Abstract
Resistance against the aphid Macrosiphum euphorbiae previously was observed in tomato and attributed to a novel gene, designated Meu-1, tightly linked to the nematode resistance gene, Mi. Recent cloning of Mi allowed us to determine whether Meu-1 and Mi are the same gene. We show that Mi is expressed in leaves, that aphid resistance is isolate-specific, and that susceptible tomato transformed with Mi is resistant to the same aphid isolates as the original resistant lines. We conclude that Mi and Meu-1 are the same gene and that Mi mediates resistance against both aphids and nematodes, organisms belonging to different phyla. Mi is the first example of a plant resistance gene active against two such distantly related organisms. Furthermore, it is the first isolate-specific insect resistance gene to be cloned and belongs to the nucleotide-binding, leucine-rich repeat family of resistance genes.
The potato aphid Macrosiphum euphorbiae is an agricultural pest of several crops, causing direct and indirect damage on tomato including transmission of phytopathogenic viruses (1, 2). A single locus, named “Meu-1,” that confers resistance against the potato aphid M. euphorbiae has been described in tomato as tightly linked to the gene Mi, which confers resistance against Meloidogyne incognita and other species of root-knot nematode (3). Mi was introduced into cultivated tomato, Lycopersicon esculentum, from its wild relative L. peruvianum (4, 5). To date, all tomato lines carrying Mi have been found to be resistant to specific isolates of the potato aphid, suggesting that Meu-1 was incorporated into tomato from L. peruvianum along with Mi and that Meu-1 corresponded to either Mi or a closely linked gene.
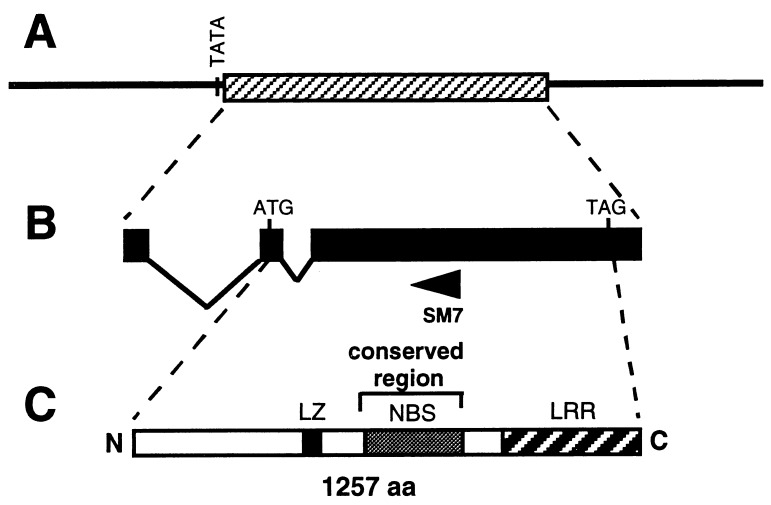
Recently, the Mi gene was isolated by a positional cloning approach (6, 7). Mi was localized to a small region of tomato chromosome 6 by identification of recombinants, and DNA sequence analysis was carried out to identify Mi candidates. Sequencing revealed two genes, Mi-1.1 and Mi-1.2, that were 95% identical to each other and encoded proteins with high similarity to previously cloned plant resistance genes. Complementation analysis showed that introduction of Mi-1.2, but not Mi-1.1, to susceptible tomato was sufficient to confer a nematode-resistant phenotype with the same spectrum of resistance as Mi (7). Mi-1.2 produces a transcript of ≈4 kb that encodes a putative protein of 1,257 amino acids (Fig. 1). This protein is a member of a family of disease resistance-associated plant proteins characterized by the presence of a nucleotide binding site and a leucine-rich repeat region (8, 9). Highest similarity to characterized resistance gene sequences is to the tomato gene Prf, which, together with Pto, is required for resistance to P. syringae carrying the avirulence gene avrPto (10).
Figure 1.
Structure of Mi-1.2. (A) The 14.7-kb genomic clone carrying the Mi-1.2 primary transcript (cross-hatched bar) and flanking sequences (line). This fragment is sufficient to confer resistance to root-knot nematodes to susceptible tomato (7). (B) Mi-1.2 transcript with exons as solid bars and the two introns as angled lines. The arrow marked SM7 represents the approximate position of the primer used to isolate the 5′ ends of transcripts expressed in leaves. (C) Predicted protein structure of Mi-1.2. LZ, the position of a possible leucine zipper; NBS, a nucleotide binding site; LRR, leucine-rich repeat region (8, 9). The NBS-containing region that is highly conserved among many plant resistance-associated proteins is shown in gray.
The cloning of Mi provided the opportunity to determine the relationship between Mi and Meu-1. Our results demonstrate that Mi also is expressed in leaves, that Mi mediates resistance in an isolate-specific manner, and that Mi and Meu-1 are, in fact, the same gene.
MATERIALS AND METHODS
Plant Materials.
Two sets of nearly isogenic tomato lines were used: CastlerockII and Moneymaker, susceptible to nematodes and aphids, and the corresponding resistant lines Sun6082 and Motelle (carrying different length introgressed regions from L. peruvianum containing Mi), respectively (3). All plants were grown in a pesticide-free greenhouse (23–25°C, 16:8 L/D). The plants were grown in 1-gallon pots in Redi-Gro soil mixture amended with slow release fertilizer. Plants were watered daily with distilled water and fertilized weekly with a foliar application of Miracle-Gro (Stern’s Miracle-Gro Products, Port Washington, NY).
RNA Gel Blot Analysis.
RNA extraction and gel blot analysis were carried out as described (7). Leaf poly(A)+ RNA was isolated from 7-week-old plants, and 5 μg/lane was fractionated in a denaturing gel. The molecular marker used was the 0.24- to 9.5-kb ladder (GIBCO/BRL).
Rapid Amplification of 5′ cDNA Ends.
Rapid amplification of 5′ cDNA ends was carried out using a Marathon cDNA Amplification kit (CLONTECH). First-strand cDNA was synthesized from 1 μg of poly(A)+ RNA isolated from leaves of 7-week-old resistant cultivar Sun6082 by using poly(T) primer. After second-strand synthesis and adapter ligation, PCR was carried out with primer SM7, 5′-GGTCAAGAGGATCAGTGTTCAGATTTCC-3′, which hybridizes to a sequence present in both Mi-1.1 and Mi-1.2 (Fig. 1) and the adapter primer. The 2-kb PCR product obtained was cloned into the vector pCR2.1 (Invitrogen).
Aphid Isolates.
Two isolates of the potato aphid M. euphorbiae were used in the bioassays, WU-3 (a red isolate) and WU-4 (a green isolate). The colonies were established from aphids collected from tomato fields on the campus of University of California, Davis in the spring and summer of 1997, respectively. Isolates were identified as M. euphorbiae by M. G. Kinsey (Department of Entomology, University of California, Davis) and were reared in the greenhouse on tomato cultivar UC82. Type specimens are filed in the Bohart Museum, University of California, Davis.
Transgenic Plants.
The susceptible line Moneymaker was transformed with a 14.7-kb tomato genomic fragment, including the Mi-1.2 coding region and 4.62 kb of the 5′ and 4.77 kb of the 3′ regulatory sequences (Fig. 1A). Primary transformants 143–11 and 143–31 carried one copy of the introduced gene and displayed nematode resistance (7). Progeny of 143–11 segregated for nematode resistance, with resistance corresponding completely to the presence of the introduced gene. The presence of Mi-1.2 in progeny of both transgenic lines was determined by PCR by using primers C1/2 (5′-CAGTGAAGTGGAAGTGATGA-3′) and C2S4 (5′-CTAAGAGGAATCTCATCACAGG-3′) (7). The size of the Mi-1.2-specific product, which corresponded to a portion of the 3′ region of the gene, was 1.6 kb.
Aphid Resistance, Survivorship Assay.
Apterous aphids of isolates WU-3 and WU-4 were collected no more than 48 h after their adult molt and were caged individually on 5-week-old plants of resistant and susceptible lines. On each plant, 3 aphids were caged on the 4th, 5th, and 6th leaves below the first fully expanded leaf. There were 4 plants per genotype/aphid combination (total of 16 plants). The plants were arranged in a complete random design in the greenhouse, and aphid survival was monitored daily until all WU3 aphids on resistant plants were dead (9 days). For each plant, the average number of days (up to 9) that the aphids lived was calculated by using the methods of Carey (11). Data for each aphid isolate were analyzed separately by using one-way ANOVA and Welch’s weighted ANOVA (sas software release ver. 6.12).
Aphid Resistance, Whole Plant Assay.
Five 5-week-old plants of each genotype were tested with aphid isolates as indicated. Plants were placed in a mesh cage with aphid-infested plants (WU3 and WU4 were tested in separate cages), and 13 days later, the number of aphids (all stages of development) on the two most infested leaflets per experimental plant was determined. The average number of aphids per leaflet per plant was analyzed by using one-way ANOVA (sas software release 6.12).
RESULTS
Mi Is Expressed in Leaves.
Mi has at least six homologues, most of which are clustered in a 650-kb region (7). Transcripts corresponding to Mi-1.1, Mi-1.2, and at least two additional homologues are present in uninduced roots of resistant tomato lines. Because potato aphids feed from leaf tissue, we examined RNA extracted from leaves for the presence of transcripts corresponding to Mi or any of its homologues. The RNA gel blot showed that 4-kb transcripts, hybridizing to a probe derived from Mi-1.1, also are expressed in leaves of resistant tomato (Fig. 2). However, no transcripts were detected from leaves of susceptible tomato. To determine whether the leaf transcripts included Mi-1.1 and/or Mi-1.2, we carried out amplification of 5′ cDNA ends using RNA isolated from resistant tomato leaves as template and a primer (SM7) whose sequence is present in both Mi-1.1 and Mi-1.2 (Fig. 1). Eight clones were partially sequenced and five were found to be homologous to Mi. Four corresponded to Mi-1.2. The fifth homologous clone was not identical to either Mi-1.1 or Mi-1.2. This result demonstrated that Mi-1.2 is expressed in leaves of resistant tomato, supporting the possibility that Meu-1 corresponds to Mi-1.2 and indicating that there is at least one additional homologue expressed in leaves whose corresponding gene has not been cloned yet.
Figure 2.
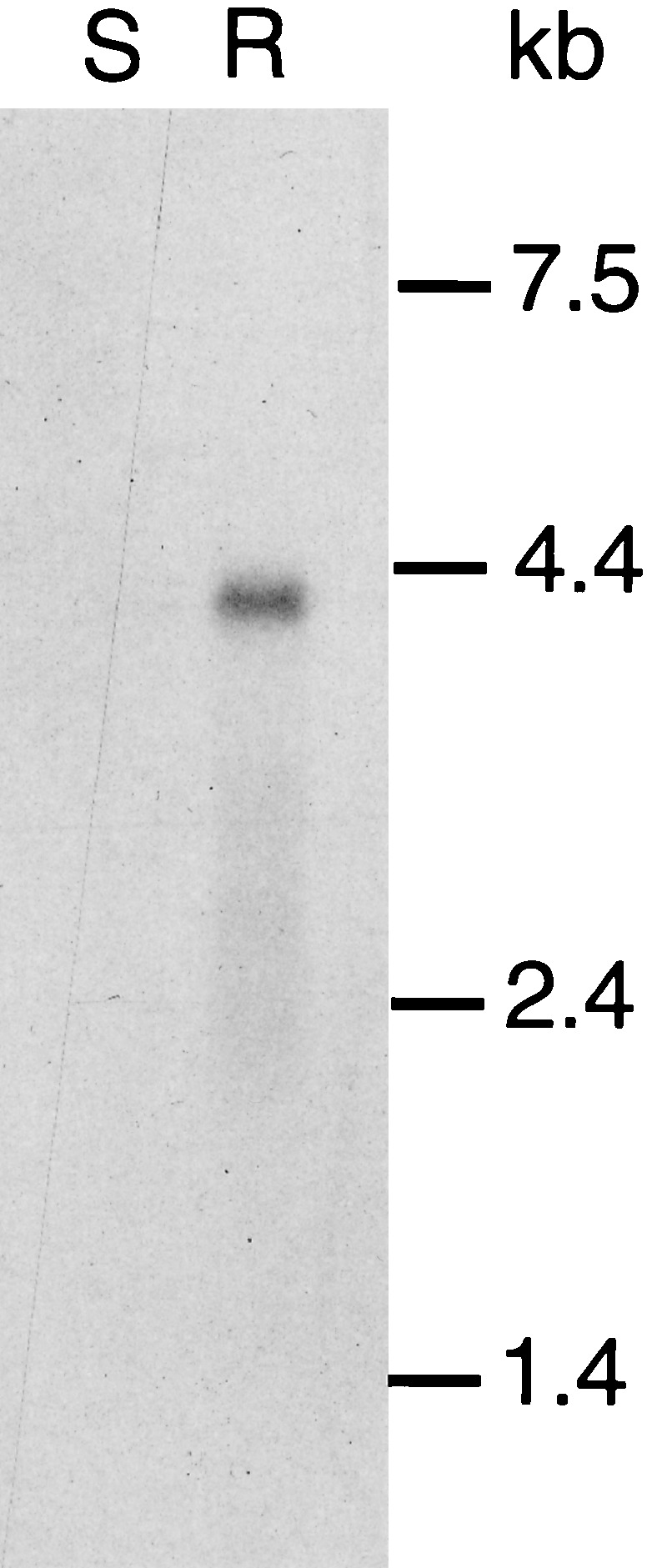
Mi expression in leaves. RNA gel blot hybridized with probe 3–3, which contains the Mi-1.1 nucleotide binding site (7). Poly(A)+ RNA of aphid-resistant tomato line Sun6082 (R) and susceptible line CastlerockII (S) was loaded 5 μg per lane.
Isolate Specificity of Aphid Resistance.
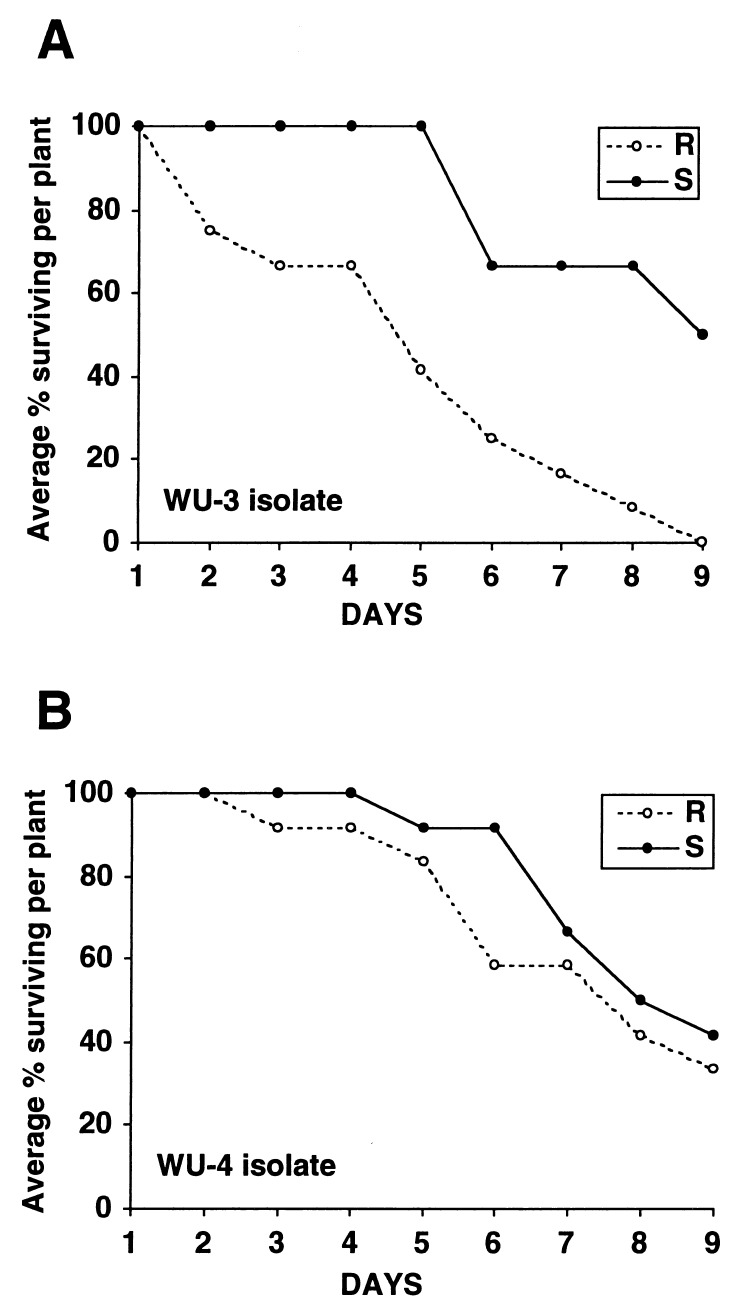
The survivorship of two aphid isolates, WU3 and WU4, was measured on resistant and susceptible nearly isogenic tomato lines, Sun6082 and CastlerockII. Survivorship of aphid isolate WU-3 was significantly lower on resistant plants than on susceptible plants (Welch’s weighted ANOVA, P = 0.0402) (Fig. 3A). After 9 days, all of the aphids on the resistant plants were dead whereas on the susceptible plants 50% were still alive. In contrast, there was no significant difference in the survivorship of WU-4 on resistant and susceptible lines (one-way ANOVA, P = 0.3626) (Fig. 3B). These data show that the aphid resistance we describe is isolate-specific and that red aphid isolate WU-3 is avirulent on the resistant cultivar whereas green aphid isolate WU-4 is not. Previous field tests showed that other green aphid populations were avirulent on the resistant tomato cultivars (3); thus, we cannot conclude that the aphid color correlates with the response.
Figure 3.
Survivorship of aphid isolates WU-3 and WU-4. Comparison of survivorship on a resistant line, Sun6082 (R), and a susceptible line, CastlerockII (S). (A) For aphid isolate WU-3, the survivorship was significantly higher on the susceptible than on the resistant tomato line (P = 0.0402). (B) For aphid isolate WU-4, there was no significant difference in survivorship on resistant and susceptible tomato lines (P = 0.3626).
Aphid Resistance on Transgenic Lines.
To determine whether the cloned gene Mi-1.2 could confer resistance to aphids, progeny of the primary transformants 143–11 and 143–31, each carrying one copy of Mi-1.2, were tested in two independent experiments using the whole plant assay.
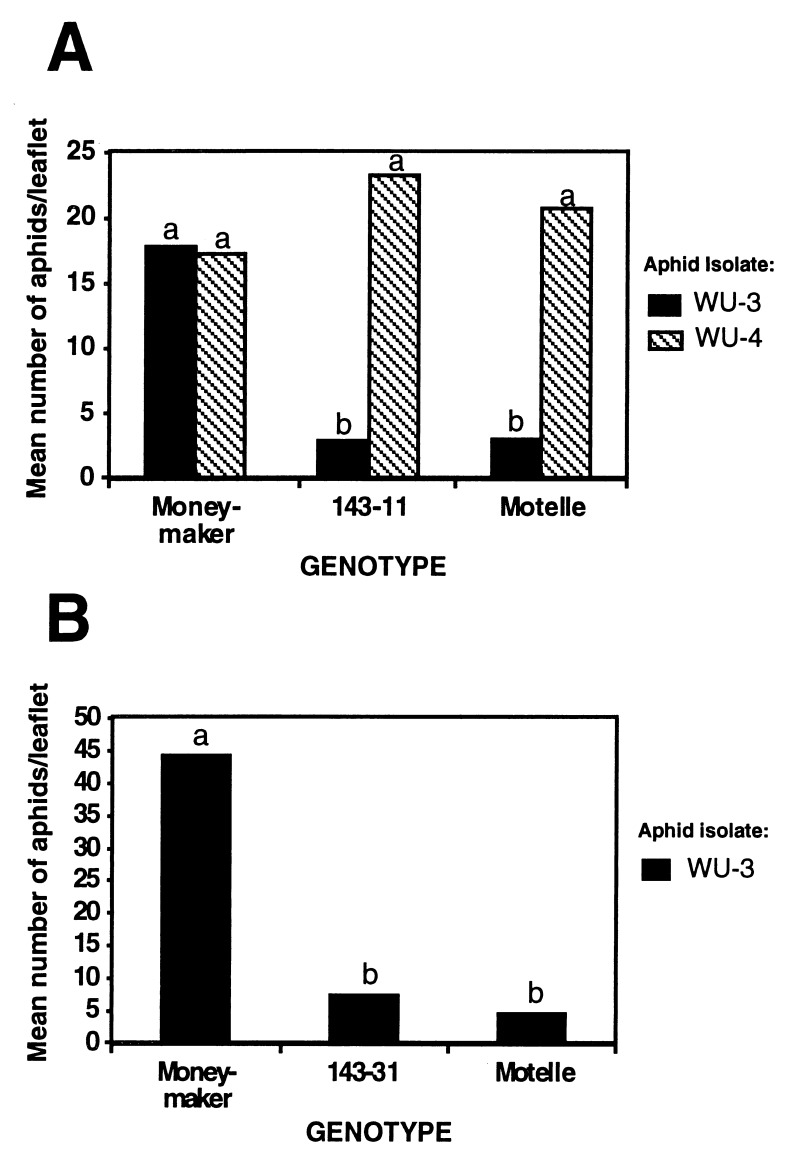
Progeny of 143–11 that carried the introduced gene were challenged with both aphids isolates, WU-3 and WU-4 (Fig. 4A). The number of WU-3 on 143–11 progeny was significantly lower than the number on the susceptible line Moneymaker (one-way ANOVA, P = 0.0107) and did not differ significantly from that found on the resistant line Motelle. Isolate WU-4 was found in similar numbers on Moneymaker, Motelle, and 143–11 progeny. There was no significant difference between aphid number on 143–11 progeny and Moneymaker (one-way ANOVA, P = 0.0649). The number of WU-3 aphids on 143–31 progeny carrying the introduced gene was similar to that found on the resistant line Motelle (Fig. 4B).
Figure 4.
Genetic complementation for aphid resistance in susceptible tomato after transformation with Mi-1.2. Comparison of the mean number of aphids per leaflet in independent experiments between two transgenic lines, 143–11 (A) and 143–31 (B), and the control lines Moneymaker (susceptible) and Motelle (resistant). Five plants per genotype were exposed to each of two different isolates of M. euphorbiae WU-3 (black bar) and WU-4 (cross-hatched bar). Significantly different means within each experiment are indicated by different letters according to Tukey HSD test (P < 0.05).
Thus, Mi-1.2 is sufficient to confer, to susceptible tomato, aphid resistance with the same specificity as the putative Meu-1 gene, supporting the conclusion that Mi-1.2 corresponds to both Mi and Meu-1. Hereafter, we will refer to the gene by its original name, “Mi.”
DISCUSSION
Our results show that Mi is expressed in leaves of resistant tomato and that this gene mediates aphid resistance in an isolate-specific manner. We conclude that Mi has dual specificity, conferring resistance against aphids and nematodes, organisms in different phyla. This demonstrates that a member of the nucleotide-binding, leucine-rich repeat family of resistance genes is able to confer isolate-specific resistance against an insect. Other members of this plant gene family mediate resistance through direct or indirect recognition of specific pathogen elicitors or avirulence gene products. These are referred to as gene-for-gene interactions (8, 9). Our finding that Mi-mediated aphid resistance is, like nematode resistance (12, 13), isolate-specific strongly suggests that the resistance we describe also functions via a gene-for-gene interaction. In this case, in which a single gene mediates resistance to two such diverse organisms, intriguing questions arise about the possible mechanism of recognition. It seems unlikely that the Mi-encoded protein interacts with identical ligands that are present in nematodes and aphids, particularly because, for both M. euphorbiae and root-knot nematodes, there is within-species variability in effectiveness of Mi-mediated resistance.
There are several other possibilities. First, Mi may be capable of mediating a response to different ligands. There is evidence that other R genes respond to more than one pathogen elicitor. The best studied example is the Arabidopsis gene RPM1, the encoded protein of which shares the same sequence motifs and general structure as Mi. This gene confers resistance to P. syringae strains carrying the unrelated avirulence genes, avrRpm1 and avrB (14, 15). Whether RPM1 physically interacts with each of these avirulence proteins is still unknown. The finding that Mi mediates resistance against pests in two diverse phyla may be a dramatic example of dual specificity of recognition by a single resistance gene. Second, Mi may not directly recognize a ligand from either pest but instead encode a component involved in the signal transduction cascade between primary receptors and the resistance response. In this case, specific receptors would trigger the same signal transduction pathway via Mi. Under this hypothesis, all components besides Mi would be present in susceptible tomato.
Whether the defense response mediated by Mi is the same for aphids and nematodes is another interesting issue. Superficially, these pests bear little resemblance to each other. One is an obligate endoparasite of roots; the other is an insect that feeds from the phloem of leaves. However, on closer examination, there are similarities in feeding between aphids and nematodes. Nematodes penetrate roots near the tip and migrate intercellularly to a feeding site in the developing vascular cylinder (16). Here, the nematodes initiate the formation of specialized feeder cells in the host from which they derive nutrients, by removing cytoplasmic fluids through the stylet, during their development as endoparasites. Aphids feed by inserting their stylets intercellularly, forming a sheath with saliva and ultimately penetrating the phloem sieve element and ingesting plant fluids (17). A mechanism that blocks ingestion of nutrients by inducing physical or chemical changes in penetrated cells or sieve elements is an example of a defense that could function against such diverse pests.
In resistant plants, nematodes do not establish feeding cells; a localized necrosis or hypersensitive response (HR) is observed near the site where feeding cells would normally be initiated, and the nematodes die or leave the roots (18–20). Whether the HR is the direct cause of resistance has not yet been determined. The mechanism of aphid resistance is unknown. The presence of Mi correlates with increased aphid mortality and reduced fecundity on resistant plants (Fig. 3A; ref. 21). Electronic monitoring experiments indicate that ingestion from phloem sieve elements is reduced significantly in resistant tomato (I.K., unpublished data). No HR has been described so far for Mi-mediated aphid resistance, but this could be because of the limited investigation. The predicted sequence of the protein encoded by Mi suggests a cytoplasmic location. If this gene product directly recognizes products of the pathogen, then these products should be present inside the plant cell, possibly injected via nematode or aphid stylets. For the aphid, the recognized molecule could be of either aphid or endosymbiont origin.
Studies of plant resistance against insects have centered mostly on chemical and morphological defenses (22–25). General defenses that are induced by previous exposure to pests and are effective against broad categories of insects also have been well investigated (26, 27). Fewer studies have addressed specific defense responses involving gene-for-gene interactions, which often include a hypersensitive response and callose deposition, a frequent mechanism of plant defense against bacterial, viral, and fungal pathogens (28). With piercing–sucking insects, in contrast to chewing insects, there is an intimate and prolonged interaction between the insect’s stylets and plant cells, providing conditions under which a specific gene-for-gene defense response could be highly effective (22).
There is evidence that other sources of resistance to piercing–sucking insects may be mediated by single resistance genes of the host/pathogen gene-for-gene resistance type. For example, the lettuce gene Nr, for resistance against the aphid Nasonovia ribisnigri, is effective only against one aphid species and, like Mi, results in greatly reduced phloem ingestion (29–31). In another example, the recently mapped gene Sd1 that confers resistance to the rosy leaf curling aphid Dysaphis devecta in apple is effective against aphids biotypes 1 and 2 but not to a third biotype (32). Triticum aestivum resistance to Hessian fly, Mayetiola destructor (Say), also has been demonstrated to be a gene-for-gene mechanism (33). Although no genes have been cloned yet, 26 resistance genes have been described as being effective against 13 biotypes of Hessian fly (34, 35). The gene-for-gene type of resistance mediated by NBS/LRR genes in insects may be a widespread and largely untapped resource for introducing important insect resistance into plants.
Acknowledgments
We thank Jorge Dubcovsky and Diego Lijavetzky for assistance with statistical analysis, Sandy Kelley, Marv Kinsey, and Christine Quintero for assistance with the aphids and greenhouse work, and Shinichiro Wachi for contributions to the DNA sequence analysis. We also thank George Bruening and Jorge Dubcovsky for helpful comments on the manuscript. This work was supported by National Science Foundation Award IBN-972 3679 to V.M.W. and D.E.U. and by National Science Foundation Cooperative Agreement BIR-8920216 to the Center for Engineering Plants for Resistance Against Pathogens (CEPRAP), a National Science Foundation Science and Technology Center.
Footnotes
References
- 1.Kok-Yokomi M L. Ph.D. thesis. Davis: University of California; 1978. [Google Scholar]
- 2.Braithwaite B M, Blake C D. Aust J Agric Res. 1961;12:1100–1107. [Google Scholar]
- 3.Kaloshian I, Lange W H, Williamson V M. Proc Natl Acad Sci USA. 1995;92:622–625. doi: 10.1073/pnas.92.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith P G. Proc Am Soc Hortic Sci. 1944;44:413–416. [Google Scholar]
- 5.Medina-Filho H P, Tanksley S D. In: Handbook of Plant Cell Culture. Evans D A, Sharp W R, Ammirato P V, Yamada Y, editors. Vol. 1. New York: MacMillan; 1983. pp. 904–923. [Google Scholar]
- 6.Kaloshian I, Yaghoobi J, Liharska T, Hontelez J, Hanson D, Hogan P, Jesse T, Wijbrandi J, Simons G, Vos P, et al. Mol Gen Genet. 1998;257:376–385. doi: 10.1007/s004380050660. [DOI] [PubMed] [Google Scholar]
- 7.Milligan, S. B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P. & Williamson, V. M. (1998) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 8.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 9.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 10.Salmeron J M, Oldroyd G E D, Rommens C M T, Scofield S R, Kim H S, Lavelle D T, Dahlbeck D, Staskawicz B J. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 11.Carey J R. Applied Demography for Biologists with Special Emphasis on Insects. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 12.Bost S C, Triantaphyllou A C. J Nematol. 1982;14:540–544. [PMC free article] [PubMed] [Google Scholar]
- 13.Jarquin-Barberena H, Dalmasso A, de Guiran G, Cardin M C. Rev Nematol. 1991;14:299–303. [Google Scholar]
- 14.Bisgrove S R, Simonich M T, Smith N M, Sattler A, Innes W R. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 16.Williamson V M, Hussey R S. Plant Cell. 1996;8:1735–1745. doi: 10.1105/tpc.8.10.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean D L, Kinsey M G. Bull Entomol Soc Am. 1984;30:26–31. [Google Scholar]
- 18.Dropkin V H. Phytopathol. 1969;59:1632–1637. [Google Scholar]
- 19.Paulson R E, Webster J M. Physiol Plant Pathol. 1972;2:227–234. [Google Scholar]
- 20.Ho J Y, Weide R, Ma H M, van Wordragen F, Lambert K N, Koornneef M, Zabel P, Williamson V M. Plant J. 1992;2:971–982. [PubMed] [Google Scholar]
- 21.Kaloshian I, Kinsey M G, Ullman D E, Williamson V M. Entomol Exp Appl. 1997;83:181–187. [Google Scholar]
- 22.Fernandes G W. Environ Entomol. 1990;19:1173–1182. [Google Scholar]
- 23.Goffreda J C, Mutschler M A. Theor Appl Genet. 1989;78:210–216. doi: 10.1007/BF00288801. [DOI] [PubMed] [Google Scholar]
- 24.Down R E, Gatehouse A M R, Hamilton W D O, Gatehouse J A. J Insect Physiol. 1996;42:1035–1045. [Google Scholar]
- 25.Gatehouse A M R, Down R E, Powell K S, Sauvion N, Rahbe Y, Newell C A, Merryweather A, Hamilton W D O, Gatehouse J A. Entomol Exp Appl. 1996;79:295–307. [Google Scholar]
- 26.Stout M J, Workman K V, Bostock R M, Duffey S S. Oecologia. 1998;113:74–81. doi: 10.1007/s004420050355. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal A A. Science. 1998;279:1201–1202. doi: 10.1126/science.279.5354.1201. [DOI] [PubMed] [Google Scholar]
- 28.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eenink A H, Groenwold R, Dieleman F L. Euphytica. 1982;31:291–300. [Google Scholar]
- 30.Eenink A H, Dieleman F L, Groenwold R. Euphytica. 1982;31:301–304. [Google Scholar]
- 31.van Helden M, Tjallingii W F, Dieleman F L. Entomol Exp Appl. 1993;66:53–58. [Google Scholar]
- 32.Roche P, Alston F H, Maliepaard C, Evans K M, Vrielink R, Dunemann F, Markussen T, Tartarini S, Brown L M, Ryder C, et al. Theor Appl Genet. 1997;94:528–533. [Google Scholar]
- 33.Hatchett J, Gallun R. Ann Entomol Soc Am. 1970;63:1400–1407. [Google Scholar]
- 34.Dweikat I, Ohm H, Mackenzie S, Patterson F, Cambron S, Ratcliffe R. Theor Appl Genet. 1994;89:964–968. doi: 10.1007/BF00224525. [DOI] [PubMed] [Google Scholar]
- 35.Dweikat I, Ohm H, Patterson F, Cambron S. Theor Appl Genet. 1997;94:419–423. doi: 10.1007/BF00224525. [DOI] [PubMed] [Google Scholar]