Abstract
Promoter-proximal stalling, a general phenomenon observed during the expression of many RNA polymerase II transcribed genes, is dependent on transcription factor IIH (TFIIH). Reactions lacking TFIIH initiated transcription, but the transcription complex encountered a block to elongation proximal to the promoter. The accumulation of promoter-proximal stalled complexes was reduced in the presence of TFIIH and efficient escape from this site also required an activator. Promoter-proximal stalled complexes could not be induced to resume elongation. Our results indicate that effective recruitment of TFIIH into transcription complexes is achieved during formation of the preinitiation complex at the promoter. The studies establish that promoter clearance is a regulated event that requires TFIIH.
The steps of the transcription cycle were dissected in studies using the bacterial RNA polymerase (RNAP) (1, 2) and recently have been extended to the eukaryotic RNAP systems (3, 4). Transcription by RNAPII requires five transcription factors (TFs; TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) that are referred to as the general transcription factors (GTFs). Stable association of RNAPII with promoter sequences requires TFIID (or TBP), TFIIB, and TFIIF (5). However, after RNAPII has stably associated with promoter sequences, two additional factors, TFIIE and TFIIH, are necessary to modulate transcription (5–7). Their recruitment is perhaps related to a unique structure that is found at the C terminus of the largest subunit of RNAPII known as the C-terminal domain (CTD) (8).
During the formation of a transcription initiation competent complex, the complex undergoes conformational changes resulting in the formation of an open complex (3, 9, 10). Open complex formation is followed by the formation of the first phosphodiester bond. RNAP can then enter into an abortive mode, producing catalytic amounts of short RNA molecules (up to nine nucleotides) (2–4). Some RNAPII molecules escape the abortive mode and enter into the productive cycle. In this case, RNAP moves away from the promoter enabling a second polymerase molecule to enter into the transcription cycle. This step is defined as promoter clearance.
Transcription by RNAPII requires the hydrolysis of the β-γ bond of ATP or dATP (11). ATP hydrolysis is required for the formation of a stable open complex (12, 3) and by a step subsequent to initiation of transcription, most likely promoter clearance (13–15). It has become evident that the ATP-dependent step is catalyzed by TFIIH. Bypassing the requirement for ATP hydrolysis also bypasses the requirement for TFIIH (16–18, 3). However, the step(s) catalyzed by TFIIH during the transcription cycle remain controversial (3, 13–15).
TFIIH is a complex factor composed of nine polypeptides and possessing four different enzymatic activities (7, 19). Studies have uncovered that TFIIH interacts with different activators, suggesting that the steps catalyzed by TFIIH during the transcription cycle are subject to regulation (7, 20). In the present studies, we found that TFIIH is not necessary, but stimulatory, for the formation of the first phosphodiester bond. TFIIH modulates clearance of RNAPII from the promoter. We found that an activator enhances the efficiency of clearance.
MATERIALS AND METHODS
Transcription Factors and Reconstituted Transcription Assays.
Transcription reactions were performed as described (21) using the pG5U50 DNA template containing five Gal4-binding sites upstream of the TATA motif of the adenovirus major late promoter. The promoter directs transcription of a 50-nucleotide U-less cassette (22).
Preinitiation complexes were assembled at 30°C for 1h on immobilized DNA (22) by incubating TFIID (160 ng) or rTBP (10 ng), TFIIA (120 ng), rTFIIB (10 ng), rTFIIF (30 ng), rTFIIE (16 ng), highly purified RNAPII (100 ng) and TFIIH (100 ng of the phenyl-Superose fraction, (21)) in a total volume of 40 μl in transcription buffer [10 mM Tris⋅HCl, pH 7.9/20 mM Hepes. pH 7.9/8% glycerol/45 mM KCl/8 mM MgCl2/5 mM (NH2)SO4/2% PEG/5 mM DTT/0.05 mM EDTA]. RNA products were extracted with phenol-chloroform and diluted with 40 μl of formamide loading dye without precipitation. Aliquots (3 μl) were loaded directly on denaturing 28% polyacrylamide/urea gels.
Preparation of the immobilized DNA template was essentially as described (22). For each transcription reaction, linearized template DNA (0.5 pmol) was biotinylated 150 bp upstream from the TATA box, and was bound to 10 μg of streptavidin-coated magnetic beads. The immobilized DNA template was washed in transcription buffer. Preinitiation complexes were assembled on the immobilized DNA template by incubating 4-fold excess of GTFs and RNAPII in a total volume of 40 μl in transcription buffer at 30°C for 1 hr. For activated transcription, Gal4-VP16 (20 ng) and coactivators PC4 (30 ng) and TFIIA (120 ng) were also incubated along with the GTFs and RNAPII. Transcription complexes were washed twice in transcription buffer containing 0.05% sarkosyl and then incubated for 30 min at 30°C with nucleotides [0.6 mM ATP/5 μM [α-32P]CTP (100 mCi/mmol; 1 Ci = 37 GBq)/0.6 mM GTP] in a final volume of 20 μl. The reactions were then placed on the magnetic stand for 1 min, and the aqueous phase was removed from the beads. The beads were repeatedly washed in transcription buffer. Following separation, the aqueous phase and the beads were resuspended in 40 μl of the formamide loading dye and 20-μl samples were loaded on the gel. Products were resolved on a 28% polyacrylamide gel (30:1 acrylamide:bisacrylamide).
Abortive initiation was performed essentially as described (23). Dinucleotide synthesis was analyzed under conditions described for reconstituted transcription assays in the presence of 500 μM ATP and 1 μM [α-32P]CTP. The reactions were followed by treatment with 10 units of calf intestinal phosphatase. Aliquots were then loaded directly on denaturing 28% polyacrylamide/urea gels. Dinucleotide priming reactions included 1 mM CpA, 1 μM [α-32P]CTP, and up to 500 μM dATP as source of energy as indicated in the figure legends. Products were quantitated using a Molecular Dynamics PhosphorImager scanner Storm 860.
RESULTS
TFIIH Stimulates Initiation of Transcription.
An assay capable of measuring the different steps of the transcription cycle (preinitiation complex formation, synthesis of the first phosphodiester bond, abortive synthesis, promoter clearance, and elongation) was developed using linear DNA molecules attached to magnetic beads as described previously (see Fig. 1 and ref. 22).
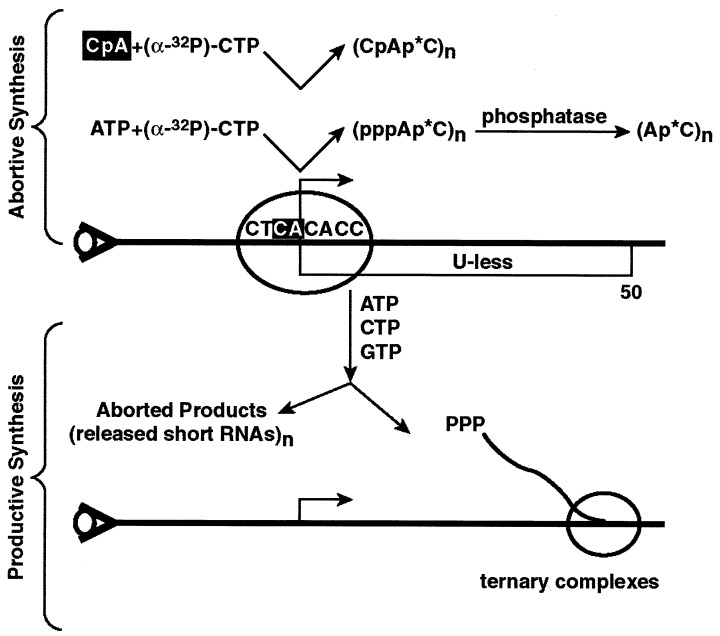
Figure 1.
Schematic representation of the DNA template containing a U-less cassette used in the experiments. The DNA sequence of the first 25 nucleotides is 5′-CTCA(+1)CACACCAACGGGCCCGAAGAGAGG(+25). Transcription complexes were formed using purified transcription factors, as described in Materials and Methods. The DNA template was biotinylated upstream of the transcription start site and was immobilized to streptavidin coated magnetic beads as described (22). Following formation of the preinitiation complexes, the complexes were washed and transcription was monitored using different assays. For abortive dinucleotide synthesis the transcription complexes were supplied with ATP and [α-32P]CTP to form the pppApC dinucleotide product. This dinucleotide product was treated with phosphatase to form the ApC. The (ApC)n denotes that the dinucleotide is produced in catalytic amounts with respect to the amount of active template molecules. In addition to dinucleotide synthesis, we also used dinucleotide priming in abortive reaction. In this protocol RNA synthesis is primed with the dinucleotide CpA that is complementary to residues −1 and +1 of the adenovirus major late promoter (as indicated in a solid box). The incorporation of [α-32P]CTP to form the trinucleotide CpApC is monitored. Reactions were also analyzed under condition allowing productive synthesis. In this case transcription was initiated by the addition of ribonucleoside triphosphates ATP, CTP, and GTP, as indicated in the figure. The partitioning of the expected RNA products is shown. The abortive products, up to nine nucleotides, are expected to be released from the template and recovered in the aqueous phase. The productive products are expected to remain in ternary complexes and therefore recovered with the beads.
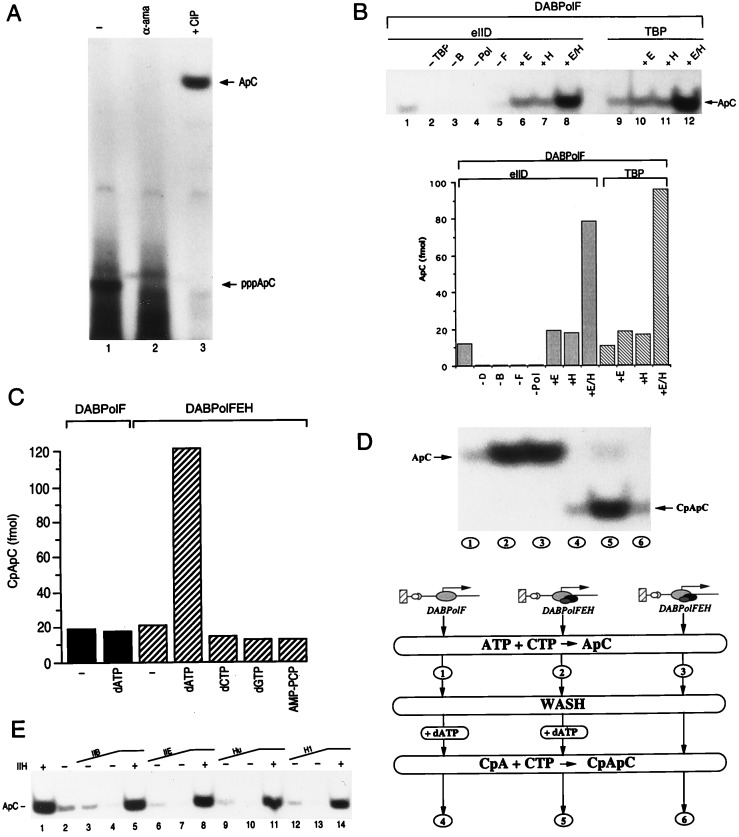
Using an abortive initiation assay, we analyze the formation of the first phosphodiester bond represented by the formation of the dinucleotide pppApC (see Fig. 1). Under these conditions, an α-amanitin-sensitive product was observed (Fig. 2A, lanes 1 and 2). Phosphatase treatment of the reaction products resulted in the conversion of the α-amanitin-sensitive product to a slower migrating band, without appreciable change in the total amount of 32P-label incorporated (Fig. 2A, lane 3, and data not shown). Phosphatase treatment was performed to insure that the reaction product migrated away from impurities present in the 32P-labeled nucleotide allowing accurate quantitation of this product.
Figure 2.
Role of TFIIH in initiation of transcription. (A) Formation of the first phosphodiester bond in an abortive initiation assay. Transcription complexes were provided with ATP and [α-32P]CTP and treated with either α-amanitin (lane 2) or phosphatase (lane 3). Arrows mark the positions of the dinucleotide. (B) Dinucleotide synthesis is dependent on TBP (lane 2), TFIIB (lane 3), TFIIF (lane 5), and RNAPII (lane 4) and is stimulated by TFIIE and TFIIH (lanes 6–8 and 10–12). Reaction were performed as indicated in A, lane 3, and the factor under analysis was omitted. (C) The stimulation of abortive initiation by TFIIH requires hydrolysis of the β-γ bond of ATP (or dATP). RNA synthesis was primed with the CpA dinucleotide. Reaction were performed in the absence (lanes 1–2) or the presence (lanes 3–7) of TFIIH and in presence of dATP (lanes 2 and 4), dCTP (lane 5), dGTP (lane 6), and AMP-PCP (lane 7). The products of abortive initiation were separated on a denaturing polyacrylamide gel and analyzed by PhosphorImager. (D) ATP hydrolysis is continuously required for the TFIIH-dependent stimulation of abortive initiation. Complete transcription reactions (lanes 2, 3, 5, and 6), or reaction lacking TFIIE and TFIIH (lanes 1 and 4) were performed on immobilized templates in the presence ATP and [α-32P]CTP (lanes 1–3), extensively washed and supplemented with the dinucleotide CpA and [α-32P]CTP (lanes 4–6) in the presence (lanes 4 and 5) or absence (lane 6) of dATP. (E) Reactions were reconstituted as in B, lane 10, but the concentration of TFIIB and TFIIE was increased 2- and 4-fold. Reaction with TFIIH were performed with the highest amount of the GTF. Histone H1 (70–140 ng) and E. coli Hu protein (0.5–1 μg) were added as indicated. Factors and DNA were preincubated for 1 hr prior to the addition of nucleotides. The amount of factor required for optimal synthesis was defined by independent titrations. The optimal concentration used is indicated in Materials and Methods.
First bond formation was dependent on TBP (supplied as recombinant or as the TFIID complex), TFIIB, TFIIF, and RNAPII (Fig. 2B). TFIIE and TFIIH were not required, in the presence of these factors, however, first bond formation was stimulated (Fig. 2B). We estimated that in the absence of TFIIE and/or TFIIH the dinucleotide was produced at ≈3% efficiency with respect to the amount of DNA added to the reaction. While this was low, it was specific as it was dependent on TBP, TFIIB and TFIIF and mutations of the TATA motif eliminated all products (Fig. 2B, and data not shown). In the presence of TFIIE and TFIIH an ≈5-fold stimulation was observed (Fig. 2B). The 3% efficiency observed in the absence of TFIIE and/or TFIIH is in close approximation with the number of DNA molecules estimated to be utilized in the reconstituted RNAPII transcription system (data not shown). Therefore, we suggest that it is only in the presence of TFIIE and TFIIH that the dinucleotide product is produced in catalytic amounts with respect to the amount of active transcription complexes. In the absence of the factors, the dinucleotide is produced in approximately stoichiometric amounts with respect to the amount of active transcription complex. The stimulation of transcription observed in the presence of TFIIE and TFIIH required energy that was specific for the hydrolysis of the β-γ bond of ATP (or dATP) (Fig. 2C). In this experiment, RNA synthesis was primed with the dinucleotide CpA and synthesis of the CpApC trinucleotide (see Fig. 1) was analyzed in the presence of dATP, or dCTP, or dGTP, or AMPPNP, a β-γ nonhydrolyzable analogue of ATP (Fig. 2C). We next analyzed whether the energy derived from ATP hydrolysis was required at each step of initiation. Complete transcription reactions, or a reaction lacking TFIIE and TFIIH were assembled on the beads as described in Fig. 1. The transcription complexes were then supplied with ATP and [α-32P]CTP and the production of the ApC dinucleotide was analyzed (Fig. 2D, lanes 1–3). Subsequently, the transcription complexes were extensively washed to remove the precursor nucleotides as well as the dinucleotide product. Reactions were next supplemented with the dinucleotide CpA and [α-32P]CTP in the presence or absence of dATP. Catalytic amounts of the trinucleotide CpApC was observed only in a reaction supplemented with dATP (lane 5), a transcription complex previously incubated with ATP, which generated catalytic amounts of the dinucleotide product (lane 3), was unable to generate catalytic amounts of the trinucleotide (lane 6). We concluded that the energy derived from the hydrolysis of the β-γ bond of the adenosine nucleotide is continuously required for the observed stimulation of abortive initiation.
Factors Binding to DNA Impose a Requirement for TFIIH During Initiation of Transcription.
Previous observations demonstrated that TFIIH in an ATP-dependent manner can overcome repression of transcription mediated by nonspecific DNA binding proteins (24). Therefore, a likely possibility to the differences observed between our findings and those of others showing a strict requirement for TFIIE and TFIIH for initiation of transcription (14), may resides in the presence of nonspecific DNA binding proteins or excess of GTFs that bind to DNA. Indeed an excess of the general transcription factors known to bind to DNA, such as TFIIB (25), TFIIE (26–27), and the large subunit of TFIIF (RAP74), inhibited the TFIIH-independent initiation of transcription (Fig. 2E and data not shown). This inhibition was overcome by TFIIH. Nonspecific DNA binding proteins such as the Escherichia coli Hu or histone H1 also inhibited the TFIIH-independent initiation of transcription. Again TFIIH overcome this inhibition (Fig. 2E).
TFIIH Is Required for the Escape of the RNAPII Complex from the Promoter.
Next, we analyzed the products of transcription reactions performed in the absence of TFIIH, but under conditions where RNAPII could synthesize a 50-nucleotide RNA product. The experimental conditions were as described in Fig. 1. Under these conditions, stalled RNAPII molecules remained bound to the template within active ternary complexes and could be recovered with the beads (Fig. 3A, lane 3). A fraction of the RNAPII entered into the abortive mode and generated short transcripts that were released and recovered in the aqueous phase (Fig. 3A, lane 4).
Figure 3.
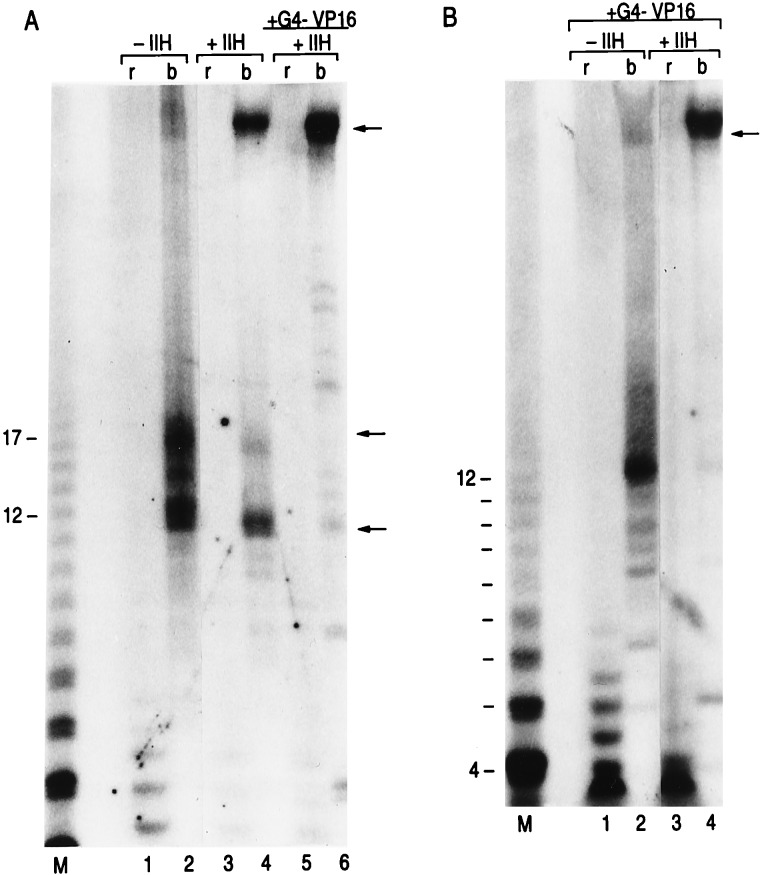
Analysis of the role of TFIIH during transcription. Transcription reactions in the presence and absence of TFIIH, as indicated on the panel, were performed using immobilized DNA template (A) or in solution (B). Reaction conditions were as described in Materials and Methods. The products of the reaction obtained in A (immobilized templates) were separated as described in Fig. 1 and analyzed by electrophoresis on denaturing polyacrylamide gels. All lanes in the panel were derived from the same gel and were expose for the same period. Arrows on the left denote aborted released products (lower arrows), as well as short RNA molecules that were recovered with the beads. The arrow on the right of each panel denotes the full-length 50-nucleotide RNA. The numbers represent the size of RNA products. Products of reactions performed in solution were directly applied to the gel. r and b at the top of the panel denote the aborted products that were released (r) and the productive products that remain bound (b) to the DNA.
When transcription reactions were performed in the absence of TFIIH (or additionally in the absence of TFIIE, data not shown), no full-length RNA was observed (Fig. 3A, compare lanes 2 and 3). Instead, the RNA transcripts recovered with the beads (ternary complexes) were short and extended predominantly from 12 to 17 nucleotides in length (Fig. 3A, lane 2). Similar short RNA molecules were present in the complete reaction, but were less abundant (compare lane 2 with 3). Also, in the absence of TFIIH, the amount of aborted RNA was increased as compared with the complete reaction (Fig. 3A, compare lane 1 with 4). All products observed were sensitive to α-amanitin (data not shown).
The results presented above establish a role for TFIIH during the transcription cycle. TFIIH (and TFIIE) is not required for initiation of transcription. In its absence, initiation of transcription takes place, but a large number of the transcription complexes are aborted. Moreover, RNAPII molecules that escape the abortive mode encounter a block to elongation when RNAPII reaches positions +12 to +17. Interestingly, this block to elongation was also observed, but to a much lesser extent, in reactions performed with all the transcription factors (Fig. 4A, compare lanes 2 and 3). The accumulation of short RNA molecules (12 to 17 nucleotides in length) was not a consequence of the assay, i.e., DNA attached to a solid support and single round transcription conditions. Similar products were observed in solution, under multiple round transcription conditions (Fig. 4B). However, because under multiple round transcription conditions, the complete reaction is more efficient than reactions lacking TFIIH (single round transcription conditions), particular products (aborted, stalled, and full length) produced in the presence and absence of TFIIH could not be compared. Stalling appears to be sequence independent, as an almost identical pattern of stalling was observed when RNAPII transcribed through G-less or U-less cassettes (data not shown). Moreover, stalling was not promoter-specific as a similar pattern was observed with the human HSP70 promoter (data not shown).
Figure 4.
Effect of Gal4-VP16 on the formation of the stalled complexes. Transcription reactions were performed as described in Fig. 3, in the presence and absence of TFIIH and Gal4-VP16, as indicated at the top of the lanes in each panel. Reactions also contained TFIID in lieu of TBP and the coactivators TFIIA and PC4, as described in Materials and Methods. Following transcription, the template-bound (b) and the aqueous (r) phases were separated and the RNA products in these fractions were analyzed on a denaturing 28% polyacrylamide/urea gel. Arrows indicate positions of stalling (12–17 nucleotides) and full-size products.
The Block to Elongation Is a Regulated Step.
Previous studies have shown that TFIIH interacts with activators (7, 20). Therefore, we investigated whether an activator could influence the formation of the stalled complexes.
The results presented in Fig. 4A illustrate that reactions performed under transcription activation conditions (with TFIID and the coactivators TFIIA and PC4) behave as above; i.e., in the absence of TFIIH, no full-length transcripts were observed and RNA molecules of 12–17 nucleotides were recovered with the beads (lane 2). In addition, aborted RNA molecules were observed in the aqueous phase (lane 1). The addition of TFIIH resulted in the production of full-length and some short transcripts (lane 4), and the amount of aborted RNAs produced were reduced. We next analyzed the effect of an activator on these products.
The addition of Gal4-VP16 to complete transcription reactions resulted in approximately a 3-fold increase in the production of the full length RNA (Fig. 4A, compare lanes 4 and 6). Importantly, the amount of short RNAs, resulting by stalling of the transcription complex, were drastically decreased (compare lanes 4 and 6). The activator had no appreciable effect in the amount of stalled complex produced on reactions lacking TFIIH (lane 2). However, whether the activator affects the ration of 12-mer vs. 17-mer remains a possibility. Again the activator, in the presence of TFIIH, suppressed the formation of promoter proximal stalled complexes and aborted products (Fig. 4B, lanes 3 and 4).
These results demonstrate that an activator can increase the production of the full-length transcript, and therefore, stimulates the efficiency of transcription. The effect of the activator under the assay conditions was low (3-fold stimulation); this is not unexpected, as transcription was performed under single round transcription conditions, and therefore, reinitiation (RNAPII loading), which is also a step regulated by activators, was not operational.
Releasing RNAPII from the Promoter-Proximal Stalling Site Requires TFIIH in the Transcription Initiation Complex.
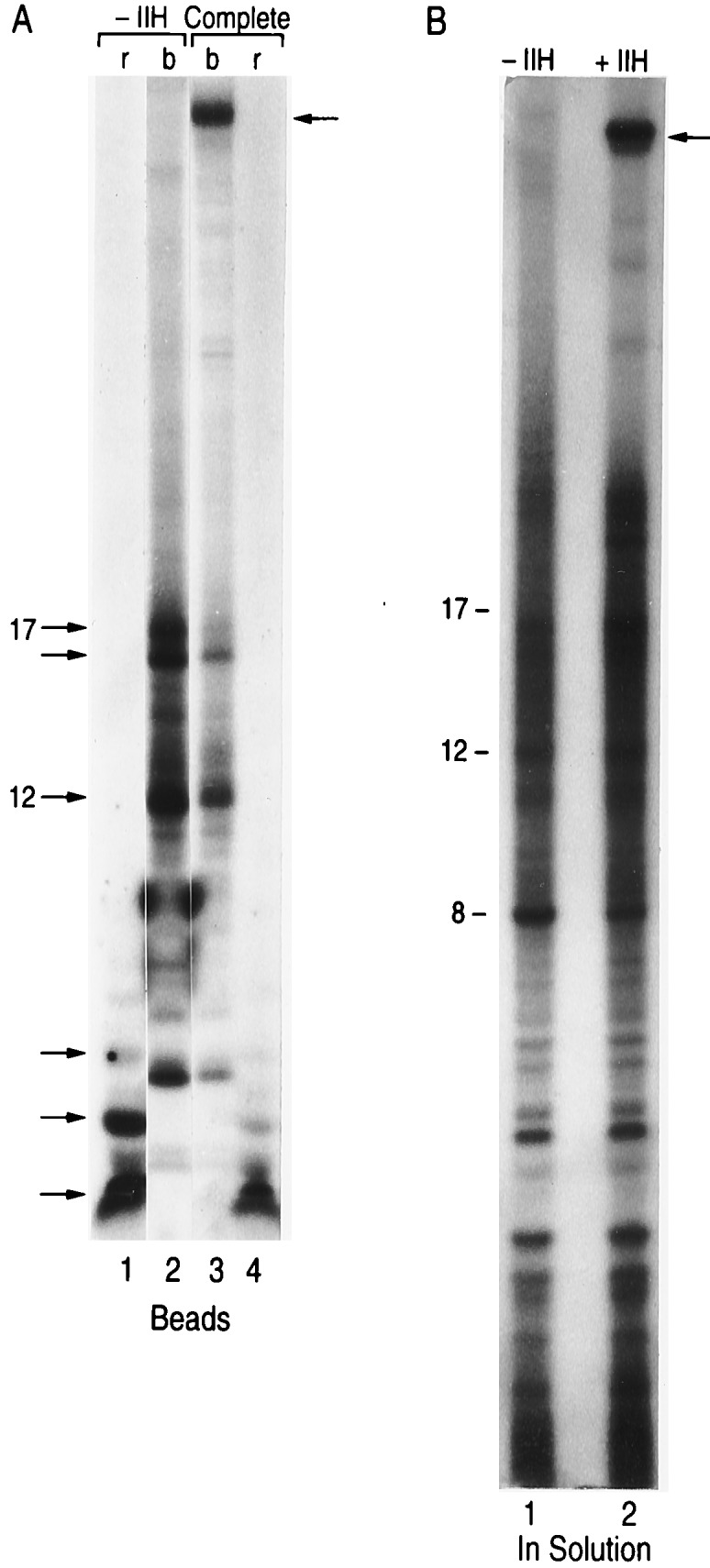
We next analyzed whether the promoter-proximal stalled complex could be chased to full-length by the addition of TFIIH. Promoter proximal stalled complexes were formed in the absence of TFIIH as described in Fig. 5, under activation conditions. The stalled complexes were extensively washed to remove nucleotides, then TFIIH with and without other transcription factors, together with ribonucleoside triphosphates were added. Under the conditions of the assay, the addition of TFIIH to the purified stalled complex was almost without effect (Fig. 5, compare lane 2 with 5, and data not shown). At best, a small percentage of the stalled complexes, apparently those smaller than 12 nucleotides in length (Fig. 5, see bracket), were elongated to ≈30 nucleotides, but no full-length 50-nucleotide transcripts was observed.
Figure 5.
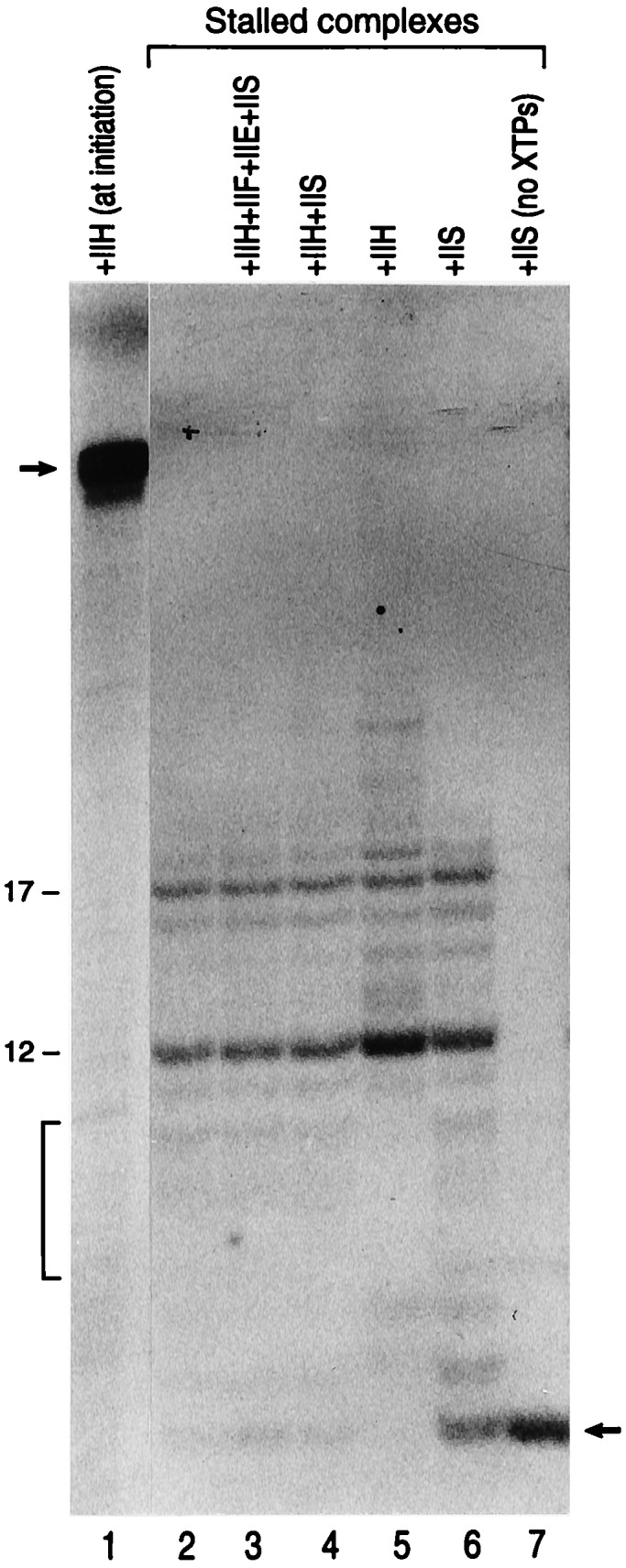
Stalled complexes cannot be chased. Transcription reactions were performed in the absence of TFIIH (lanes 2–7) or with TFIIH (lane 1) as described in the figure. Subsequent to transcription, the beads were extensively washed with transcription buffer (in the absence of nucleotides). Reactions performed in the absence of TFIIH were mixed and divided into six identical aliquots and were supplemented with nucleotides and different factors, as indicated on the top of the panel. Products were separated by electrophoresis on a polyacrylamide-urea gel as described in Materials and Methods. Numbers on the left denote the 12- and 17-nucleotide RNA products. The bracket indicates small RNAs that may have chased to larger products after addition of TFIIH (see lane 5). The arrow (top of panel) denotes the 50-nucleotide RNA, whereas the arrow on the right side denotes the four-nucleotide RNA obtained in reactions treated with TFIIS in the absence of nucleotides. Chasing of the stalled complexes could not be observed under the conditions of this experiment, or conditions of other experiments in which the stalled complexes were supplemented with HeLa cell nuclear extracts or different combination of factors.
In light of the above finding indicating that the stalled complexes could not be chased by the addition of TFIIH, we studied whether the promoter proximal stalled complexes represent “normal” stalled ternary complexes. Studies have demonstrated that the elongation factor TFIIS, in the absence of ribonucleoside triphosphates, induces a 3′ to 5′ endonuclease activity of RNAPII that is specific for RNA in ternary complexes (28, 29). Thus, the isolated promoter-proximal ternary complexes were treated with TFIIS in the absence of nucleotides. Under these conditions the RNA on the ternary complexes disappeared with the concomitant accumulation of a shorter 4-nucleotide RNA (lane 7). The addition of TFIIS together with nucleotides resulted in decreased TFIIS-induced nuclease activity, but was unable to stimulate the stalled complexes to resume elongation (lane 6). The addition of TFIIS together with TFIIH (lane 4), or with other transcription factors known to affect elongation (TFIIF), or known to recruit TFIIH to the transcription initiation complex (TFIIE), was also without effect (lanes 3 and 4). One explanation of these results is that TFIIH needs to be incorporated into the transcription complex during the formation of the transcription initiation complex or prior to the formation of the promoter-proximal stalled complexes (15). These findings may be related to studies performed with purified RNAPII and poly(dC)-tailed template indicating that a block to elongation was observed when the RNAPII reached +15 (30).
DISCUSSION
In this study, we have analyzed the function of TFIIH during transcription using an assay capable of dissecting the early steps of the transcription cycle. Our findings demonstrate that TFIIH is not necessary, but stimulates the formation of the first phosphodiester bond. However, we found that TFIIH is required for the escape of the transcription complexes from the promoter; i.e., promoter clearance. TFIIH also affects the efficiency of the transcription cycle. In its presence, a larger number of RNAPII molecules entered into the productive transcription cycle. More importantly, our findings establish that the TFIIH-mediated step is regulated by activators, consistent with studies demonstrating that TFIIH can interact with a variety of activators (7, 20).
Similar conclusions were reached by Goodrich and Tjian (13) using an abortive initiation assay coupled to dinucleotide priming. However, studies of Dvir et al. (14), using an approach similar to the one described by Goodrich and Tjian (13), suggested that TFIIH is required for initiation of transcription. We thought the discrepancies may reside in the conditions of the transcription assays and therefore, we analyzed the components of the transcription reaction for their ability to support initiation of transcription in the absence of TFIIH. Our studies uncovered that excess amount of GTFs capable of binding to DNA, inhibited TFIIH-independent initiation of transcription. Studies of others have demonstrated that TFIIH can overcome repression of transcription mediated by nonspecific DNA binding proteins (24). Thus, we suggest that the differences between the studies of Dvir et al. (14) and those of Goodrich and Tjian (13) and our observations presented here, resides in the presence of nonspecific DNA binding proteins or the concentration of GTFs in the reaction that impose a requirement for TFIIH. It is, however, important to emphasize that our findings demonstrating that TFIIH is required for promoter escape are in complete agreement with studies of Dvir et al. (15, 31, 32).
Interestingly, studies of Timmers and coworkers (3) using a highly purified transcription system uncovered that TFIIH, together with ATP hydrolysis, is necessary for the formation of a stable open complex. Similar findings were obtained by Gralla and coworker using extracts (12). These findings are in agreement with our own results (data not shown), and with the experiments demonstrating a stimulation of initiation of transcription in the presence of TFIIH and ATP hydrolysis (Fig. 2). Our results established that in the absence of TFIIH initiation of transcription was effective, yet the amount of product obtained in the abortive reactions appears stoichiometric with the number of template molecules capable of forming productive transcription complexes. This finding was surprising as it has been well established that the abortive reaction produces catalytic amounts of products (2, 4). Catalytic amounts of the initiated product was attained only under conditions of stable open complex formation, i.e., in the presence of TFIIH and ATP hydrolysis. Importantly, however, the initiated product (dinucleotide) produced in the absence of TFIIH was not a dead-end product. Transcription complexes lacking TFIIH initiated transcription, but encountered a block to elongation, between 12 to 17 nucleotides, resulting in the accumulation of promoter-proximal stalled complexes. Importantly, promoter-proximal stalled complexes were also observed in reactions performed in the presence of TFIIH, but in this case, stalled complexes were due to a transitory pausing site. In the presence of an activator and TFIIH, pausing was drastically decreased. Stalling at these sites appears to be sequence independent, as an almost identical pattern of stalling was observed when RNAPII transcribed through G-less or U-less cassettes.
We observed that RNAPII complexes already stalled (due to the absence of TFIIH), could not be stimulated to resume elongation by the addition of TFIIH (with or without other transcription factors, Fig. 5). The inability of TFIIH (and other transcription factors) to induce the promoter-proximal stalled complexes to resume elongation is apparently due to the properties of the promoter-proximal stalled complex. This complex responded to TFIIS-induced RNAPII endonuclease activity (Fig. 5), which is specific for ternary complexes (28, 29). Yet, we speculate that this complex is in transition. During the initial steps of RNA synthesis, the moving RNAPII undergoes conformational changes that appear to be required to attain a mature elongation complex (3, 9). The mature elongation complex does not contain TFIIH, which leaves the transcription complex approximately after the formation of the thirtieth phosphodiester bond (22). The promoter-proximal stalled complexes have not yet cleared the promoter and require the action of TFIIH. We hypothesize that the recruitment of TFIIH into the transcription complex is mediated during preinitiation complex formation, or prior to the formation of the twelfth phosphodiester bond (15). TFIIH, which is also involved in nucleotide excision repair, can be incorporated into elongation complexes that are stalled at lesions, yet the mechanism by which TFIIH is recruited to RNAPII stalled at a lesion is unclear and requires a set of factors apparently devoted to this function, i.e., CSA and CSB (33, 34).
Our findings also have implications for previous studies indicating that activators influence the efficiency of elongation in vivo (20, 35). These previous studies analyzed the effect of activators on transcription elongation as a function of the distribution of RNAPII on genes using nuclear run-on experiments. Interestingly, a direct correlation between activators capable of stimulating elongation (higher density of RNAPII at the 3′-end of the gene) and their ability to interact with TFIIH was established (20). These studies, however, did not distinguish promoter clearance from elongation. Indeed, elongation was defined as “all nucleotide addition steps after initiation” which includes promoter clearance (20). In light of our findings demonstrating that TFIIH is required for promoter clearance and that TFIIH does not travel with RNAPII in vitro (22), we believe that those studies measured, in part, the ability of TFIIH to catalyze promoter clearance.
The TFIIH-dependent phenomenon observed in the studies presented here, specifically the accumulation of promoter-proximal stalled RNAPII complexes, is reminiscent of studies performed in vivo demonstrating that RNAPII that initiate transcription are paused in a region proximal to the promoter. This phenomenon appears to be general as it has been observed in a number of genes in different species (36–38). An important question is whether the promoter-proximal stalled complex observed in vivo is a consequence of the inability to recruit TFIIH (or an active form of the factor) to the initiation complex, and whether this complex ever engages in productive transcription. We suggest that the concentration of TFIIH in vivo is limiting and that TFIIH is recruited to the transcription initiation complex by activators.
Acknowledgments
We thank Drs. Don Luse and John Lis for helpful discussions throughout the course of this study and for communicating results prior to publication and members of the Reinberg laboratory for helpful suggestions. We also thank Drs. Richard Ebright, Mike Hampsey, John Lis, Don Luse, George Orphanides, Marc Timmers, and Lynne Vales for helpful comments on the manuscript. This work was supported by grants from the National Institute of Health (GM-37120) and from the Howard Hughes Medical Institute to D.R.
ABBREVIATIONS
- TF
transcription factor
- RNAP
RNA polymerase
- GTF
general transcription factor
References
- 1.Chamberlin M J. Annu Rev Biochem. 1974;43:721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- 2.McClure W R. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 3.Holstege F C P, Fiedler U, Timmers H T M. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luse D S, Jacob G A. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 5.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 6.Flores O, Ha I, Reinberg D. J Biol Chem. 1990;264:8913–8921. [PubMed] [Google Scholar]
- 7.Drapkin R, Reinberg D. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 8.Dahmus M E. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 9.Luse D S, Kochel T, Kuempel E, D, Coppola J A, Cai H. J Biol Chem. 1987;262:289–297. [PubMed] [Google Scholar]
- 10.Jiang Y, Yan M, Gralla J D. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunick D, Zandomani R, Ackerman S, Weinmann R. J Biol Chem. 1982;270:6798–6807. [Google Scholar]
- 12.Jiang Y, Gralla J D. J Biol Chem. 1995;270:1277–1281. doi: 10.1074/jbc.270.3.1277. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich J A, Tjian R. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 14.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 15.Dvir A, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 17.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 18.Tantin D, Carey M. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 19.Svejstrup J Q, Vichi P, Egly J M. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 20.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maldonado E, Drapkin R, Reinberg D. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 22.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 23.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Nature (London) 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 24.Stelzer G, Goppelt A, Lottspeich F, Meisterernst M. Mol Cell Biol. 1994;14:4712–4721. doi: 10.1128/mcb.14.7.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinberg D, Roeder R. J Biol Chem. 1987;262:3331–3337. [PubMed] [Google Scholar]
- 26.Maxon M E, Tjian R. Proc Natl Acad Sci USA. 1994;91:9529–9533. doi: 10.1073/pnas.91.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuldell N H, Buratowski S. Mol Cell Biol. 1997;17:5288–5298. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reines D, Ghanouni P, Li Q Q, Mote J., Jr J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- 29.Izban M G, Luse D S. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 30.Sluder A E, Price D H, Greenleaf A L. J Biol Chem. 1988;263:9917–9925. [PubMed] [Google Scholar]
- 31.Dvir A, Tan S, Conaway J W, Conaway R C. J Biol Chem. 1997;272:28175–28178. doi: 10.1074/jbc.272.45.28175. [DOI] [PubMed] [Google Scholar]
- 32.Dvir A, Conaway R C, Conaway J W. J Biol Chem. 1996;271:23352–23356. doi: 10.1074/jbc.271.38.23352. [DOI] [PubMed] [Google Scholar]
- 33.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 34.Tantin D, Kansal A, Carey M. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 36.Bentley D L, Groudine M. Nature (London) 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 37.Lis J T, Wu C. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. New York: Raven; 1994. pp. 459–475. [Google Scholar]
- 38.Akhtar A, Faye G, Bentley D. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]