Abstract
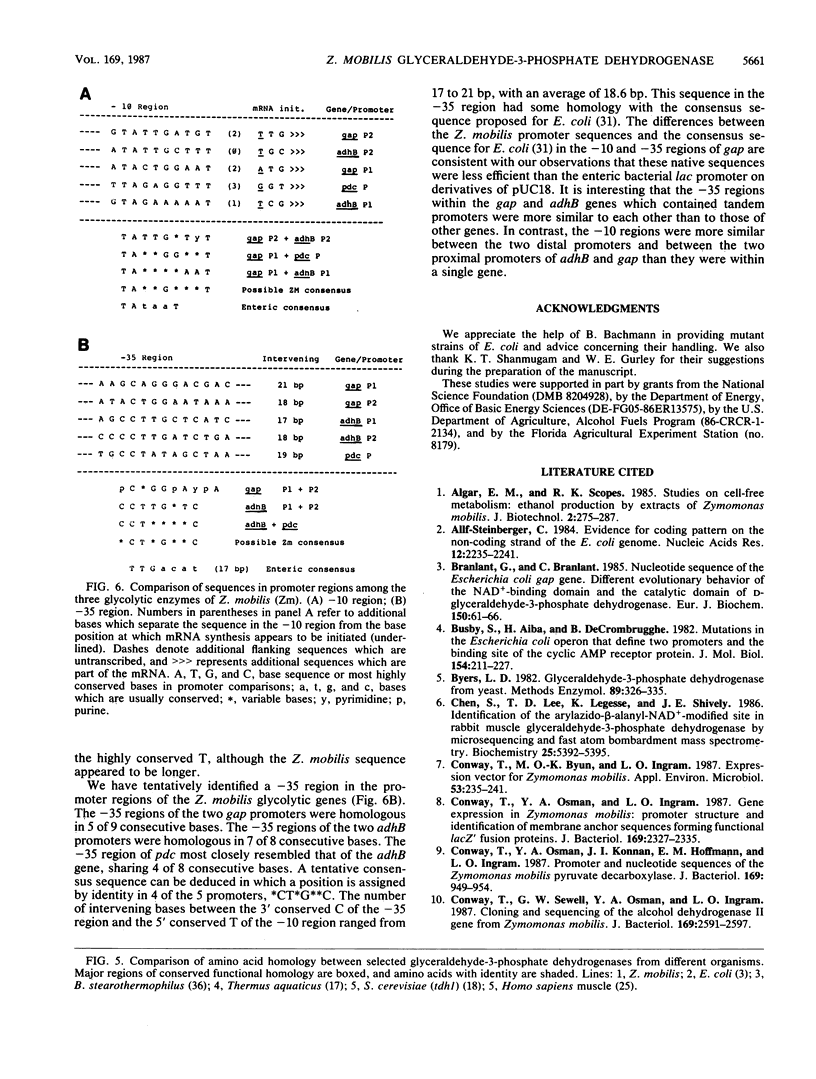
The gene encoding glyceraldehyde-3-phosphate dehydrogenase was isolated from a library of Zymomonas mobilis DNA fragments by complementing a deficient strain of Escherichia coli. It contained tandem promoters which were recognized by E. coli but appeared to function less efficiently than the enteric lac promoter in E. coli. The open reading frame for this gene encoded 337 amino acids with an aggregate molecular weight of 36,099 (including the N-terminal methionine). The primary amino acid sequence for this gene had considerable functional homology and amino acid identity with other eucaryotic and bacterial genes. Based on this comparison, the gap gene from Z. mobilis appeared to be most closely related to that of the thermophilic bacteria and to the chloroplast isozymes. Comparison of this gene with other glycolytic enzymes from Z. mobilis revealed a conserved pattern of codon bias and several common features of gene structure. A tentative transcriptional consensus sequence is proposed for Z. mobilis based on comparison of the five known promoters for three glycolytic enzymes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alff-Steinberger C. Evidence for a coding pattern on the non-coding strand of the E. coli genome. Nucleic Acids Res. 1984 Mar 12;12(5):2235–2241. doi: 10.1093/nar/12.5.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant G., Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1985 Jul 1;150(1):61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- Busby S., Aiba H., de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J Mol Biol. 1982 Jan 15;154(2):211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- Byers L. D. Glyceraldehyde-3-phosphate dehydrogenase from yeast. Methods Enzymol. 1982;89(Pt 500):326–335. doi: 10.1016/s0076-6879(82)89059-9. [DOI] [PubMed] [Google Scholar]
- Chen S., Lee T. D., Legesse K., Shively J. E. Identification of the arylazido-beta-alanyl-NAD+-modified site in rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by microsequencing and fast atom bombardment mass spectrometry. Biochemistry. 1986 Sep 23;25(19):5391–5395. doi: 10.1021/bi00367a007. [DOI] [PubMed] [Google Scholar]
- Conway T., Byun M. O., Ingram L. O. Expression Vector for Zymomonas mobilis. Appl Environ Microbiol. 1987 Feb;53(2):235–241. doi: 10.1128/aem.53.2.235-241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Ingram L. O. Gene expression in Zymomonas mobilis: promoter structure and identification of membrane anchor sequences forming functional lacZ' fusion proteins. J Bacteriol. 1987 Jun;169(6):2327–2335. doi: 10.1128/jb.169.6.2327-2335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes E. A., Ribbons D. W., Rees D. A. Sucrose utilization by Zymomonas mobilis: formation of a levan. Biochem J. 1966 Mar;98(3):804–812. doi: 10.1042/bj0980804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek K. M., Ingram L. O. Magnesium limitation and its role in apparent toxicity of ethanol during yeast fermentation. Appl Environ Microbiol. 1986 Nov;52(5):975–981. doi: 10.1128/aem.52.5.975-981.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hillman J. D., Fraenkel D. G. Glyceraldehyde 3-phosphate dehydrogenase mutants of Escherichia coli. J Bacteriol. 1975 Jun;122(3):1175–1179. doi: 10.1128/jb.122.3.1175-1179.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking J. D., Harris J. I. D-glyceraldehyde-3-phosphate dehydrogenase. Amino-acid sequence of the enzyme from the extreme thermophile Thermus aquaticus. Eur J Biochem. 1980 Jul;108(2):567–579. doi: 10.1111/j.1432-1033.1980.tb04752.x. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Nowak K., Wolny M., Banaś T. The complete amino acid sequence of human muscle glyceraldehyde 3-phosphate dehydrogenase. FEBS Lett. 1981 Nov 16;134(2):143–146. doi: 10.1016/0014-5793(81)80587-x. [DOI] [PubMed] [Google Scholar]
- Olsen K. W. Structural basis for the thermal stability of glyceraldehyde-3-phosphate dehydrogenases. Int J Pept Protein Res. 1983 Oct;22(4):469–475. doi: 10.1111/j.1399-3011.1983.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman Y. A., Ingram L. O. Mechanism of ethanol inhibition of fermentation in Zymomonas mobilis CP4. J Bacteriol. 1985 Oct;164(1):173–180. doi: 10.1128/jb.164.1.173-180.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M. C., Lazar G., Goodman H. M. Evidence in favor of the symbiotic origin of chloroplasts: primary structure and evolution of tobacco glyceraldehyde-3-phosphate dehydrogenases. Cell. 1986 Oct 10;47(1):73–80. doi: 10.1016/0092-8674(86)90367-3. [DOI] [PubMed] [Google Scholar]
- Swings J., De Ley J. The biology of Zymomonas. Bacteriol Rev. 1977 Mar;41(1):1–46. doi: 10.1128/br.41.1.1-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Carne A. F., Runswick M. J., Bridgen J., Harris J. I. D-glyceraldehyde-3-phosphate dehydrogenase. Complete amino-acid sequence of the enzyme from Bacillus stearothermophilus. Eur J Biochem. 1980 Jul;108(2):549–565. doi: 10.1111/j.1432-1033.1980.tb04751.x. [DOI] [PubMed] [Google Scholar]