Abstract
To evaluate the physiological significance of cyclic electron flow around photosystem (PS) I, we used a reverse genetic approach to focus on 11 chloroplast genes that encode homologs of mitochondrial complex I subunits (ndhA-K). Since their discovery, the exact function of the respiratory components in plant chloroplasts has been a matter of discussion. We disrupted one of these genes (ndhB) in tobacco by chloroplast transformation. Analysis of the transient increase in chlorophyll fluorescence after actinic light illumination and the redox kinetics of P700 (reaction center chlorophylls of PS I) suggest that the cyclic electron flow around PS I is impaired in the ndhB-deficient transformants. Transformants grew normally in a greenhouse, suggesting that the cyclic electron flow around PS I mediated by ndh gene products is dispensable in tobacco under mild environmental conditions.
Photosynthetic electron flow provides the first stable products of photosynthesis: NADPH and ATP. Despite the importance of this electron flow, a fundamental problem remains unsolved; that is, how an appropriate balance between the production of NADPH and ATP is maintained. To answer this question, the contributions of the Q cycle, cyclic electron flow around photosystem (PS) I, and pseudocyclic electron flow (water-water cycle) in chloroplast energetics must be evaluated quantitatively (1). There is little doubt that cyclic electron flow around PS I provides extra ATP in some cellular processes, such as N2 fixation in cyanobacterial heterocysts (2) and CO2 concentration in cyanobacterial and C4 photosynthesis (3–8). However, it is unclear whether this cyclic electron flow contributes to the supply of ATP during steady-state photosynthesis in nonspecialized photosynthetic cells of higher plants (1, 9, 10).
Although molecular biological dissection using a reverse genetic approach is an effective means to evaluate the physiological significance of cyclic electron flow around PS I, it has not been attempted because of a lack of information about the genes responsible for the electron flow. However, the discovery of an ndhB-deficient mutant of Synechocystis PCC6803 that lacked cyclic electron flow around PS I led to the idea that electron flow is mediated by the respiratory complex, NAD(P)H dehydrogenase, in cyanobacteria (4–8).
Eleven ndh genes encoding homologs of mitochondrial complex I subunits are also present in the chloroplast genome of higher plants (11, 12). Although respiratory function is limited to the mitochondria, a respiratory complex, NAD(P)H dehydrogenase, may catalyze cyclic electron flow around PS I in chloroplasts, as in cyanobacteria. However, the existence of NAD(P)H dehydrogenase-mediated electron flow in higher plants is still a matter of controversy (1, 13), because genes for the crucial flavoprotein subunits have not yet been identified (14). Moreover, physiological evidence alone has been insufficient to show an NAD(P)H dehydrogenase-mediated pathway for cyclic electron flow around PS I in higher plants because of the existence of an alternative, ferredoxin (Fd)-dependent, antimycin A-sensitive pathway (15–19).
In this study, we disrupted ndhB in tobacco by using a plastid transformation technique (20) to provide direct evidence from a higher plant for the mediation of cyclic electron flow around PS I by chloroplast ndh gene products, i.e., a putative NAD(P)H dehydrogenase complex. We also evaluated the contribution of this pathway to the ATP pool during steady-state photosynthesis under mild environmental conditions.
MATERIALS AND METHODS
Vector Construction.
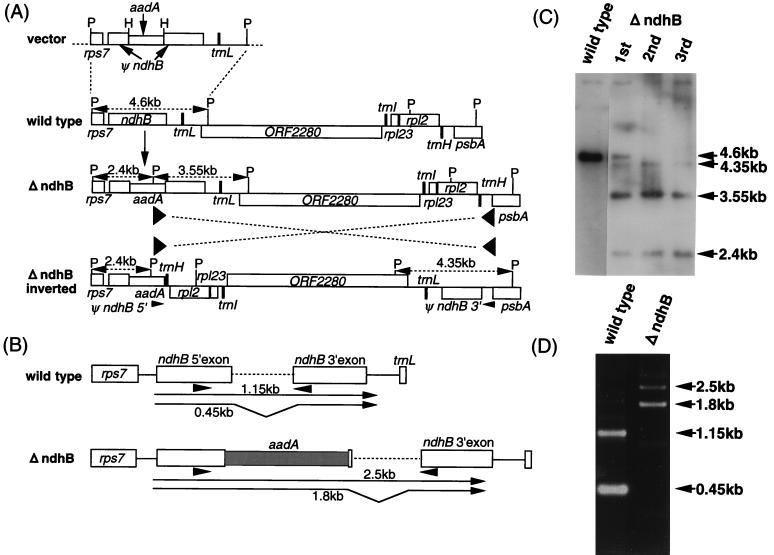
A chimeric aadA gene, which consists of the aadA coding region (21) controlled by the plastid ribosomal RNA operon promoter, the rbcL ribosome binding site, and the psbA terminator, was constructed essentially as reported by Svab and Maliga (20). The 4.6-kb PstI fragment (95,353–99,988 or 142,546–147,181 in ref. 22) containing the ndhB gene isolated from Nicotiana tabacum cv. Xanthi chloroplast DNA was cloned into pBluescript II SK+ (Stratagene), which had a deletion from EcoRI to XhoI in the multicloning site. The chimeric aadA gene was inserted at the HindIII site present in the 5′ exon of ndhB in the same orientation as ndhB (Fig. 1A, vector).
Figure 1.
Gene disruption of ndhB by plastid transformation. (A) The 5′ exon of ndhB was disrupted by insertion of the aadA cassette at the unique HindIII site (vector). Disrupted ndhB (ψndhB) was incorporated into the wild-type genome by two homologous recombination events (ΔndhB). The PstI fragment sizes detected by Southern hybridization are shown. Sequence inversion occurred via the duplicated psbA terminator sequences indicated by the arrowheads (ΔndhB inverted). HindIII and PstI sites are indicated by H and P, respectively. (B) Schematic representation of the transcripts in wild-type and ΔndhB tobacco chloroplasts. Arrowheads indicate the positions of the PCR primers. Reverse transcription–PCR fragment sizes are indicated. (C) Total cellular DNA extracted from the wild-type and transformed tobacco plants after the first, second, and third rounds of spectinomycin selection was digested with PstI, then analyzed by Southern hybridization with a 4.6-kb PstI fragment as the probe. (D) Total cellular RNAs extracted from wild-type and ΔndhB plants were used to synthesize cDNA. For reverse transcription–PCR, the primers used are as indicated in B.
Transformation of Plastid Genome.
Tobacco plants (N. tabacum cv. Xanthi) were grown as aseptic shoot cultures on Murashige-Skoog (MS) medium (23) containing 3% sucrose. Leaves were used for bombardments according to the method described by Svab and Maliga (20). Transgenic calli and shoots were selected on RMOP medium (24) containing spectinomycin (500 μg/ml). Resistant shoots were rooted on MS medium. Leaf discs were made from leaves of rooted plants and further selected by spectinomycin to obtain homoplasmic transformants. Homoplasmic transformants were rooted on MS media, then transferred to soil and cultured in a greenhouse.
Analysis of DNA and RNA.
Total cellular DNA was extracted from spectinomycin-resistant clones (calli containing regenerated shoots) after the first screening, and from leaves of rooted plants after the second and third screenings. DNA digested with PstI was electrophoresed through a 0.7% agarose gel and transferred to Hybond N+ (Amersham). The Southern blot then was probed with the 4.6- kb PstI fragment containing ndhB.
Total cellular RNA was extracted from leaves of plants aseptically grown on Murashige-Skoog medium. RNA (5 μg) was converted to first-strand cDNA by using random hexamers as primers. To eliminate contamination from double-stranded genomic DNA, cDNA preparations were treated with EcoRI, which cleaves the ndhB gene within its intron. Reverse transcription–PCR was carried out by using the following primers: 5′-ATGGCTATAACAGAGTTTCTCTT-3′ and 5′-AAGCAGCTACTTTCGAAGTAAC-3′.
Monitoring of the Transient Increase in Chlorophyll Fluorescence.
Cyclic electron flow around PS I was monitored by the transient increase of dark-level chlorophyll fluorescence after actinic light (AL) illumination (20 μmol m−2⋅s−1 for 5 min) by using intact leaves from plants grown in a greenhouse (25). A pulse-amplitude-modulation chlorophyll fluorometer (Waltz, Effeltrich, Germany) was equipped with an ED101 emitter-detector unit. The maximum yield of chlorophyll fluorescence was induced by an 800-ms pulse of saturating white light. During illumination with AL, the 800-ms saturating pulse was applied every 1 min to monitor nonphotochemical quenching.
Measurement of the Redox Kinetics of P700.
Changes in the levels of P700+, assessed by monitoring absorbance at 820 nm was measured by using pulse-amplitude-modulation with emitter-detector units ED800T (26). Far-red light (FR; above 720 nm, 6.8 W m−2) and AL (900 μmol m−2⋅s−1 for 2 min) were applied to leaves via the multibranched fiberoptic system that was equipped with the detector.
RESULTS
Because an ndhB-deficient mutation in Synechocystis PCC6803 causes the inactivation of cyclic electron flow (4), we chose the corresponding gene, ndhB, from tobacco chloroplasts as a target for insertional inactivation by using a plastid transformation technique (20). A PstI fragment carrying ndhB disrupted by the insertion of the chimeric aadA gene (Fig. 1A, vector) was introduced into tobacco leaves by particle bombardment (20). The chloroplast genome of the transformants from the first round of spectinomycin screening consisted of a mixture of wild-type and transformed genome copies (Fig. 1 A and C). Subsequently, one transformed clone was used in the second and third rounds of screening to obtain the homoplasmic transformant (ΔndhB). Southern analysis showed that two cycles of screening were enough to sort out the functional ndhB gene (Fig. 1 A and C). Although the two major signals (2.4 and 3.55 kb) suggest that the two homologous recombination events led to the insertion of the aadA gene (Fig. 1A, ΔndhB), the minor signal (4.35 kb) produced was unexpected (Fig. 1C). This signal is ascribed to a sequence inversion via the short inverted repeat sequence generated by the introduction of the psbA terminator sequence (from the chimeric aadA gene) into the vicinity of the endogenous psbA sequence (Fig. 1A, ΔndhB inverted). This hypothesis was confirmed by PCR amplification using primers complementary to the 3′ exon of ndhB and the psbA coding region, respectively (data not shown). This inversion did not inactivate any gene, rather it modified the overall genome structure. The transformed genome was stable and never replaced by the wild-type genome in shoot cultures grown on spectinomycin-free medium or in progeny from repetitive backcrossing with wild-type tobacco pollens.
The aadA cassette also was inserted between rbcL and accD as described by Svab and Maliga (20). This line, 4Y26, in which no endogenous genes were disrupted, was used as a control for aadA expression in chloroplasts.
Reverse transcription–PCR showed the absence of a functional ndhB transcript in ΔndhB (Fig. 1 B and D). The 0.45-kb fragment, which originates from the spliced transcript of the intact ndhB gene, was absent in ΔndhB. The 1.15-kb fragment in the wild type is the unspliced precursor of the ndhB transcript. The larger transcripts detected in ΔndhB are precursors that were not processed at the aadA terminator (spliced 1.8 kb and unspliced 2.5 kb). Thus, ndhB was completely disrupted in ΔndhB.
Because ndhB is monocistronically transcribed (11), it is unlikely that the expression of neighboring genes was modified by the insertion of the aadA cassette into ndhB. The psbA terminator in the aadA cassette functions as a processing site rather than as a transcriptional termination site (Fig. 1D). The endogenous ndhB terminator should function to terminate the chimeric transcription derived from the ndhB and aadA cassette promoters.
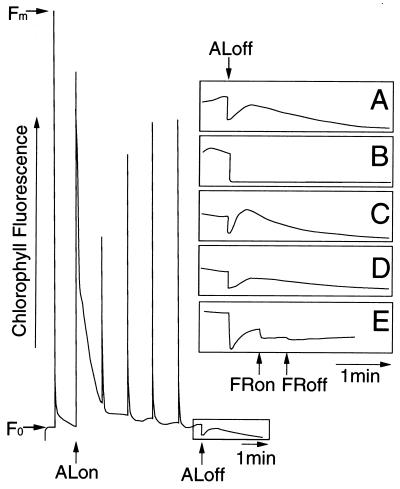
To determine whether ndh gene products function in the electron flow from NAD(P)H to plastoquinone (PQ) in higher plants as in cyanobacteria (5–8), we compared electron transfer from the stromal donors to PQ of the wild-type and ΔndhB plants. The electron flow was monitored by the transient increase in chlorophyll fluorescence (apparent Fo) after AL illumination, which represents electron donation to the intersystem chain from the electron donors accumulated in the stroma during AL illumination (25). The transient rise in chlorophyll fluorescence in the dark was caused by the reduction of PQ, because it was quenched by FR (Fig. 2E), as has been reported (26). Because AL illumination at higher intensity gave more nonphotochemical quenching and influenced chlorophyll fluorescence after AL illumination by relaxation of thylakoid energization, we used AL at a low intensity (20 μmol m−2⋅s−1 for 5 min) in our assay. The wild-type leaves showed a transient increase in chlorophyll fluorescence in the dark after termination of AL (Fig. 2 A and C), which was larger in fully expanded leaves than in immature leaves. The increase in chlorophyll fluorescence was absent in immature leaves of ΔndhB (Fig. 2B), indicating that the cyclic electron flow is missing. In contrast, fully expanded leaves of ΔndhB showed a small increase in the fluorescence, which was much less than that of the wild-type plants (Fig. 2D). Because ndhB was completely inactivated (Fig. 1), this small rise in chlorophyll fluorescence in mature leaves should be caused by different electron flow, such as that in the Fd-mediated pathway. In mature leaves of ΔndhB, electron flow may function through this pathway. There were no differences in the fluorescence patterns of the control line, 4Y26, and the untransformed plants (data not shown), indicating that inactivation of electron donation to the intersystem chain is not caused by aadA expression in chloroplasts.
Figure 2.
Analysis of the transient increase in chlorophyll fluorescence (apparent Fo) after termination of AL illumination. Chlorophyll fluorescence of tobacco leaves was monitored by using a pulse-amplitude-modulation chlorophyll fluorometer. The box indicates the transient increase in chlorophyll fluorescence after 5-min AL illumination (20 μmol m−2⋅s−1). (A–E) Close-ups of the corresponding regions of wild-type (A, C, and E) and ΔndhB (B and D) leaves. (A and B) Immature leaves (50 mm long). (C–E) Mature leaves (130 mm long). (E) FR (above 720 nm, 6.8 W m−2 for 30 s) was applied 30 s after the AL termination. Fo and Fm stand for minimum and maximal fluorescent yield, respectively.
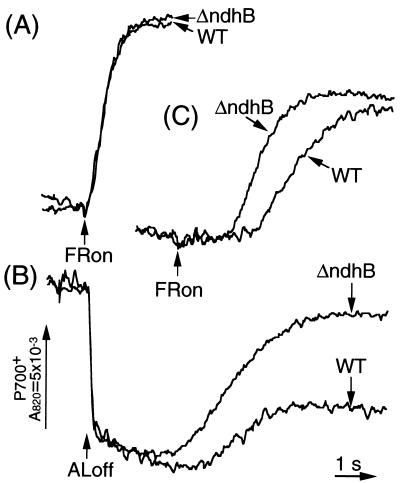
To confirm that electron donation to the intersystem chain from the stromal electron donors functions in cyclic electron flow around PS I, we compared the oxidation kinetics of P700 by FR between wild-type and ΔndhB leaves (Fig. 3). In dark-adapted leaves, P700 was rapidly oxidized by FR both in wild-type and ΔndhB plants within 1 s (Fig. 3A). In Fig. 3B, leaves were illuminated by AL (900 μmol m−2⋅s−1 for 2 min) under a background of FR to store electrons in the stromal pool, as well as in the intersystem carriers. After termination of AL illumination, P700+ was transiently reduced by electrons from the PQ pool, and thereafter P700 was reoxidized by background FR. The reoxidation of P700 was much faster in ΔndhB leaves than in the wild-type leaves (Fig. 3B), indicating that the electron from the stromal pool (FR-dependent cyclic electron flow around PS I) is reduced in ΔndhB leaves. In ΔndhB leaves, however, oxidation kinetics after AL illumination was not identical to that after the multiple-turnover light illumination, which saturated the intersystem carriers with electrons (data not shown). This result indicates that alternative cyclic electron flow from the stromal pool to the intersystem, such as Fd-mediated pathway, must be remaining in ΔndhB leaves. In Fig. 3C, leaves were illuminated by AL without background FR. Subsequently, FR was turned on 10 s after the AL termination, when chlorophyll fluorescence started to increase in the dark (Fig. 2). Oxidation of P700 also proceeded more rapidly in ΔndhB than in wild-type plants (Fig. 3C), indicating that the rate of PQ reduction by the stromal electron pool is slower in ΔndhB (Fig. 2). These data support the result that electrons in the stromal pool are transferred to P700 through PQ in the wild-type leaves (Fig. 2E). We conclude that the putative NAD(P)H dehydrogenase complex encoded by chloroplast ndh genes mediates the electron flow from the stromal donors to PQ in cyclic electron flow around PS I.
Figure 3.
The redox kinetics of P700 in young mature leaves (80 mm long) of the wild-type (WT) and ΔndhB plants. (A) Oxidation kinetics of P700 by FR in dark-adapted leaves. (B) Redox kinetics of P700 after termination of AL illumination (900 μmol m−2⋅s−1 for 2 min) under a background of FR. (C) Oxidation kinetics of P700 10 s after the AL termination.
To evaluate the contribution of cyclic electron flow around PS I to the ATP pool during steady-state photosynthesis, nonphotochemical quenching was compared between the wild-type and ΔndhB plants under various light intensities (20–2,000 μmol m−2⋅s−1). However, no significant differences in nonphotochemical quenching was observed (data not shown), indicating that cyclic electron flow mediated by NAD(P)H dehydrogenase does not contribute to thylakoid energization. Furthermore, ΔndhB plants grew and set seeds normally under greenhouse conditions. No differences in visible phenotypes was detected between the ΔndhB and wild-type plants. The cyclic electron flow mediated by NAD(P)H dehydrogenase appears to be dispensable in tobacco under the growth conditions used.
DISCUSSION
In this study, we demonstrate that the putative NAD(P)H dehydrogenase complex containing the ndhB gene product mediates electron flow from the stromal electron donors to PQ in cyclic electron flow around PS I in higher plants. Enigmatic occurrence of the respiratory component, NAD(P)H dehydrogenase, in chloroplasts was explained by its participation in photosynthetic electron flow. Our results suggest that electron flow in chloroplsts is similar to that of cyanobacteria (5–8), in which photosynthetic and respiratory electron flow occurs in the same membranes (27). High similarity of amino acid sequences between chloroplast and cyanobacterial ndh genes (60–80%) suggests that cyclic electron flow around PS I mediated by NAD(P)H dehydrogenase in chloroplasts originated from the respiratory electron flow in cyanobacteria.
The endosymbiotic origin of chloroplasts also implies the existence of respiratory electron flow in chloroplasts (chlororespiration). Chlororespiratory electron flow was first identified in the green alga Chlamydomonas reinhardtii (28, 29) and was suggested to also occur in higher plants (30). The scheme of chlororespiration consists of the reduction of the PQ pool by NAD(P)H, followed by an oxygen-dependent oxidation, resulting in the formation of a pH gradient across the thylakoid membranes. The terminal oxidase for this reaction has yet to be characterized. The electron flow catalyzed by NAD(P)H dehydrogenase may function in chlororespiratory electron flow in the dark. However, RNA editing of ndhD is suppressed to a very low level in nonphotosynthetic cells and activated by light in leaves, indicating that the gene product should function in photosynthetic cells in the light (31).
To evaluate the effect of ndhB gene disruption on cyclic electron flow around PS I, we compared chlorophyll fluorescence and redox kinetics of P700 between wild-type and ΔndhB plants. Although the gene disruption was complete (Fig. 1), impairment of the electron flow from the stromal donors to the intersystem chain was incomplete (Figs. 2 and 3). Fd-mediated cyclic electron flow, catalyzed by the still unidentified ferredoxin-PQ reductase (FQR) may contribute the remaining electron flow (1, 15–19). Although PQ is reduced by Fd via cyt b-559(Fd) in the Fd-mediated pathway (18), NAD(P)H dehydrogenase may allow the entry of electrons directly from NAD(P)H (13, 19). Physiological significance of redundant pathways with different electron donors has not been addressed thus far.
Considering the transient increase in chlorophyll fluorescence after AL illumination, the contribution of the Fd-mediated pathway to total cyclic electron flow is small and undetectable in immature leaves (Fig. 2). In contrast, analysis of the redox kinetics of P700 showed that the Fd-mediated pathway contributed more significantly (Fig. 3). We believe this discrepancy to be caused by different assay conditions. In the chlorophyll fluorescence assay, electron flow from the stromal pool to PQ was determined in the dark. In the P700 assay, however, electron flow was analyzed under a background of FR, in which FR-dependent cyclic electron flow around PS I may function.
What is the physiological significance of cyclic electron flow around PS I mediated by NAD(P)H dehydrogenase in higher plants? Complete disruption of ndhB did not influence the energization of thylakoid membranes. Our results are consistent with the observation that the electron flow is predominantly linear under mild environmental conditions in C3 plants (32). The NAD(P)H dehydrogenase pathway scarcely contributed to the ATP pool coupled to linear electron flow during steady-state photosynthesis. This result was supported by the visible phenotype of ΔndhB plants, which was indistinguishable from that of the wild-type plants under greenhouse conditions. In contrast, an ndhB-deficient mutation leads to a requirement for increased CO2 concentration during growth in cyanobacteria (4). In cyanobacteria, requirement for ATP may be much higher during steady-state photosynthesis because of the energization of the concentrating machinery for inorganic carbon. This idea is supported by the fact that cyclic electron flow around PS I is much more enhanced in bundle-sheath cells in C4 photosynthetic plants (3, 33).
It has been proposed that cyclic electron flow around PS I functions in adaptation to environmental stresses (10). First, cyclic electron flow could adjust the productive ratio of NADPH/ATP depending on the requirements of biochemical reactions in the stroma. Second, the proton gradient generated across the thylakoid membranes prevents overreduction of the intersystem by means of down-regulating PS II (34). Because electron flow is predominately linear under mild environmental conditions, cyclic electron flow around PS I scarcely contributes to the down-regulation of PS II. Under environmental stresses such as drought, however, linear electron flow cannot produce a sufficient proton gradient because of the lack of electron acceptors for PS I. Because cyclic electron flow around PS I does not require electron acceptors, it may function to down-regulate PS II under stress conditions. The ΔndhB plants presently are being analyzed for their tolerance to various environmental stresses.
Acknowledgments
We are deeply grateful to Dr. R. Bressan for his critical reading of the manuscript. We also thank Mrs. P. Yamada for her help in the preparation of the manuscript and Dr. R. Winz and Dr. R. Prieto for their helpful discussion.
ABBREVIATIONS
- PS
photosystem
- Fd
ferredoxin
- AL
actinic light
- FR
far-red light
- PQ
plastoquinone
References
- 1.Bendall D S, Manasse R S. Biochim Biophys Acta. 1995;1229:23–38. [Google Scholar]
- 2.Wolk C P, Ernst A, Elhai J. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, the Netherlands: Kluwer; 1994. pp. 769–823. [Google Scholar]
- 3.Asada K, Heber U, Schreiber U. Plant Cell Physiol. 1993;34:39–50. [Google Scholar]
- 4.Ogawa T. Proc Natl Acad Sci USA. 1991;88:4275–4279. doi: 10.1073/pnas.88.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi H, Endo T, Schreiber U, Asada K. Plant Cell Physiol. 1992;33:1099–1105. [Google Scholar]
- 6.Mi H, Endo T, Schreiber U, Ogawa T, Asada K. Plant Cell Physiol. 1992;33:1233–1237. [Google Scholar]
- 7.Mi H, Endo T, Schreiber U, Ogawa T, Asada K. Plant Cell Physiol. 1994;35:163–173. [Google Scholar]
- 8.Mi H, Endo T, Ogawa T, Asada K. Plant Cell Physiol. 1995;36:661–668. [Google Scholar]
- 9.Fork D C, Herbert S K. Photosynth Res. 1993;36:149–168. doi: 10.1007/BF00033035. [DOI] [PubMed] [Google Scholar]
- 10.Heber U, Walker D. Plant Physiol. 1992;100:1621–1626. doi: 10.1104/pp.100.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsubayashi T, Wakasugi T, Shinozaki K, Yamaguchi-Shinozaki K, Zaita N, Hidaka T, Meng B Y, Ohto C, Tanaka M, Kato A, et al. Mol Gen Genet. 1987;210:385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- 12.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al. Nature (London) 1986;322:572–574. [Google Scholar]
- 13.Guedeney G, Corneille S, Cuiné S, Peltier G. FEBS Lett. 1996;378:277–280. doi: 10.1016/0014-5793(95)01473-x. [DOI] [PubMed] [Google Scholar]
- 14.Fearnley I M, Walker J E. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 15.Tagawa K, Tsujimoto H Y, Arnon D I. Proc Natl Acad Sci USA. 1963;49:567–572. doi: 10.1073/pnas.49.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heber U, Egneus H, Hanck U, Jensen M, Köster S. Planta. 1978;143:41–49. doi: 10.1007/BF00389050. [DOI] [PubMed] [Google Scholar]
- 17.Mills J D, Slovacek R E, Hind G. Biochim Biophys Acata. 1978;504:298–309. doi: 10.1016/0005-2728(78)90178-0. [DOI] [PubMed] [Google Scholar]
- 18.Miyake C, Schreiber U, Asada K. Plant Cell Physiol. 1995;36:743–748. [Google Scholar]
- 19.Endo T, Mi H, Shikanai T, Asada K. Plant Cell Physiol. 1997;38:1272–1277. [Google Scholar]
- 20.Svab Z, Maliga P. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldschmidt-Clermont M. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murashige T, Skoog F. Physiol Plant. 1962;15:493–497. [Google Scholar]
- 24.Klein T M, Harper E C, Svab Z, Sanford C, Fromm M E, Maliga P. Proc Natl Acad Sci USA. 1988;85:8502–8505. doi: 10.1073/pnas.85.22.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber U, Schliwa U, Bilger W. Photosynth Res. 1986;10:53–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]
- 26.Asada K, Heber U, Schreiber U. Plant Cell Physiol. 1992;33:927–932. [Google Scholar]
- 27.Scherer S. Trends Biochem Sci. 1990;15:458–462. doi: 10.1016/0968-0004(90)90296-n. [DOI] [PubMed] [Google Scholar]
- 28.Bennoun P. Proc Natl Acad Sci USA. 1982;79:4352–4356. doi: 10.1073/pnas.79.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peltier G, Schmidt G W. Proc Natl Acad Sci USA. 1991;88:4791–4795. doi: 10.1073/pnas.88.11.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garab G, Lajkó F, Mustárdy L, Márton L. Planta. 1989;179:349–358. doi: 10.1007/BF00391080. [DOI] [PubMed] [Google Scholar]
- 31.Hirose T, Sugiura M. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbinson J, Gentry B, Baker N R. Photosynth Res. 1990;25:213–224. doi: 10.1007/BF00033162. [DOI] [PubMed] [Google Scholar]
- 33.Kubicki A, Funk E, Westhoff P, Steinmüller K. Planta. 1996;199:276–281. [Google Scholar]
- 34.Horton P, Ruban A V, Walters R G. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]