C-terminal complex reveals that binding of Pals1 deforms the PDZ binding pocket, thereby bypassing the enforcement of C-terminal binding, and presumably decoupling binding to Cdc42. These results reveal a conformational plasticity of PDZ domains that can be exploited for both recognition and regulation.
Results
A small, internal Par-6 PDZ ligand in Pals1/Sdt
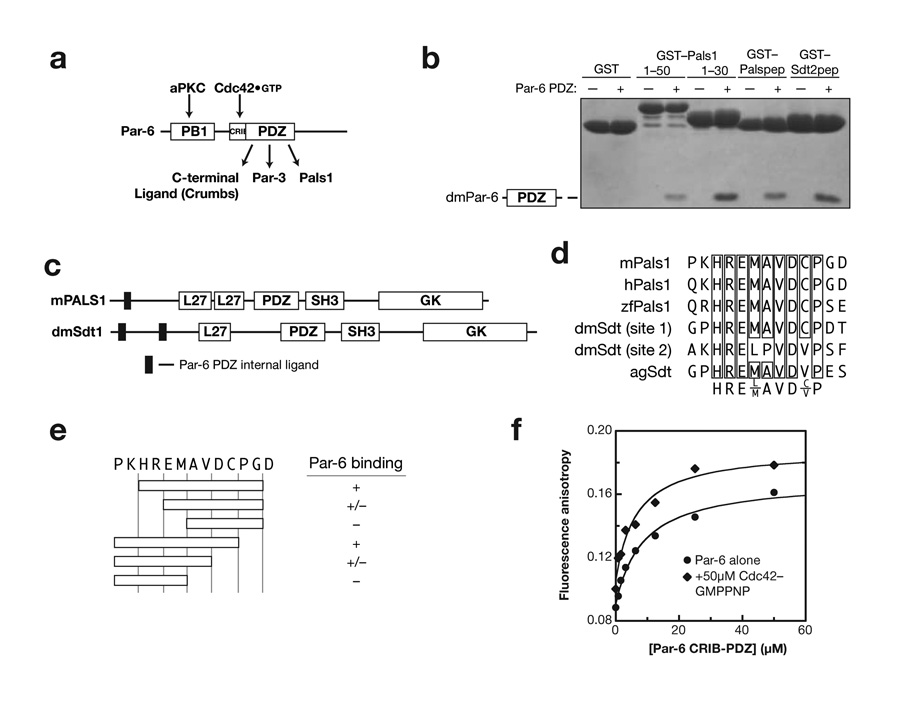
Par-6 has been shown to bind Pals1 through an interaction with the Par-6 PDZ domain (Fig. 1a)10. In order to examine the mechanism of this interaction, we identified the minimal components of Pals1, and its Drosophila homologue Stardust (Sdt) necessary for binding. As shown in Figure 1, Pals1 and Sdt have domain structures typical of other members of the MAGUK (membrane associated guanylate kinase) protein family with PDZ and SH3-GK protein modules13. The region of Pals1 that binds to Par-6 has previously been found to be in a region N-terminal to these domains10. Further deletion analysis of the Pals1 N-terminus reveals that a small, eight-residue motif is sufficient for binding to the Par-6 PDZ domain (Fig. 1b). Two similar motifs are present in Sdt, both of which also bind to the Par-6 PDZ (Fig. 1b,c). The non-redundant sequence database contains several sequences related to the eight-residue Pals1 sequence from several metazoan organisms. Alignment of these sequences reveals a core motif in the Pals1/Sdt Par-6 ligand sequence is highly conserved (Fig. 1d). As shown in Figure 1e, this core motif is necessary and sufficient for binding to Par-6.
Figure 1.
Identification of Par-6 PDZ binding motifs in Pals1 and Stardust. (a) Schematic of Par-6 domain structure and domain ligands (PB1: Phox and Bem1; CRIB: Cdc42/Rac Interactive Binding; PDZ: PSD-95, Discs Large, ZO-1). (b) Deletion analysis of Pals1 and Sdt reveals a short, internal Par-6 PDZ ligand. GST fusions of various murine Pals1 or Drosophila Stardust fragments were tested for their ability to bind Par-6 using glutathione agarose affinity chromatography (Palspep: Pals1 residues 29 to 40; Sdt2pep: Sdt residues 375 to 388). (c) Domain structure of Pals1 and Sdt showing the relationship of the Par-6 PDZ internal ligands to the MAGUK protein domains (L27: Lin-2, Lin-7; SH3: Src Homology 3; GK: Guanylate Kinase Homology). (d) Alignment of Pals1/Sdt Par-6 ligand sequences showing conserved core sequence. (e) Deletion analysis of Pals1/Sdt Par-6 ligand sequences. The conserved core sequence is required for binding to the Par-6 PDZ domain. (f) The Rho GTPase Cdc42 does not regulate Pals1 binding to the Par-6 PDZ domain. A rhodamine labeled peptide with the Pals1 sequence was used to measure binding to the Par-6 CRIB-PDZ fragment. The higher anisotropy of the Cdc42 complex at saturation occurs because of its higher molecular weight. The binding curves are for Kd’s of 6 and 8 μM for Cdc42-bound and free Par-6, respectively.
The small size of the Pals1/Sdt internal ligand indicates that its binding mode differs from that of the nNOS - syntrophin internal interaction. Because the nNOS β-finger is structurally coupled to the adjacent nNOS PDZ domain, over 100 residues from nNOS are required for binding to the syntrophin PDZ9. The eight residues sufficient for the Pals1 - Par-6 interaction are more similar to canonical C-terminal interactions that require approximately six residues5. However, the region of Pals1 that binds to Par-6 does not occur at its C-terminus, and therefore must utilize a different mechanism of recognition.
We also find that binding of the Pals1/Sdt internal ligand is independent of the interaction of Par-6 with the Rho GTPase Cdc42. Cdc42 binds to a CRIB (Cdc42/Rac Interactive Binding) motif adjacent to the Par-6 PDZ domain and increases the affinity of the PDZ domain for C-terminal ligands ∼13-fold12. Previous qualitative data with a large fragment of Pals1 indicated that Cdc42 does not affect the Par-6 - Pals1 interaction12. To test the possible coupling of Cdc42 and Pals1 binding in a quantitative fashion, we synthesized the Pals1 binding sequence with an NH2-terminal rhodamine and measured binding using fluorescence anisotropy. As shown in Figure 1e, Cdc42 binding to Par-6 has no effect on the affinity of Par-6 for Pals1.
The small size of the Pals1 ligand, which is inconsistent with the formation of a specialized structure, and the lack of Cdc42 regulation of Pals1 binding leads to several questions about the mechanism of Par-6 recognition of this ligand. In particular, how does the short Pals1 sequence satisfy the steric restraints of the PDZ binding pocket? Also, given that Pals1 and C-terminal ligands compete for binding to Par-612, how is it that the Par-6 - Pals1 interaction is independent of Cdc42 binding?
Crystal Structure of the Par-6 PDZ-Sdt/Pals1 Complex
In order to address the mechanism of internal recognition by Par-6 - Pals1/Sdt, we determined the structure of the Par-6 PDZ in complex with the minimal core motif using X-ray crystallography. The Drosophila Par-6 PDZ domain (residues 156-255) was co-crystallized with the Sdt/Pals1 derived peptide Ac-YPKHREMAVDCP-CONH2 (representing amino acids 29 to 40 from Pals1) and formation of crystals was dependent on the presence of peptide. Crystals grew in the space group R3 with one complex per asymmetric unit and diffracted to 2.5Å (diffraction was somewhat anisotropic - see methods). Phases were determined by molecular replacement using the PDZ domain from the Par-6 PDZ-C-terminal peptide complex (PDB code 1RZX) as a search model. The Pals1 peptide was clearly visible in the initial electron density maps. The refined model contains residues 156-255 from Par-6, residues 30-40 of Pals1, and 5 waters and has an R-factor of 21% and an Rfree of 25%. Simulated annealing omit maps for both the bound peptide and carboxylate binding loop are shown in Supplementary Figure 1.
In the structure of the Par6-Pals1 complex, the Pals1 ligand occupies the same binding site as C-terminal ligands. C-terminal ligands bind in a groove that is formed between strand 2 and helix 2 of the PDZ domain3,4. The H-R-E-M-A-V portion of Pals1 lies in this same groove corresponding to the equivalent residues of the C-terminal ligand (V-K-E-S-L-V-COOH). This is similar to the nNOS-syntrophin internal PDZ interaction in which a sequence in the nNOS beta-finger mimics C-terminal ligands (this sequence is known as the “pseudopeptide”) and occupies the same binding site as C-terminal ligands (Hillier et al., 1999). The most critical residues for Par-6 PDZ C-terminal ligands are the P0 valine (the C-terminal residue), the P-1 leucine, and the P-3 aspartic acid12. Although the P0 valine is conserved in Pals1, the P-1 leucine is replaced by an alanine. Previous alanine scanning studies have shown that an alanine at this position abrogates binding in the context of the C-terminal ligand12 indicating that, although the general features of pseudopeptide recognition in the Sdt/Pals1 internal ligand are similar, there are notable differences.
Internal Recognition Through PDZ Domain Plasticity
Binding of Pals1 induces a conformational change in the carboxylate-binding loop of the Par-6 PDZ domain. In comparison to the structure of the Par-6 PDZ in complex with a C-terminal ligand, the overall fold of the PDZ domain is the same (Fig. 2a). However, two loops that connect the β1-β2 strands and connecting the β2-β3 strands contain significant deviations between the two structures, with distances between backbone atoms of over 3 Å, and up to 7 Å in the carboxylate-binding loop. The altered conformation of the carboxylate-binding loop occurs between residues 163 and 171 (note that these residues make no significant crystal contacts). Residue 171 is the first residue of the “GLGF” loop, a set of conserved residues in PDZ domains that make up the base of the carboxylate-binding loop. However, in Par-6 this residue is a proline rather than a glycine. Mutation of this proline in Par-6 to the canonical glycine leads to disruption of regulated C-terminal binding but does not alter internal binding12. The peptide backbone of this loops forms contacts with the carboxylate from C-terminal ligands and therefore plays a central role in enforcing C-terminal binding3,4. The other loop that differs between the two complexes is a large loop connecting strands 2 and 3. As this loop makes extensive inter-domain contacts due to crystal packing and does not significantly contact the PDZ ligand, it is likely that the difference in this loop is not important for internal recognition.
Figure 2.
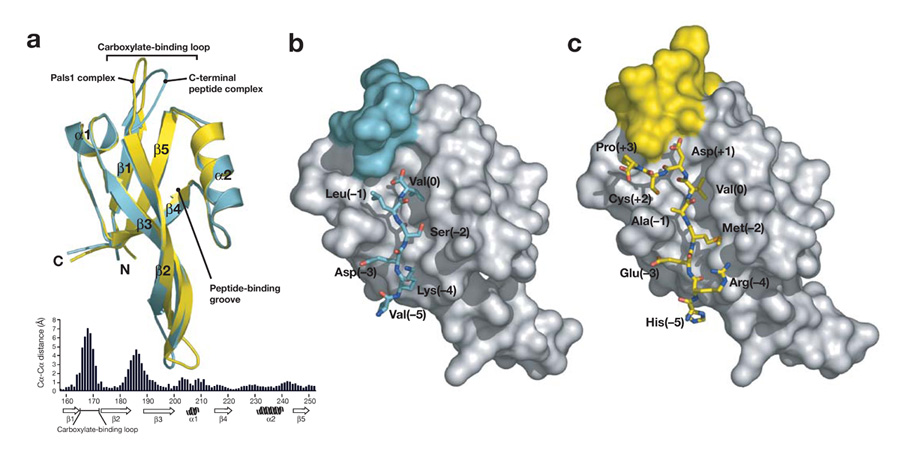
Comparison of Par-6 PDZ internal and C-terminal binding. (a) Comparison of Par-6 PDZ domains from the Pals1 complex and the C-terminal peptide complex (PDB code: 1RZX). The Pals1 complex is shown in yellow and the C-terminal complex in cyan with the secondary structure elements labeled. The ligands have been removed for clarity. The distance between equivalent Cα atoms in the C-terminal peptide and Pals1 bound Par-6 PDZ domains is plotted below the structures. (b). Par-6 PDZ in complex with a carboxy-terminal peptide. Binding of C-terminal ligands is enforced by the carboxylate-binding loop (colored cyan on the PDZ surface) through a steric mechanism in which residues that would extend past the P0 residue would clash with the loop. (c) Par-6 PDZ in complex with an internal peptide from Pals1. When bound to the Pals1 peptide, the carboxylate-binding loop (colored yellow on the PDZ surface) is deformed, allowing it to extend past the P0 residue.
The conformational change in the Par-6 PDZ carboxylate-binding loop allows the Pals1 ligand to bypass the requirement for a C-terminus. As shown in Figure 2b, the carboxylate-binding loop caps the PDZ binding pocket, preventing binding of ligands that continue past the P0 residue. This loop has a similar conformation in free PDZ domain structures4. The conformational change that occurs in the carboxylate-binding loop significantly alters this region of the binding pocket, however (Fig. 2b,c; Fig. 3). A lysine from the carboxylate binding loop (lysine 165) is positioned closer to the aspartic acid side chain (Fig. 3) and the interaction between these two residues is critical for Pals1 binding (see below). Additionally, hydrogen bonding interactions that take place between the carboxylate and the PDZ backbone (Fig. 3a), are now replaced with backbone-backbone interactions in the Par-6 Pals1 complex (Fig. 3b).
Figure 3.
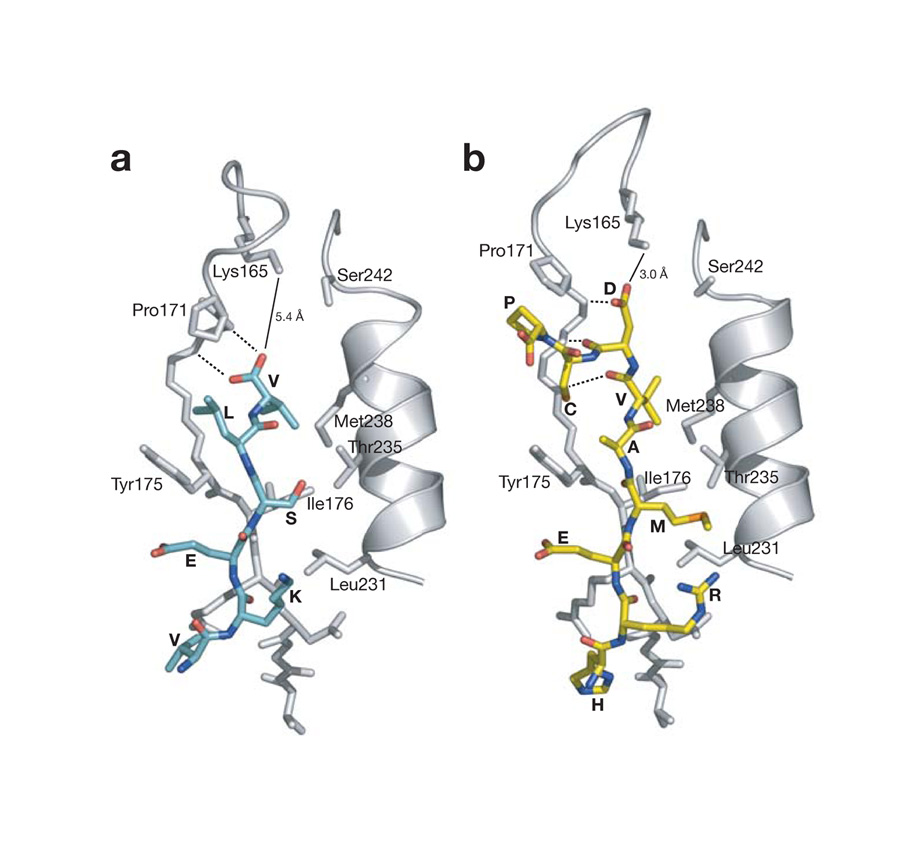
Critical interactions in Pals1 internal PDZ binding. (a) Par-6 PDZ - C-terminal ligand interactions. The peptide-binding pocket from the C-terminal peptide - Par-6 PDZ complex (PDB code: 1RZX) is shown. Peptide residues are labeled by their amino acid and PDZ domain residues are labeled by amino acid and sequence number. The distance between the C-terminus and lysine 165 is shown (solid line) along with interactions between the carboxylate and the PDZ backbone (dashed lines). (b) Par-6 PDZ - Pals1 internal ligand interactions. The interactions between the PDZ domain and peptide are shown as in (a).
The rearrangement of the Par-6 PDZ carboxylate-binding loop to allow for Pals1 binding differs from nNOS-syntrophin. In the nNOS-syntrophin interaction, the structure of the syntrophin PDZ domain is not altered when nNOS binds7. Instead, the ligand adapts to the requirements enforced by the syntrophin PDZ carboxylate-binding loop by forming a sharp turn that avoids the steric constraints imposed by the loop.
The ability of Pals1 to bind internally does not result solely from the residues that follow the P0 valine. A peptide containing the sequence of the C-terminal ligand, followed by the residues from Pals1 that follow the P0 valine (D-C-P) does not bind to Par-6 (data not shown) indicating that there is an interplay between residues before and after the P0 valine. This interplay arises because of differences in the way the P-1 binding pocket is utilized in the two types of ligands (see below).
Determinants of Internal and Cdc42 Regulated Binding
A critical question in understanding Par-6 PDZ internal vs. C-terminal recognition is why the internal ligand can deform the carboxylate-binding loop whereas the C-terminal ligand cannot. In other words, why is a C-terminus required in one class of ligands, but not for internal ligands? To answer this question, we generated a series of substitutions in the Pals1 sequence and tested their ability to bind to the Par-6 PDZ domain.
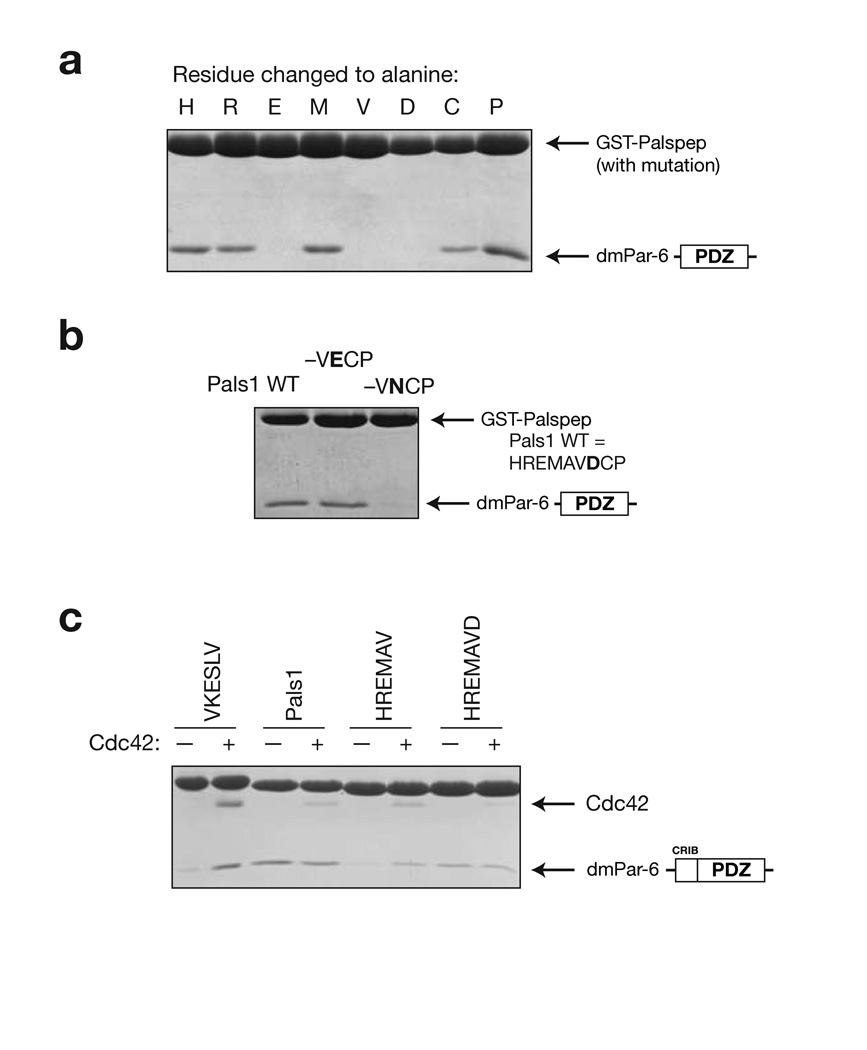
Alanine scanning of the Pals1 internal ligand reveals a different pattern of required residues from Par-6 PDZ C-terminal ligands. We had previously identified the C-terminal ligand -VKESLV-COOH as a Par-6 PDZ ligand12, and the C-terminus of the transmembrane protein Crumbs (Crb) with the sequence -EERLI-COOH has also been shown to bind11. From this data, the pattern of required residues for Par-6 PDZ C-terminal ligands is X-E-X-L-V/I - COOH (where X indicates that the residue can be replaced by alanine without significantly altering binding). We generated alanine substitutions of the Pals1/Sdt ligand and tested their ability to bind the Par-6 PDZ domain in a qualitative pull-down assay (Fig. 4a). These results indicate that the pattern of required residues in the Pals1 ligand is X-E-X-A-V-D-X-. While C-terminal ligands require a large hydrophobic residue at the P-1 position for binding, the Pals1 internal ligand has an alanine at this position. This is because the residues following the P0 valine (particularly the P+2 cysteine) occupy the binding site that P-1 leucine occupies in C-terminal ligands.
Figure 4.
Analysis of Par-6 PDZ ligand binding determinants. (a) Alanine scanning of Pals1 ligand. GST-fusions of peptide sequences with an alanine substituted at each position along the Pals1 sequence were tested for their ability to bind the Par-6 PDZ domain. (b) Evaluation of the importance of the P+1 residue in Pals1. In the structure of the Par-6 Pals1 complex, the P+1 aspartic acid forms a salt bridge with a conserved lysine in the PDZ domain. GST fusions of mutants in this position were tested for their ability to bind the Par-6 PDZ domain. Mutation to an asparagine abrogates binding indicating that the salt bridge is critical for binding. (c) Identifying the determinants of Cdc42 regulation. As has been previously shown, Cdc42 binding to the CRIB-PDZ fragment of Par-6 alters its affinity for carboxy terminal ligands. GST-fusions of several Pals1 truncations were made to identify the determinants of Cdc42 regulation. When the P+1 residue is removed (HREMAV sequence), Cdc42 binding causes an increase in peptide binding. The presence of this residue (HREMAVD sequence) causes Cdc42 to no longer affect peptide binding, presumably because the carboxylate-binding loop is deformed.
In the structure of the Par-6 PDZ - Pals1 complex, the P+1 aspartic acid appears to form a salt bridge with a conserved lysine in the carboxylate-binding loop. This lysine forms interactions with the C-terminal carboxylate through a series of water molecules in C-terminal ligands4. As the alanine scanning showed that this aspartic acid is crucial for binding, we made further mutations in this residue to test the requirement for a carboxylate at this position. As shown in Figure 4b, removal of the carboxylic acid by mutation of the aspartic acid to asparagine completely disrupts binding. Taken with the alanine scanning results that show that residues following this residue are not critical, we conclude that the P+1 aspartic acid is a key determinant for internal binding. The ion pairing interaction with a conserved PDZ lysine side chain may be important for stabilizing the deformed conformation of the carboxylate-binding loop.
A key difference between binding of C-terminal and internal ligands is that C-terminal ligands are regulated by Cdc42 whereas internal ligands are not. To investigate the sequence determinants of the two ligands that lead to this difference, we made a series of deletions in the Pals1 ligand and tested their ability to bind Par-6 in a Cdc42 regulated manner. As shown in Figure 4c, a ligand that is truncated to the P0 valine binds with higher affinity in the presence of Cdc42. However, a ligand that extends one residue beyond this position to the P+1 aspartic acid binds to the Par-6 PDZ domain in a Cdc42 independent manner. This suggests that Cdc42 regulated binding is dependent on the normal conformation of the carboxylate binding loop and that the deformation of this loop by the Pals1 ligand decouples the two binding sites.
Discussion
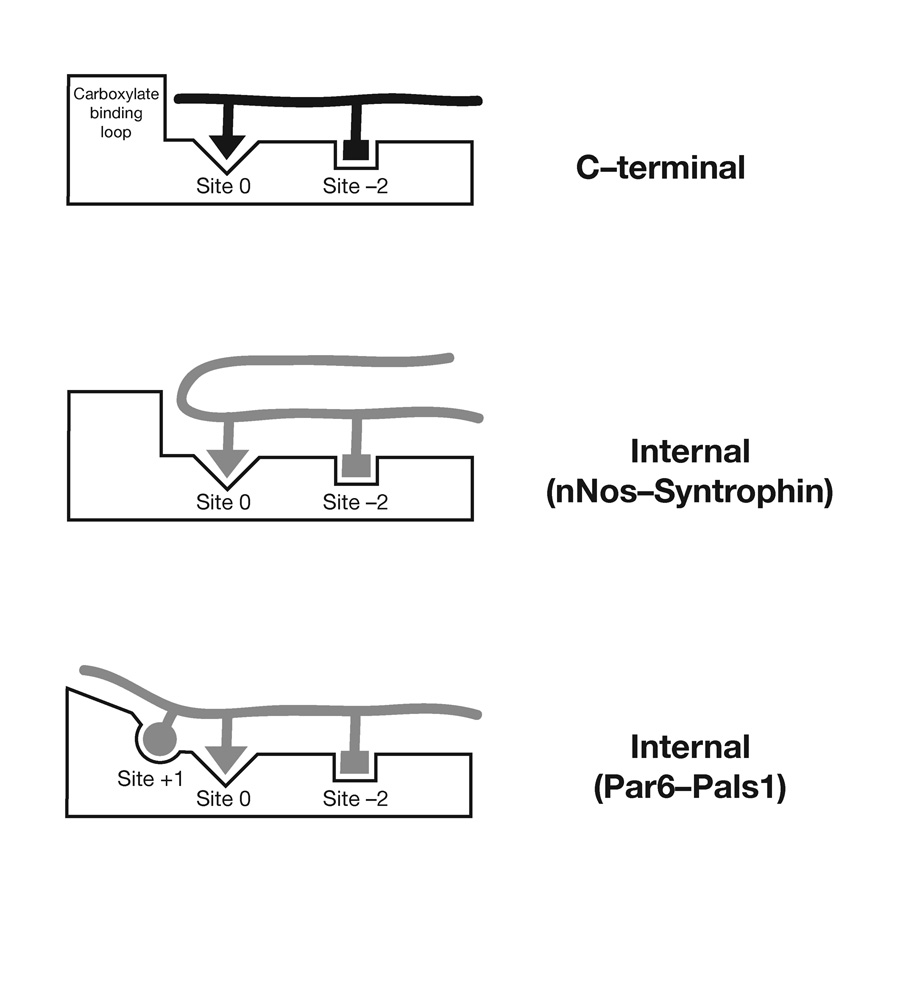
PDZ domains are typically selective for C-terminal ligands, a restraint imposed by the conformational properties of their binding pocket (Fig. 5). Internal interactions that bypass the requirement for a C-terminus have been thought to be restricted to ligands that form specialized conformations that adapt to the properties of the PDZ binding pocket3,7 (Fig. 5). We have demonstrated that the interaction of Par-6 with Pals1/Sdt utilizes an internal PDZ - peptide interaction that deforms the PDZ carboxylate-binding loop (Fig. 5). This mechanism of internal binding indicates that internal PDZ ligands do not require the specific conformational constraints of the nNOS β-finger, suggesting that internal ligands may be more common than previously thought3. Comparison of PDZ sequences to the Par-6 PDZ domain does not reveal any obvious differences from other PDZ domains that would suggest that it is unique in its ability to bind Pals1-type internal ligands. Further work will be required to establish whether the mode of internal binding described here is a property specific to the Par-6 PDZ domain, or if a large subset of PDZ domains can utilize this mechanism.
Figure 5.
Modes of PDZ C-terminal and Internal Recognition. In PDZ C-terminal ligand recognition the carboxylate-binding loop enforces C-terminal binding by preventing extension past the P0 residue. In the β-finger internal PDZ recognition mode of recognition, used by nNOS-syntrophin7 and presumably disulfide containing ligands8, a sharp turn in the ligand allows it to bypass the steric requirement imposed by the carboxylate-binding loop. The Pals1 - Par-6 PDZ interaction represents a new type of internal interaction in which the carboxylate-binding loop is deformed to allow for residues past the P0 residue. An interaction with the P+1 residue is critical for this mode of recognition.
In the first structure of a PDZ domain with a C-terminal ligand4, the authors noted some plasticity in the carboxylate-binding loop upon ligand binding. The mode of internal sequence recognition described here may be an extension of this mechanism. However, it is clear that, at least in the case of the Par-6 - Pals1 interaction, specific interactions beyond the P0 residue are required in order to take advantage of carboxylate-binding loop plasticity. It is likely that the deformation of the carboxylate-binding loop is energetically costly, and additional interactions are required to compensate for this.
The plasticity of the Par-6 PDZ domain may be important for regulation of the large number of Par-6 binding partners, which includes Cdc42, Par-3, Pals1/Sdt, Crumbs and Lgl. We have shown that these ligands represent a complex set of cooperative and competitive binding partners. For example, binding of the C-terminal ligand Crumbs and the internal Pals1/Sdt are competitive with each other12, while the Pals1/Sdt PDZ domain itself has been shown to bind to Crumbs14. All three of these proteins are present in epithelial cells10,15, so understanding the partitioning of possible complexes, and the role of these complexes in establishing and maintaining cell polarity will require further in vitro and in vivo analysis.
We have shown previously C-terminal ligand binding to the Par-6 PDZ is regulated by Cdc4212. Here we show that deformation of the Par-6 PDZ by Pals1 not only allows for internal recognition, but also renders Pals1 binding independent of Cdc42. These results suggest that coupling between Cdc42 and C-terminal ligand binding occurs predominantly through the carboxylate-binding loop, although other structural changes occur upon Cdc42 binding12. As Cdc42 binds to a region directly adjacent to the carboxylate-binding loop, the loop is well positioned to transduce Cdc42 binding to the peptide-binding pocket.
In summary, we have identified an internal ligand for the Par-6 PDZ domain and shown that it utilizes a distinct mode of recognition from previously studied internal interactions. Rather than the adapting to the specific conformation of the PDZ binding pocket, the Par-6 PDZ domain is deformed to bypass the requirement for chain termination. We have also shown that the two different PDZ conformations have different regulatory properties, with only C-terminal ligands being regulated by Cdc42. These results provide a framework for understanding diverse PDZ-ligand interactions.
Methods
Protein Expression and Purification
We purified polyhistidine fusions using Ni-NTA resin followed by incubation with TEV protease to remove the histidine tag. Tag cleavage was followed by further purification using ion exchange chromatography to achieve a final purity of > 99% as measured by SDS-PAGE and/or MALDI-TOF mass spectrometry.
In Vitro Binding Assays
For GST pull-down assays, we adsorbed the GST fusion onto glutathione agarose beads and washed with binding buffer (10 mM HEPES, 100 mM NaCl, 2 mM DTT, 0.05% (v/v) Triton, pH 7.5). We then added ligands at the indicated concentration and incubated at room temperature for 15 minutes. We removed unbound protein by washing three times with a large excess of binding buffer. After elution of GST fusions and bound proteins by addition of SDS loading buffer, we visualized the results of the binding assays by SDS-PAGE.
Rhodamine labeled Pals1 peptide for fluorescence anisotropy was prepared using FMOC solid phase synthesis. We coupled the rhodamine (rhodamine B, Sigma) to the peptide NH2-terminal amine as the last step of the synthesis before the peptide was cleaved from the resin. We purified the peptide by reverse phase HPLC and verified its identity by mass spectrometry. Fluorescence anisotropy binding assays were performed on an ISS PC1 fluorimeter and the data was fit to an equation describing a bimolecular binding reaction with peptide concentration below the Kd of the interaction.
Crystallization and Structure Determination
The Par-6 PDZ domain (from Drosophila, residues 156-255) was crystallized by hanging drop vapor diffusion at 15°C using 26% (w/v) PEG6000, 100 mM HEPES pH 7.1 as a mother liquor. We mixed the Par-6 PDZ domain, concentrated to 10 mg/ml, with a 3-fold molar excess of peptide representing Pals1 29-40 (NH2-terminal acetylated and with a COOH-terminal amide). Crystals grew in space group R3 with cell constants of a=b=63.1Å, c=99.3Å and one PDZ - Pals1 peptide complex per asymmetric unit.
Crystals were cooled in liquid nitrogen using mother liquor + 10% (v/v) glycerol as a cryoprotectant. Diffraction data was collected at beamline 8.2.2 at the Advanced Light Source using an ADSC Q315 CCD detector. Crystals diffracted somewhat anisotropically, ranging between 2.5 and 2.7 Å. Data reduction was carried out using the HKL2000 software package16. Crystallographic data statistics are shown in Table I.
Table 1.
X-Ray Diffraction Data Collection and Refinement Statistics
| Space Group | R3 |
| Cell dimensions (Å) | a = b = 63.1, c = 99.3 |
| Resolution range (Å) | 30 - 2.5 |
| Completeness (%) | 95.7 (74.4)1 |
| R linear (%) | 4.2 (11.3) |
| I/σ | 56 (8) |
| Unique reflections | 4870 (378) |
| Rcryst | 0.216 |
| Rfree | 0.254 |
| Ramachandran Statistics (non-glycine) | 88% core 12% allowed 0% disallowed |
| RMS deviation from ideal values | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.2 |
| Solvent Molecules | 5 |
| Average B-factor | 85 (after TLS refinement), 72 (before TLS) |
Highest resolution shell (2.50 - 2.59 Å)
The coordinates of the Par-6 PDZ domain from the Par-6 PDZ - C-terminal peptide complex (PDB code 1RZX) were used as a search model for molecular replacement using the program AMORE17. A single solution was found that provided initial phases that were further improved with rigid body optimization using CNS18. Initial electron density maps showed clear density in the peptide-binding pocket. The initial model was refined with alternate cycles of positional and restrained B factor refinement followed by manual rebuilding using composite simulated annealing omit maps in O (Jones). To further improve the model, additional TLS refinement using refmac19 was used utilizing the same set of test reflections as in CNS. Two TLS groups were defined that included the PDZ domain and the Pals1 ligand respectively. TLS and restrained refinement resulted in a final model with an R of 0.216 and Rfree of 0.254 and excellent geometry (Table I). Additional information about the model, including simulated annealing electron density maps, B-factor distributions and real space correlation coefficients can be found in supplementary figures 1-4.
Supplementary Material
Acknowledgments
We thank A. Berglund, T. Stevens, B. Volkman, and members of the Prehoda Lab for helpful comments and suggestions. We thank the support staff at beamline 8.2.1 at the Advanced Light Source for technical assistance. This work was supported by grants from the American Heart Association, National Institutes of Health (GM068032), and a Damon Runyon Scholar Award to K.E.P.
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.van Ham M, Hendriks W. PDZ domains-glue and guide. Mol. Biol. Rep. 2003;30:69–82. doi: 10.1023/a:1023941703493. [DOI] [PubMed] [Google Scholar]
- 3.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell. Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 4.Doyle DA, et al. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 5.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 6.Harris BZ, Lau FW, Fujii N, Guy RK, Lim WA. Role of electrostatic interactions in PDZ domain ligand recognition. Biochemistry. 2003;42:2797–27805. doi: 10.1021/bi027061p. [DOI] [PubMed] [Google Scholar]
- 7.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 8.Gee SH, et al. Cyclic peptides as non-carboxyl-terminal ligands of syntrophin PDZ domains. J Biol. Chem. 1998;273:21980–21987. doi: 10.1074/jbc.273.34.21980. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson KS, Hillier BJ, Lim WA, Bredt DS. PSD-95 assembles a ternary complex with the N-methyl-D-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J Biol. Chem. 1999;274:27467–27473. doi: 10.1074/jbc.274.39.27467. [DOI] [PubMed] [Google Scholar]
- 10.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 11.Lemmers C, et al. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol. Biol. Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 13.McGee AW, et al. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol. Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]
- 14.Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1) Gene. 2003;302:21–29. doi: 10.1016/s0378111902010843. [DOI] [PubMed] [Google Scholar]
- 15.Roh MH, et al. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 2002;157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Navaza J. Implementation of molecular replacement in AMoRe. Acta. Crystallogr. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta. Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.