Abstract
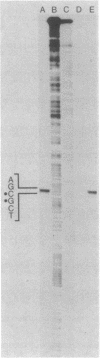
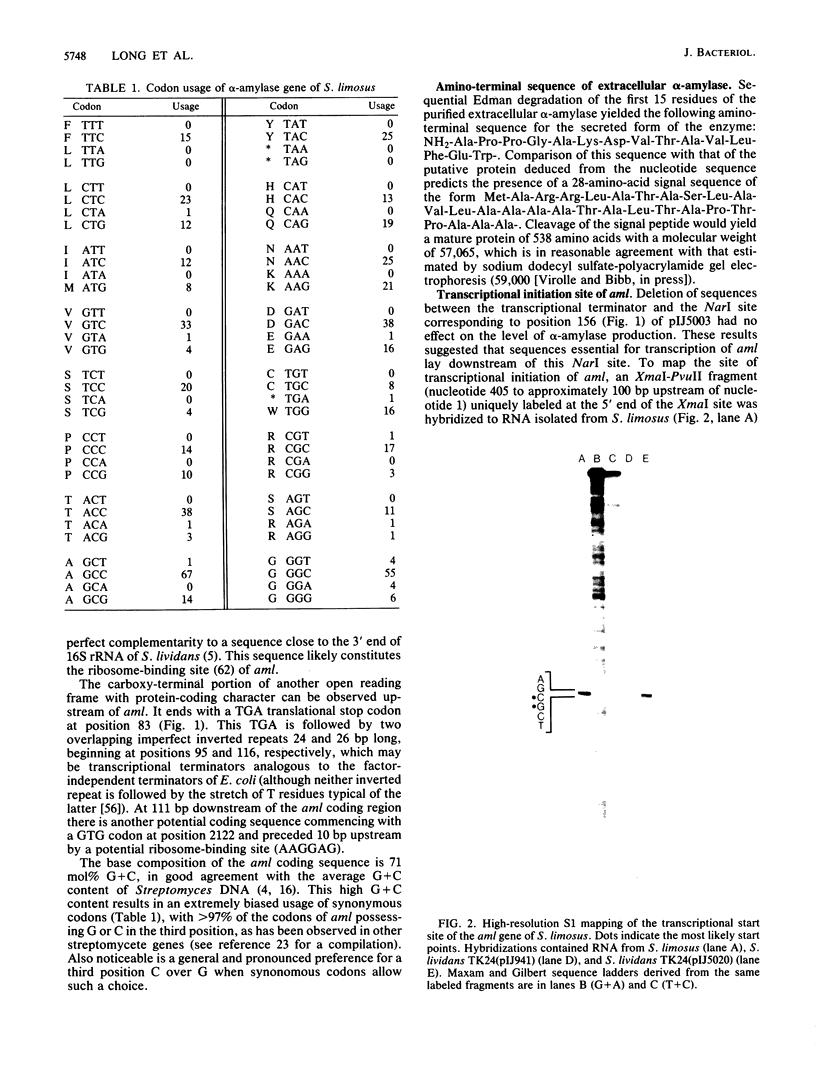
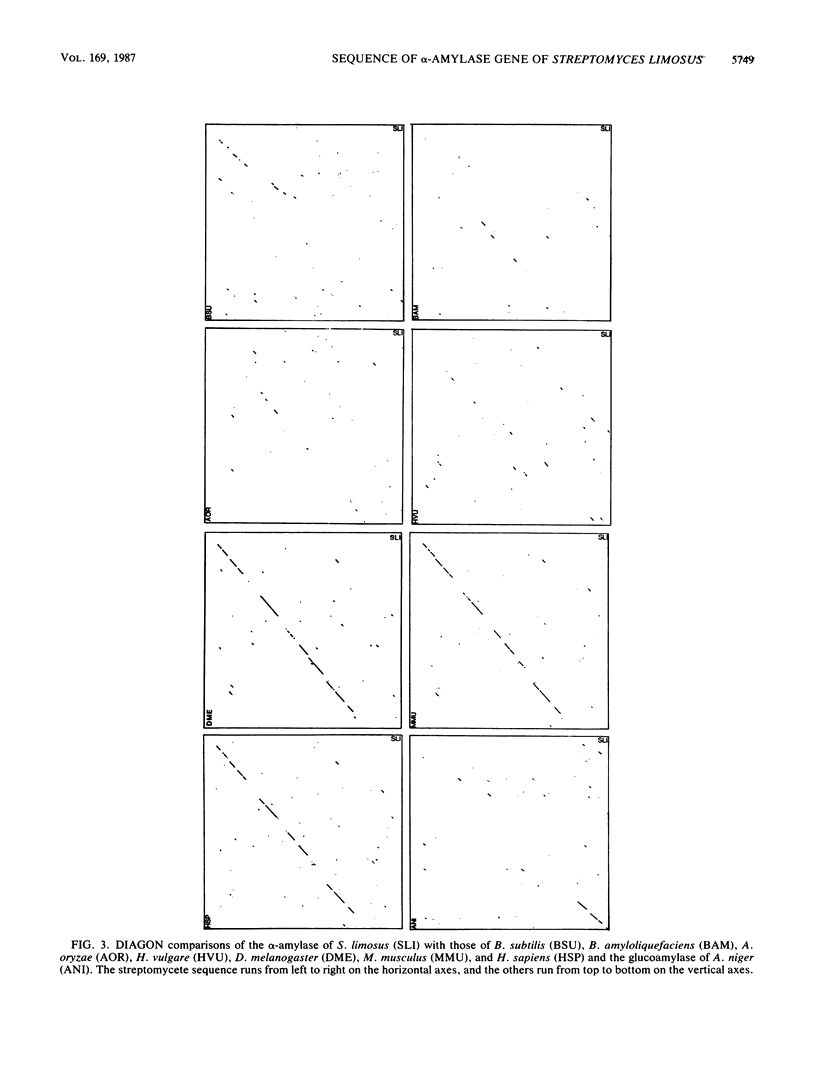
The nucleotide sequence of the coding and regulatory regions of the alpha-amylase gene (aml) of Streptomyces limosus was determined. High-resolution S1 mapping was used to locate the 5' end of the transcript and demonstrated that the gene is transcribed from a unique promoter. The predicted amino acid sequence has considerable identity to mammalian and invertebrate alpha-amylases, but not to those of plant, fungal, or eubacterial origin. Consistent with this is the susceptibility of the enzyme to an inhibitor of mammalian alpha-amylases. The amino-terminal sequence of the extracellular enzyme was determined, revealing the presence of a typical signal peptide preceding the mature form of the alpha-amylase.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahmsén L., Moks T., Nilsson B., Hellman U., Uhlén M. Analysis of signals for secretion in the staphylococcal protein A gene. EMBO J. 1985 Dec 30;4(13B):3901–3906. doi: 10.1002/j.1460-2075.1985.tb04164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschauer H., Vértesy L., Braunitzer G. Die Sequenz des alpha-Amylaseinhibitors Hoe-467 A (alphs-Amylaseinaktivator Hoe-467 A) aus Streptomyces tendae 4158. Hoppe Seylers Z Physiol Chem. 1981 Apr;362(4):465–467. [PubMed] [Google Scholar]
- Benigni R., Petrov P. A., Carere A. Estimate of the genome size by renaturation studies in Streptomyces. Appl Microbiol. 1975 Aug;30(2):324–326. doi: 10.1128/am.30.2.324-326.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Jones G. H., Joseph R., Buttner M. J., Ward J. M. The agarase gene (dag A) of Streptomyces coelicolor A3(2): affinity purification and characterization of the cloned gene product. J Gen Microbiol. 1987 Aug;133(8):2089–2096. doi: 10.1099/00221287-133-8-2089. [DOI] [PubMed] [Google Scholar]
- Boel E., Hjort I., Svensson B., Norris F., Norris K. E., Fiil N. P. Glucoamylases G1 and G2 from Aspergillus niger are synthesized from two different but closely related mRNAs. EMBO J. 1984 May;3(5):1097–1102. doi: 10.1002/j.1460-2075.1984.tb01935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer P. H., Hickey D. A. The alpha-amylase gene in Drosophila melanogaster: nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res. 1986 Nov 11;14(21):8399–8411. doi: 10.1093/nar/14.21.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987 Aug;209(1):101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- Béguin P., Cornet P., Aubert J. P. Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J Bacteriol. 1985 Apr;162(1):102–105. doi: 10.1128/jb.162.1.102-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S. Engineering for protein secretion in gram-positive bacteria. Methods Enzymol. 1987;153:507–516. doi: 10.1016/0076-6879(87)53075-0. [DOI] [PubMed] [Google Scholar]
- Chapon C., Raibaud O. Structure of two divergent promoters located in front of the gene encoding pullulanase in Klebsiella pneumoniae and positively regulated by the malT product. J Bacteriol. 1985 Nov;164(2):639–645. doi: 10.1128/jb.164.2.639-645.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Débarbouillé M., Shuman H. A., Silhavy T. J., Schwartz M. Dominant constitutive mutations in malT, the positive regulator gene of the maltose regulon in Escherichia coli. J Mol Biol. 1978 Sep 15;124(2):359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- Freundlieb S., Boos W. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986 Feb 25;261(6):2946–2953. [PubMed] [Google Scholar]
- Hagenbüchle O., Bovey R., Young R. A. Tissue-specific expression of mouse-alpha-amylase genes: nucleotide sequence of isoenzyme mRNAs from pancreas and salivary gland. Cell. 1980 Aug;21(1):179–187. doi: 10.1016/0092-8674(80)90125-7. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann O., Vértesy L., Braunitzer G. The primary structure of alpha-amylase inhibitor Z-2685 from Streptomyces parvullus FH-1641. Sequence homology between inhibitor and alpha-amylase. Biol Chem Hoppe Seyler. 1985 Dec;366(12):1161–1168. doi: 10.1515/bchm3.1985.366.2.1161. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Hoshiko S., Makabe O., Nojiri C., Katsumata K., Satoh E., Nagaoka K. Molecular cloning and characterization of the Streptomyces hygroscopicus alpha-amylase gene. J Bacteriol. 1987 Mar;169(3):1029–1036. doi: 10.1128/jb.169.3.1029-1036.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Kishida H., Okanishi M. Molecular cloning of a xylanase gene from Streptomyces sp. No. 36a and its expression in streptomycetes. J Antibiot (Tokyo) 1986 Jul;39(7):985–993. doi: 10.7164/antibiotics.39.985. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. I. Los Alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982 Jan 11;10(1):183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kendall K., Cullum J. Cloning and expression of an extracellular-agarase from Streptomyces coelicolor A3(2) in Streptomyces lividans 66. Gene. 1984 Sep;29(3):315–321. doi: 10.1016/0378-1119(84)90060-x. [DOI] [PubMed] [Google Scholar]
- Kluh I. Amino acid sequence of hog pancreatic alpha-amylase isoenzyme I. FEBS Lett. 1981 Dec 28;136(2):231–234. doi: 10.1016/0014-5793(81)80624-2. [DOI] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- MacDonald R. J., Crerar M. M., Swain W. F., Pictet R. L., Thomas G., Rutter W. J. Structure of a family of rat amylase genes. Nature. 1980 Sep 11;287(5778):117–122. doi: 10.1038/287117a0. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Wong H. C., Ting Y. E., Van Arsdell J. N., Chang S. Control of lysogeny and immunity of Bacillus subtilis temperate bacteriophage SP beta by its d gene. J Bacteriol. 1986 Sep;167(3):952–959. doi: 10.1128/jb.167.3.952-959.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondou F., Shareck F., Morosoli R., Kluepfel D. Cloning of the xylanase gene of Streptomyces lividans. Gene. 1986;49(3):323–329. doi: 10.1016/0378-1119(86)90368-9. [DOI] [PubMed] [Google Scholar]
- Murai H., Hara S., Ikenaka T., Goto A., Arai M., Murao S. Amino acid sequence of protein alpha-amylase inhibitor from Streptomyces griseosporeus YM-25. J Biochem. 1985 Apr;97(4):1129–1133. doi: 10.1093/oxfordjournals.jbchem.a135157. [DOI] [PubMed] [Google Scholar]
- Murphy N., McConnell D. J., Cantwell B. A. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase. Nucleic Acids Res. 1984 Jul 11;12(13):5355–5367. doi: 10.1093/nar/12.13.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide T., Emi M., Nakamura Y., Matsubara K. Corrected sequences of cDNAs for human salivary and pancreatic alpha-amylases [corrected]. Gene. 1984 May;28(2):263–270. doi: 10.1016/0378-1119(84)90265-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Gutierrez C., Schwartz M. Essential and nonessential sequences in malPp, a positively controlled promoter in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1201–1208. doi: 10.1128/jb.161.3.1201-1208.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1984 Jun 25;259(12):7577–7583. [PubMed] [Google Scholar]
- Robbins P. W., Wirth D. F., Hering C. Expression of the Streptomyces enzyme endoglycosidase H in Escherichia coli. J Biol Chem. 1981 Oct 25;256(20):10640–10644. [PubMed] [Google Scholar]
- Rogers J. C. Conserved amino acid sequence domains in alpha-amylases from plants, mammals, and bacteria. Biochem Biophys Res Commun. 1985 Apr 16;128(1):470–476. doi: 10.1016/0006-291x(85)91702-4. [DOI] [PubMed] [Google Scholar]
- Rogers J. C., Milliman C. Isolation and sequence analysis of a barley alpha-amylase cDNA clone. J Biol Chem. 1983 Jul 10;258(13):8169–8174. [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J. DNA sequence analysis by primed synthesis. Methods Enzymol. 1980;65(1):560–580. doi: 10.1016/s0076-6879(80)65060-5. [DOI] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takkinen K., Pettersson R. F., Kalkkinen N., Palva I., Söderlund H., Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [PubMed] [Google Scholar]
- Uhlén M., Guss B., Nilsson B., Gatenbeck S., Philipson L., Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984 Feb 10;259(3):1695–1702. [PubMed] [Google Scholar]
- Vértesy L., Oeding V., Bender R., Zepf K., Nesemann G. Tendamistat (HOE 467), a tight-binding alpha-amylase inhibitor from Streptomyces tendae 4158. Isolation, biochemical properties. Eur J Biochem. 1984 Jun 15;141(3):505–512. doi: 10.1111/j.1432-1033.1984.tb08221.x. [DOI] [PubMed] [Google Scholar]
- Vértesy L., Tripier D. Isolation and structure elucidation of an alpha-amylase inhibitor, AI-3688, from Streptomyces aureofaciens. FEBS Lett. 1985 Jun 3;185(1):187–190. doi: 10.1016/0014-5793(85)80767-5. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. T., Goodfellow M., Alderson G., Wellington E. M., Sneath P. H., Sackin M. J. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983 Jun;129(6):1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- Yamashita I., Suzuki K., Fukui S. Nucleotide sequence of the extracellular glucoamylase gene STA1 in the yeast Saccharomyces diastaticus. J Bacteriol. 1985 Feb;161(2):567–573. doi: 10.1128/jb.161.2.567-573.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]