Abstract
Modes of signaling in fibroblasts can differ substantially depending on whether these cells are in their natural three-dimensional environment compared to artificial two-dimensional culture conditions. Although studying cell behavior in two-dimensional environments has been valuable for understanding biological processes, questions can be raised about their in vivo physiological relevance. This review focuses on some of our research involving fibroblast behavior in cell-derived three-dimensional matrices. Specifically, we examine how these matrices affect cell morphology, adhesion, proliferation, and signaling compared to two-dimensional substrates. We stress the importance of controls for three-dimensional matrix studies and discuss cancer as an example in which altered three-dimensional matrices can influence fibroblast signaling. Studying cells in three-dimensional microenvironments can lead to the design of more physiologically relevant conditions for assaying drug responses and deciphering biological mechanisms.
Keywords: fibroblast, fibronectin, three-dimensional matrix, integrin signaling, Rho GTPases
1. Introduction
1.1 Functions of the extracellular matrix
The extracellular matrix (ECM) is composed of a three-dimensional (3D) scaffold of collagen, fibronectin (FN), and other proteins, interlaced with proteoglycans. The ECM promotes signaling and provides structural support to the cells and tissues of the body. It also serves as a reservoir for growth factors, cytokines, enzymes, and other diffusible molecules. The ECM can be deposited by both epithelial and mesenchymal cells to create an environment best suited for their function. Fibroblasts and other mesenchymal cells, however, are the most important players in ECM remodeling. Studying these cells in model 3D matrices allows us to better understand signaling responses as they would normally occur in vivo. This approach can lead to the generation of more accurate cell-based assays for probing drug function or for engineering of suitable biomaterials to influence cell proliferation, cell motility, and cell fate [1, 2].
1.2 Integrins and the extracellular matrix protein fibronectin
Fibroblasts remodel the ECM primarily through integrin receptors and coreceptors. The integrins comprise a family of 8 β and 18 α subunits that form 24 distinct, noncovalently bound, heterodimeric receptors. Most integrins can bind to more than one ligand, but each receptor appears to have distinct biological roles as assessed by knockout mouse studies [3]. Since integrins lack intrinsic enzymatic activity, they transmit information about the changing extracellular environment via numerous intracellular signaling and structural proteins. Early integrin-mediated adhesion is thought to lead to the activation of focal adhesion kinase (FAK) and other tyrosine kinases. These signals are further propagated via various pathways, including activation of the Rho GTPases: Rac, Cdc42, and Rho [4]. These GTPases in conjunction with many other proteins play major roles in regulating cell behavior in response to the changing extracellular matrix milieu.
FN is a major matrix component often associated with biological processes involving ECM remodeling. Some of these processes include tissue repair and regeneration, stromatogenesis, and fibrosis. Integrins such as α5β1 and αvβ3 are known to interact with FN via the arginine-glycine-aspartic acid (RGD) binding domain [3]. Both of these integrins require the RGD binding sequence for maximal activity, but α5β1, which is often the primary receptor for FN, also requires the nearby synergy site [5-7]. By controlling the ability of cells to interact with this synergy site, cells can switch which of these integrins is preferentially used for migration in a 3D matrix [8]. Additionally, the binding of β1 or β3 integrins to a ligand can mediate transdominant inhibition of the other integrin [9-12].
1.3 Cell-derived 3D matrix
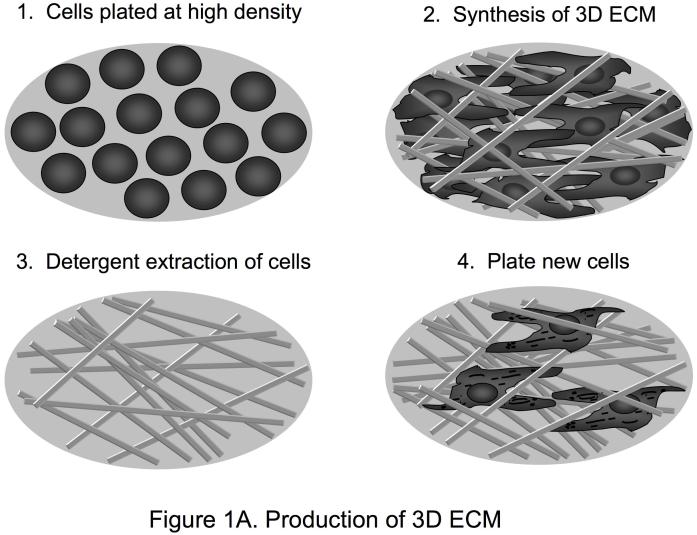
Natural 3D matrices can have varying degrees of both compositional and structural heterogeneity, depending on the cells and environment in which they are formed. Artificial or synthetic cellular environments are now being used to mimic and model 3D matrix in vivo situations. Although 3D collagen gels have provided many valuable insights into the roles of three-dimensionality and extracellular matrix tension in cell function [13], 3D microenvironments in living animals obviously contain many other ECM proteins besides collagen I, as well as growth factors. A newer, alternative approach beyond the use of 3D collagen gels involves using natural, cell-derived 3D extracellular matrices [14, 15]. 3D cell cultures that generate 3D matrices, e.g. fibroblasts generating matrices 15 μm thick, or cryostat sections of tissues can be extracted with detergent at alkaline pH to remove cellular contents. This treatment leaves behind a 3D ECM that is intact and cell-free (Figure 1A). These 3D cell-derived matrices are comprised of fibronectin with varying levels of collagen, heparan sulfate proteoglycan and laminin. These matrices are highly fibrillar and contain spaces originally occupied by the cells that generated the matrix. They are also pliable as time-lapse movies show that migrating cells can readily deform the 3D ECM [16].
Figure 1.
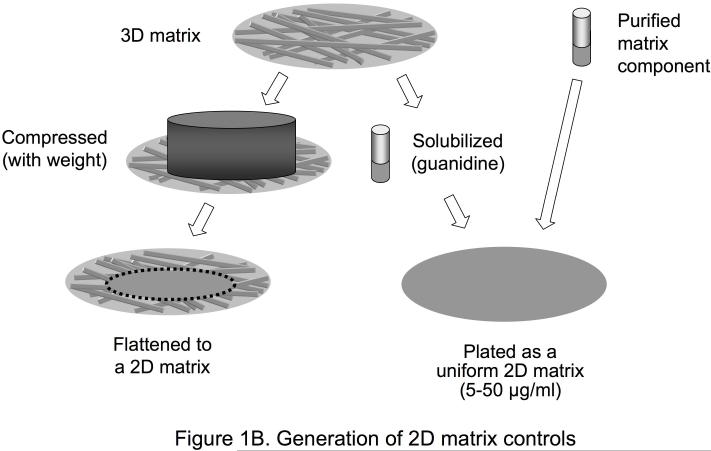
Production of 3D ECM and 2D matrix controls. A. This illustration describes the method for generating tissue culture surfaces coated with cell-derived three-dimensional matrices. This method can be adapted to many cell types. Using NIH 3T3 fibroblasts, matrices 7-15 μm thick composed mainly of fibronectin fibrillar lattices can be obtained. First, fibroblasts are plated and maintained in culture in a confluent state. Five to nine days later, the matrices are denuded of cells using detergent, and cellular debris is removed. The 3D matrices can be washed and stored for periods of up to two to three weeks at 4°C before plating cells on them to examine cellular responses. B. Various methods can be used to obtain appropriate controls for use in 3D ECM studies. To obtain 2D controls with the same molecular composition as the 3D matrix, the 3D matrix can be solubilized in guanidine and plated as a 2D matrix, or the 3D matrix can be mechanically compressed using a known weight applied to a given area. Alternatively, a purified matrix component such as fibronectin can be solubilized and plated as a 2D matrix.
Since natural 3D matrices are generally less rigid, or more pliable, than 2D matrices [17], cells within a 3D matrix can have differences in signaling responses due to modulation of both integrin activity and downstream effectors such as Rho GTPases and myosin II [18]. It is therefore important to take into account differences in substrate rigidity when generating and using artificial or synthetic 3D cellular microenvironments. For example, culturing NIH 3T3 fibroblasts and rat kidney cells on 3D polyamide nanofibers compared to two-dimensional culture surfaces induces activation of Rac [19], whereas human primary fibroblasts have decreased Rac activation when plated on a more pliable cell-derived 3D matrix compared to a 2D substrate coated with FN [16]. These signaling differences could result from differences in substrate properties and/or different cell types, but they demonstrate that placing cells in different 3D microenvironments can lead to differences in signaling responses.
1.4 Controls for 3D ECM studies
As reviewed below, comparisons of 2D tissue culture and 3D ECM conditions identify a number of morphological, signaling, and behavioral differences in cellular responses. Appropriate controls for these comparisons are important, since it is difficult to compare 2D with 3D systems. Controls can include compressing the 3D ECM to produce a flat 2D substrate that contains all the same proteins, solubilizing the 3D ECM in guanidine and plating it on 2D surfaces, and coating 2D surfaces with individual components, such as FN, known to be present in the 3D matrices (Figure 1B). Additional 3D controls include comparisons with 3D collagen gels and reconstituted basement membrane extracts, for example, Matrigel, as well as attempting to generate 3D fibrils using purified FN. In all cases, these controls have replicated few if any of the following characteristic responses to cell-derived 3D ECM [14].
2. Biological responses of fibroblasts to 3D cell-derived matrices compared to 2D substrates
2.1 Cell attachment and migration on cell-derived 3D matrix
A comparison of human primary fibroblasts grown within 3D matrices to those grown on 2D regular tissue culture surfaces (with or without coating the substrate with matrix proteins) revealed striking differences in behavior and in several aspects of signaling [14]. Initial cell adhesion, i.e. cell attachment assayed after 10 minutes, is enhanced 6-fold for human fibroblasts interacting with the 3D cell-derived matrix compared to flat substrates of fibronectin, collagen, laminin, or even 3D collagen gels. Flattening of the 3D matrices or solubilization and coating on glass surfaces resulted in much lower adhesion equal to that occurring on purified ECM components.
Assays for fibroblast migration in 3D matrices indicate that cell migration path lengths are markedly enhanced because of both a modest stimulation of cell migration velocity by ∼50% [14] and a substantial increase in directionality or persistence of migration [16]. This increased persistence is due to altered Rac activity (see below).
2.2 Integrin response to 3D matrix
Although one might predict that the set of integrins used in 3D microenvironments would be complex due to the presence of multiple molecules in 3D matrices, there was found to be an unexpected dependence solely on the α5β1 integrin [14]. In contrast, the attachment of human fibroblasts to purified 2D FN is dependent on both this major FN integrin receptor and the αVβ3 general-purpose Arg-Gly-Asp integrin receptor, as shown by inhibition of fibroblast adhesion to purified 2D FN with antibodies to both types of integrins (unpublished observations). The mechanism of this striking integrin specificity in 3D is not yet clear, but we speculate that it may result from transdominant inhibition of αVβ3 by α5β1 triggered by the high levels of integrin binding to FN in the form of a pliable fibrillar matrix. Interestingly, a recent study has suggested that a 3D matrix can promote α5β1 integrin activation [20, 21].
In fact, the molecular composition of the cell adhesion complexes that form on 3D compared to 2D substrates differ. The most striking difference in these “3D-matrix adhesions” is the absence of the αVβ3 integrin, which is a characteristic component of focal adhesions but is absent from the “3D matrix adhesions” formed on fibrillar 3D matrices. Another interesting difference between cell adhesions on 2D versus 3D surfaces is the involvement of FN. Focal adhesions can form on surfaces lacking FN, whereas 3D matrix adhesions appear to be intimately associated with fibronectin-containing fibrils [14].
2.3 Effect of 3D matrix on proliferation rate and cell morphology
Besides altered cell adhesion, human fibroblasts cultured in 3D matrices have twice the proliferation rate, which is remarkable considering that FN is an excellent substrate for fibroblast growth and proliferation. Analogous to adhesion to 3D ECM, this enhancement of fibroblast proliferation is also dependent primarily on the α5β1 integrin, according to an antibody inhibition analysis [14].
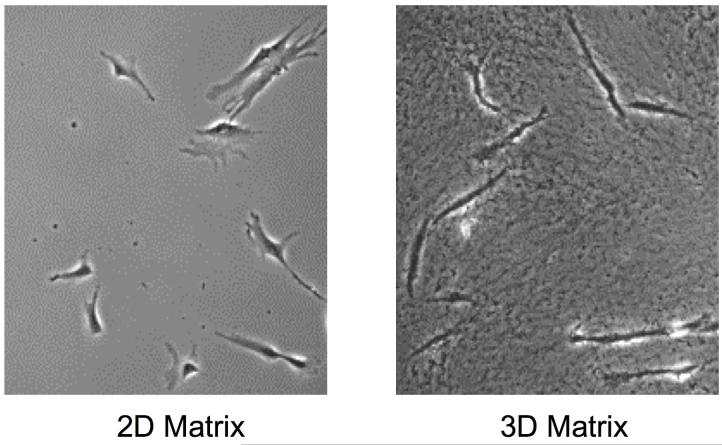
Fibroblasts cultured within 3D cell-derived matrices also have an elongated, spindle-shaped morphology characteristic of fibroblasts in vivo (Figure 2). Although a similar morphological change has been known for many decades to occur after culturing fibroblasts in collagen gels [22], the speed of this acquisition of a markedly elongated and/or arborized shape is much faster in cell-derived 3D matrices [14]. The mechanism underlying this difference in morphology remains to be clarified. Fibroblast morphology in 3D collagen gels needs to be examined in the presence and absence of FN to determine the role of FN versus collagen in this alteration. Interestingly, simultaneous experimental apposition of flat surfaces to which FN or collagen has been attached to both ventral and dorsal surfaces of fibroblasts results in an elongated cell shape; the authors suggest that loss of polarity may contribute to this effect [23].
Figure 2.
Differences in cell morphology between cells on a 2D matrix and in a fibrillar 3D matrix. Fibroblasts cultured in a cell-derived 3D matrix appear elongated and spindle-shaped compared to cells plated on a 2D matrix, which possess characteristic fan-shaped lamellae. On a 2D matrix, an artificial cell polarity is created between the upper and lower surfaces of the cell. No such polarity exists within the 3D matrix since the cells are completely surrounded by extracellular matrix proteins. Note the fibrillar appearance of the 3D matrix.
3. Signaling in 3D compared to 2D microenvironments
3.1 Protein kinase activity in 3D matrix
Various forms of signal transduction can differ markedly, slightly, or even not at all in their response to 3D versus 2D interactions. The tyrosine phosphorylation of FAK at residue 397 has been studied intensively in 2D settings as an early step in integrin signaling. Remarkably, phosphorylation of this kinase in cell adhesions is virtually absent from human fibroblasts in the cell-derived 3D matrix. Instead, phosphorylation appears to be confined to spot-like focal complexes or mini focal adhesions, and overall signaling at this well-known signaling site is reduced by more than four-fold [14]. In puzzling contrast, tyrosine phosphorylation of the focal adhesion protein paxillin at tyrosine 31 is not significantly altered. This discrepancy might be due to dependence of FAK signaling on matrix or substrate rigidity [24]. Conversely, activation of the MAP kinase ERK1/2 is enhanced about 25%, which may be associated with the enhanced proliferation rate [14]. Overall, these findings strongly suggest that the significant number of signaling studies published to date using cells grown on flat 2D substrates should be reevaluated under arguably more physiological 3D conditions. While the principles of signaling established using regular tissue culture are likely to remain valid, the extent, nature, and regulation of specific signaling responses of cells may well differ in 3D microenvironments.
3.2 Rho GTPases and 3D matrix
A particularly instructive effect of 3D versus 2D environments on signaling is seen with Rho family GTPases. Although overall activation levels of Rho and Cdc42 are unchanged, Rac activation is reduced by ∼30%. Although this alteration might seem minor, it has major effects on fibroblast migratory behavior [16]. Levels of Rac activation can be manipulated in living cells using RNA interference [25] or by genetic means [16]. A series of analyses demonstrates that a decrease in overall Rac activity by 30% or greater results in a switch of the mode of cell migration from relatively random to more directionally persistent. This doubling of the persistence of migration appears to be caused by suppression of lateral or peripheral lamellae of migrating cells. Cells with slightly reduced Rac levels have only a single leading edge and consequently fail to migrate in other directions, because they do not activate lateral lamellae as the new leading edge advances [16]. This “Rac switch” for persistence of migration is found in multiple cell types including human epithelial and tumor cells. Figure 3 summarizes this Rac switch, where a modest reduction in overall Rac activation levels in a 3D microenvironment results in a marked alteration in the mode of cell migration, with enhanced persistence of migration.
Figure 3.
The Rac switch. Fibroblasts migrating in a 3D microenvironment have moderately lower total levels of Rac activation, which results in fewer peripheral lamellae and greater directionality and persistence of migration. The reduction in overall Rac levels does not affect the localization of Rac to the leading edge, which represents a general mechanism for stimulating migration using leading lamellae.
Chemotaxis is a well-studied phenomenon by which an external soluble factor induces directional migration. It is generally thought to depend on a phosphatidylinositol 3′-kinase (PI3K) and PIP3-dependent signaling system, and inhibition of PI3K is known to inhibit chemotaxis [26, 27]. In striking contrast, the Rac-dependent switch in directional persistence described above is not dependent on PI3K. Nevertheless, a modest reduction in Rac that promotes more directional migration can cooperate with an external chemotactic signal to enhance the effectiveness of human fibroblast chemotactic migration [16]. A simple interpretation is that once chemotaxis defines a direction of migration, Rac-regulated persistence promotes efficiency of overall cell migration.
4. Other signaling pathways, ECM pliability, and cancer
We predict that further examination of other signaling pathways will identify yet additional signaling systems affected by 3D versus 2D microenvironments. Although no general principles for predicting which pathways will be affected are known yet, it is possible that examination of the dozens of other known signaling pathways will identify key elements. One important physical feature is likely to be the markedly increased pliability of fibrillar 3D matrices compared to flat, rigid 2D substrates. In fact, rigidifying 3D matrices by chemical crosslinking results in changes in the types of cell adhesion structures from 3D matrix adhesions to structures that resemble focal adhesions on rigid substrates [14]. Future studies should ideally examine the effect of graded changes in rigidity of 3D fibrillar matrices on fibroblast signaling, cell morphology, and biological responses.
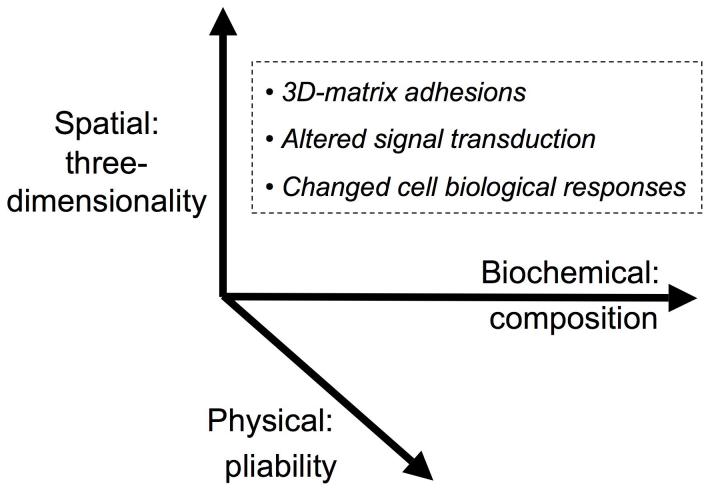
Figure 4 summarizes the major microenvironmental factors affecting cellular function. The biochemical composition of the surrounding ECM can substantially affect cellular responses, as can ECM pliability. In addition, however, three-dimensionality of the ECM also appears to be important for fibroblast responses, affecting the type of cell adhesions, the nature of signal transduction, and various biological responses.
Figure 4.
Factors in the microenvironment that regulate cell responses. At least three major classes of microenvironmental factors affect cellular behavior. The biochemical composition of the ECM, e.g. its content of collagen versus other molecules, plays a well-known role. In addition, however, physical parameters of the matrix, particularly its pliability, can have major effects on cellular responses. A third parameter involves spatial cues, particularly whether a matrix is two-dimensional (providing a unidirectional source of polarity) or three-dimensional. Each of these classes of microenvironmental factors can affect the type of cell adhesion, e.g. 3D-matrix adhesions compared to focal adhesions, can have different effects on various signal transduction pathways, and can change cell morphology, proliferation, migration, and differentiation.
A biological condition in which the normal 3D matrix is known to be altered is cancer, which is often associated with desmoplasia. This latter phenomenon, in which the matrix becomes stiffer and denser, is likely to have direct effects on cell functions. For example, a stiffer matrix is known to produce alterations in signaling [24]. Interestingly, stromal fibroblasts isolated from tumor tissue can generate a 3D ECM that differs from that produced by normal fibroblasts. This altered ECM can cause normal fibroblasts to mimic tumor fibroblasts through the induction of α-smooth muscle actin and desmin [28]. The relationships between 3D ECM, pliability, signaling, and cancer are currently under intense investigation [21].
5. Conclusions
Many researchers continue to discover differences in cell signaling when comparing cells in 3D microenvironments to 2D matrices. While much has been done, the enormous diversity and complexity of the extracellular matrix milieu that cells encounter assures us that our work is just beginning.
In this review, we have discussed our work with cell-derived 3D matrices and research from other groups demonstrating differences in cell behavior in 3D verses 2D microenvironments. Signaling and adhesion structures observed in 2D cultures may represent exaggerated stages of dynamic in vivo situations. Consequently, cells may react differently to external perturbations when 2D cultures are used as a tool to test drug responses or to assay biological function. Using ex vivo 3D microenvironments instead of 2D culture conditions may save time and expense in the long run by more accurately modeling in vivo biological responses.
Acknowledgments
The authors would like to thank Kazue Matsumoto for supplying images of cells in 3D matrices, and Andrew Doyle and Catherine Galbraith for valuable comments and suggestions, and Harry Grant for excellent proofreading. Research support was provided by the Intramural Research Program of the NIH, NIDCR, to K.M.Y.
Abbreviations
- ECM
extracellular matrix
- FN
fibronectin
- 3D
three-dimensional
- 2D
two-dimensional
- FAK
focal adhesion kinase
- MAP kinase
mitogen-activated protein kinase
- ERK 1/2
extracellular signal-regulated kinase 1/2
- PI3K
phosphatidylinositol 3′-kinase
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- [2].Schindler M, Nur EKA, Ahmed I, Kamal J, Liu HY, Amor N, Ponery AS, Crockett DP, Grafe TH, Chung HY, Weik T, Jones E, Meiners S. Living in three dimensions: 3D nanostructured environments for cell culture and regenerative medicine. Cell Biochem Biophys. 2006;45:215–227. doi: 10.1385/CBB:45:2:215. [DOI] [PubMed] [Google Scholar]
- [3].Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- [4].DeMali KA, Wennerberg K, Burridge K. Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol. 2003;15:572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- [5].Aota S, Nagai T, Yamada KM. Characterization of regions of fibronectin besides the arginine-glycine-aspartic acid sequence required for adhesive function of the cell-binding domain using site-directed mutagenesis. J Biol Chem. 1991;266:15938–15943. [PubMed] [Google Scholar]
- [6].Nagai T, Yamakawa N, Aota S, Yamada SS, Akiyama SK, Olden K, Yamada KM. Monoclonal antibody characterization of two distant sites required for function of the central cell-binding domain of fibronectin in cell adhesion, cell migration, and matrix assembly. J Cell Biol. 1991;114:1295–1305. doi: 10.1083/jcb.114.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Obara M, Kang MS, Yamada KM. Site-directed mutagenesis of the cell-binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell. 1988;53:649–657. doi: 10.1016/0092-8674(88)90580-6. [DOI] [PubMed] [Google Scholar]
- [8].Mao Y, Schwarzbauer JE. Accessibility to the Fibronectin Synergy Site in a 3D Matrix Regulates Engagement of alpha 5beta 1 versus alpha vbeta 3 Integrin Receptors. Cell Commun Adhes. 2006;13:267–277. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- [9].Blystone SD, Graham IL, Lindberg FP, Brown EJ. Integrin alpha v beta 3 differentially regulates adhesive and phagocytic functions of the fibronectin receptor alpha 5 beta 1. J Cell Biol. 1994;127:1129–1137. doi: 10.1083/jcb.127.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Blystone SD, Slater SE, Williams MP, Crow MT, Brown EJ. A molecular mechanism of integrin crosstalk: alphavbeta3 suppression of calcium/calmodulin-dependent protein kinase II regulates alpha5beta1 function. J Cell Biol. 1999;145:889–897. doi: 10.1083/jcb.145.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Diaz-Gonzalez F, Forsyth J, Steiner B, Ginsberg MH. Transdominant inhibition of integrin function. Mol Biol Cell. 1996;7:1939–1951. doi: 10.1091/mbc.7.12.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ly DP, Zazzali KM, Corbett SA. De novo expression of the integrin alpha5beta1 regulates alphavbeta3-mediated adhesion and migration on fibrinogen. J Biol Chem. 2003;278:21878–21885. doi: 10.1074/jbc.M212538200. [DOI] [PubMed] [Google Scholar]
- [13].Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- [14].Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- [15].Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- [16].Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17:524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- [18].Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- [19].Nur EKA, Ahmed I, Kamal J, Schindler M, Meiners S. Three dimensional nanofibrillar surfaces induce activation of Rac. Biochem Biophys Res Commun. 2005;331:428–434. doi: 10.1016/j.bbrc.2005.03.195. [DOI] [PubMed] [Google Scholar]
- [20].Mao Y, Schwarzbauer JE. Stimulatory effects of a three-dimensional microenvironment on cell-mediated fibronectin fibrillogenesis. J Cell Sci. 2005;118:4427–4436. doi: 10.1242/jcs.02566. [DOI] [PubMed] [Google Scholar]
- [21].Larsen M, Artym VV, Green JA, Yamada KM. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol. 2006;18:463–471. doi: 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- [22].Elsdale T, Bard J. Collagen substrata for studies on cell behavior. J Cell Biol. 1972;54:626–637. doi: 10.1083/jcb.54.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beningo KA, Dembo M, Wang YL. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci U S A. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Endo Y, Even-Ram S, Pankov R, Matsumoto K, Yamada KM. Inhibition of rho GTPases by RNA interference. Methods Enzymol. 2006;406:345–361. doi: 10.1016/S0076-6879(06)06025-3. [DOI] [PubMed] [Google Scholar]
- [26].Haugh JM, Codazzi F, Teruel M, Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- [28].Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol. 2005;167:475–488. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]