Abstract
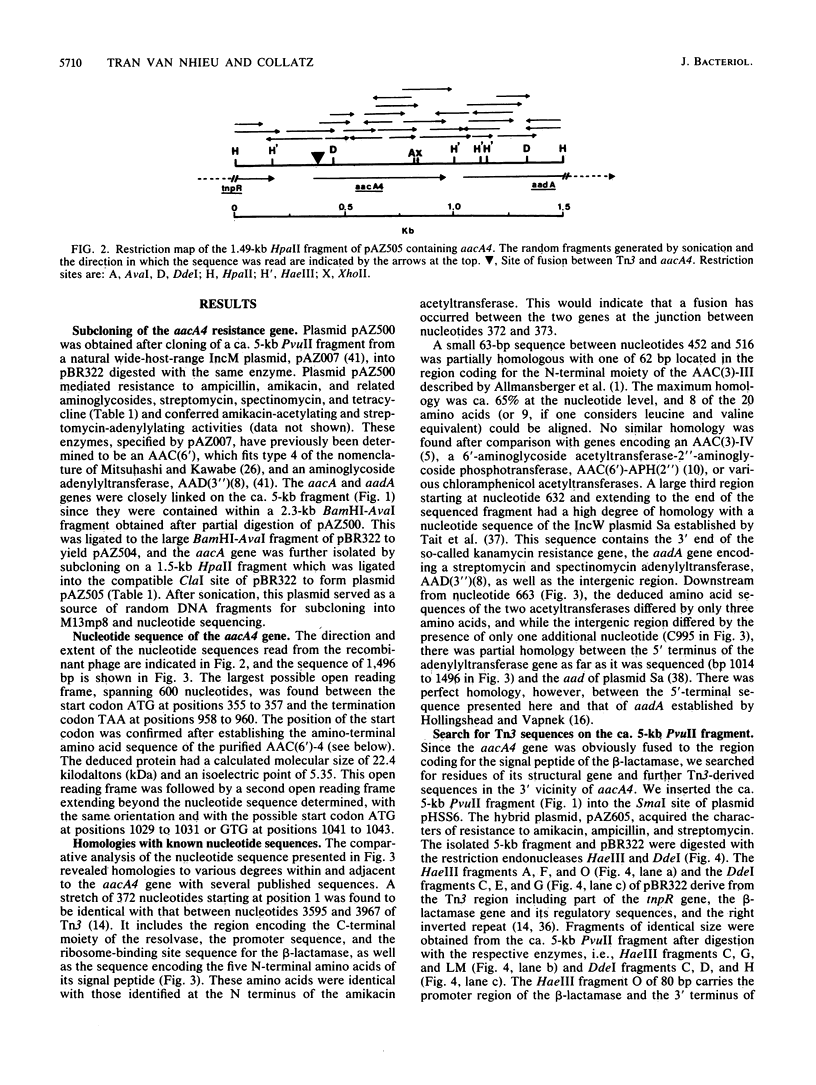
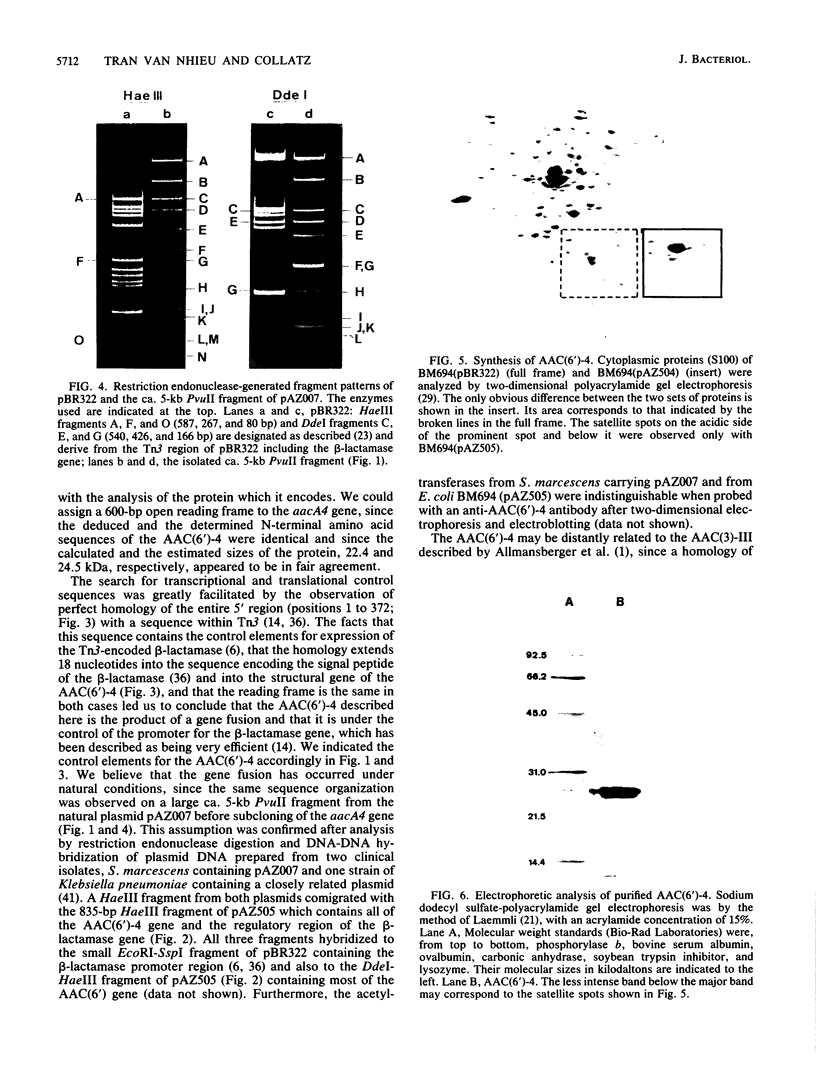
The gene aacA4 encoding an aminoglycoside 6'-N-acetyltransferase, AAC(6')-4, was cloned from a natural multiresistance plasmid, and its nucleotide sequence was determined. The gene was 600 base pairs (bp) long, and the AAC(6')-4 had a calculated molecular size of 22.4 kilodaltons and an isoelectric point of 5.35. The sequence of the 17 N-terminal amino acids was determined from the purified enzyme. The AAC(6')-4 gene was part of a resistance gene cluster, and its expression was under the control of the regulatory sequences of the beta-lactamase encoded by Tn3. The five N-terminal amino acids were identical to those of the signal peptide of the Tn3-encoded beta-lactamase, and the entire 5' region of aacA4, as far as it was sequenced (354 bp, including the promoter and the ribosome-binding site sequences), was identical to that of the beta-lactamase gene. This led us to presume an in vivo fusion between the beta-lactamase and the acetyltransferase genes. The latter was followed, in a polycistronic arrangement, by an aminoglycoside 3",9-adenylyltransferase gene, aadA, with an intergenic region of 68 bp. At a distance of ca. 1.3 kilobases in the 3' direction, we found remnants of a second Tn3-like element specifying an active beta-lactamase. At their 5' extremities, the two incomplete copies of Tn3, which were in tandem orientation, were interrupted within the resolvase gene. We speculate that Tn3-related sequences have played a role in the process of selection and dissemination of the AAC(6')-4 gene, which specifies resistance to amikacin and related aminoglycosides.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allmansberger R., Bräu B., Piepersberg W. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol Gen Genet. 1985;198(3):514–520. doi: 10.1007/BF00332949. [DOI] [PubMed] [Google Scholar]
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Enzymatic acetylation of aminoglycoside antibiotics by Escherichia coli carrying an R factor. Biochemistry. 1971 May 11;10(10):1787–1796. doi: 10.1021/bi00786a009. [DOI] [PubMed] [Google Scholar]
- Betts R. F., Valenti W. M., Chapman S. W., Chonmaitree T., Mowrer G., Pincus P., Messner M., Robertson R. Five-year surveillance of aminoglycoside usage in a university hospital. Ann Intern Med. 1984 Feb;100(2):219–222. doi: 10.7326/0003-4819-100-2-219. [DOI] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Bräu B., Pilz U., Piepersberg W. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol Gen Genet. 1984;193(1):179–187. doi: 10.1007/BF00327434. [DOI] [PubMed] [Google Scholar]
- Crameri R., Davies J. E. Increased production of aminoglycosides associated with amplified antibiotic resistance genes. J Antibiot (Tokyo) 1986 Jan;39(1):128–135. doi: 10.7164/antibiotics.39.128. [DOI] [PubMed] [Google Scholar]
- Davies J., Smith D. I. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol. 1978;32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Ferretti J. J., Gilmore K. S., Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6'-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986 Aug;167(2):631–638. doi: 10.1128/jb.167.2.631-638.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. S., Fitch W. M. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol Biol Evol. 1983 Dec;1(1):57–66. doi: 10.1093/oxfordjournals.molbev.a040298. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Davies J. Characterization of the plasmids comprising the "R factor" R5 and their relationships to other R plasmids. Plasmid. 1980 May;3(3):260–277. doi: 10.1016/0147-619x(80)90040-2. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Giles I. G., Akhtar M. The sequence of an antibiotic resistance gene from an antibiotic-producing bacterium. Homologies with transposon genes. FEBS Lett. 1983 Aug 22;160(1-2):67–71. doi: 10.1016/0014-5793(83)80937-5. [DOI] [PubMed] [Google Scholar]
- Hollingshead S., Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985 Jan;13(1):17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Kawabe H., Naito T., Mitsuhashi S. Acetylation of amikacin, a new semisynthetic antibiotic, by Pseudomonas aeruginosa carrying an R factor. Antimicrob Agents Chemother. 1975 Jan;7(1):50–54. doi: 10.1128/aac.7.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H., Naito T., Nakagawa S., Fujisawa K. I. BB-K 8, a new semisynthetic aminoglycoside antibiotic. J Antibiot (Tokyo) 1972 Dec;25(12):695–708. doi: 10.7164/antibiotics.25.695. [DOI] [PubMed] [Google Scholar]
- Koshland D., Sauer R. T., Botstein D. Diverse effects of mutations in the signal sequence on the secretion of beta-lactamase in Salmonella typhimurium. Cell. 1982 Oct;30(3):903–914. doi: 10.1016/0092-8674(82)90295-1. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Gerbaud G., Courvalin P. Translocation of sequences encoding antibiotic resistance from the chromosome to a receptor plasmid in Salmonella ordonez. Mol Gen Genet. 1981;182(3):390–408. doi: 10.1007/BF00293927. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine J. F., Maslow M. J., Leibowitz R. E., Pollock A. A., Hanna B. A., Schaefler S., Simberkoff M. S., Rahal J. J., Jr Amikacin-resistant gram-negative bacilli: correlation of occurrence with amikacin use. J Infect Dis. 1985 Feb;151(2):295–300. doi: 10.1093/infdis/151.2.295. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Wiedemann B. Amikacin resistance mediated by multiresistance transposon Tn2424. J Bacteriol. 1983 Aug;155(2):755–760. doi: 10.1128/jb.155.2.755-760.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M. M., de Jongh C. A., Schimpff S. C., Tillman G. L. Long-term amikacin use. Effects on aminoglycoside susceptibility patterns of gram-negative bacilli. JAMA. 1982 Sep 10;248(10):1199–1202. doi: 10.1001/jama.248.10.1199. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Price K. E., Kresel P. A., Farchione L. A., Siskin S. B., Karpow S. A. Epidemiological studies of aminoglycoside resistance in the U.S.A. J Antimicrob Chemother. 1981 Jul;8 (Suppl A):89–105. doi: 10.1093/jac/8.suppl_a.89. [DOI] [PubMed] [Google Scholar]
- Radika K., Northrop D. B. Purification of two forms of kanamycin acetyltransferase from Escherichia coli. Arch Biochem Biophys. 1984 Aug 15;233(1):272–285. doi: 10.1016/0003-9861(84)90626-x. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Kumada T., Hsieh W. C., Chung H. Y., Chong Y., Hare R. S., Miller G. H., Sabatelli F. J., Howard J. Comparison of aminoglycoside resistance patterns in Japan, Formosa, and Korea, Chile, and the United States. Antimicrob Agents Chemother. 1985 Aug;28(2):282–288. doi: 10.1128/aac.28.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tait R. C., Rempel H., Rodriguez R. L., Kado C. I. The aminoglycoside-resistance operon of the plasmid pSa: nucleotide sequence of the streptomycin-spectinomycin resistance gene. Gene. 1985;36(1-2):97–104. doi: 10.1016/0378-1119(85)90073-3. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. E., Roberts M., Woloj M., Crosa J. H. Molecular cloning of amikacin resistance determinants from a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1986 Aug;30(2):315–320. doi: 10.1128/aac.30.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Van Nhieu G. T., Goldstein F. W., Pinto M. E., Acar J. F., Collatz E. Transfer of amikacin resistance by closely related plasmids in members of the family Enterobacteriaceae isolated in Chile. Antimicrob Agents Chemother. 1986 May;29(5):833–837. doi: 10.1128/aac.29.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. On the hydrophobic nature of signal sequences. Eur J Biochem. 1981 May 15;116(2):419–422. doi: 10.1111/j.1432-1033.1981.tb05351.x. [DOI] [PubMed] [Google Scholar]