Abstract
Smad4 plays a pivotal role in signal transduction of the transforming growth factor β superfamily cytokines by mediating transcriptional activation of target genes. Hetero-oligomerization of Smad4 with the pathway-restricted SMAD proteins is essential for Smad4-mediated transcription. We provide evidence that SMAD hetero-oligomerization is directly required for the Smad4 C-terminal domain [Smad4(C)] to show its transcriptional transactivating activity; this requirement obtains even when Smad4(C) is recruited to promoters by heterologous DNA-binding domains and in the absence of the inhibitory Smad4 N-terminal domain. Defined mutations of GAL4 DNA-binding domain fusion of Smad4(C) that disrupt SMAD hetero-oligomerization suppressed transcriptional activation. Importantly, we found that an orphan transcriptional activator MSG1, a nuclear protein that has strong transactivating activity but apparently lacks DNA-binding activity, functionally interacted with Smad4 and enhanced transcription mediated by GAL4 DNA-binding domain-Smad4(C) and full-length Smad4. Transcriptional enhancement by MSG1 depended on transforming growth factor β signaling and was suppressed by Smad4(C) mutations disrupting SMAD hetero-oligomerization or by the presence of Smad4 N-terminal domain. Furthermore, Smad4(C) did not show any detectable transactivating activity in yeast when fused to heterologous DNA-binding domains. These results demonstrate additional roles of SMAD hetero-oligomerization in Smad4-mediated transcriptional activation. They also suggest that the transcriptional-activating activity observed in the presence of Smad4 in mammalian cells may be derived, at least in part, from endogenously expressed separate transcriptional activators, such as MSG1.
Cytokines belonging to the transforming growth factor β (TGFβ) superfamily exert biological effects through transcriptional activation of target genes. In vertebrate cells, signaling from the ligand-activated membrane receptor serine/threonine kinases to nuclear targets is mediated by the SMAD family signal transducer proteins. One of the SMAD proteins, Smad4, plays a unique and pivotal role in SMAD-mediated transcriptional activation (reviewed in ref. 1). By using Smad4-deficient cells, it has been demonstrated that transcriptional activation by fusion proteins of the GAL4 DNA-binding domain (GAL4DB) with Smad1 or Smad2 observed in mammalian cells depends on the presence of endogenous Smad4 (2). The C-terminal domain of Smad4 [Smad4(C)] is involved in both transcriptional activation and hetero-oligomerization with the pathway-restricted SMAD proteins (2–9), whereas the N-terminal domain [Smad4(N)] inhibits both of these Smad4(C) functions through direct physical interaction with Smad4(C) (4, 10). SMAD hetero-oligomerization has been proposed to be essential for releasing the transcriptional-activating domain located in Smad4(C) from autosuppression by Smad4(N) (11) and/or recruiting Smad4 from cytosol to nucleus (2, 12, 13), whereas molecular details of these proposed mechanisms are unclear.
MSG1 is a 27-kDa nuclear protein that strongly activates transcription but apparently lacks DNA-binding activity (14, 15). Although the biological significance of MSG1 has not yet been elucidated, its possible involvement in cell differentiation or development has been proposed based on its tissue-restricted and developmentally restricted expression (14, 16–18).
Based on a hypothesis that MSG1 may function to associate with DNA-binding protein(s) and contribute to transcriptional activation, we screened for possible MSG1-interacting proteins by the yeast two-hybrid system (19) and isolated Smad4(C) as a candidate MSG1-interacting protein. In this report, we show that in mammalian cells MSG1 enhances transcriptional activation mediated by GAL4DB-Smad4(C) as well as by full-length Smad4 in a manner that depends on TGFβ signaling and SMAD hetero-oligomerization. We also found that basal transcriptional activation mediated by GAL4DB-Smad4(C) detected in the absence of MSG1 cotransfection also depended on SMAD hetero-oligomerization. In yeast, Smad4(C) did not show any detectable transactivating activity. These results demonstrate additional essential roles of SMAD hetero-oligomerization in activating transcription mediated by Smad4. They also suggest possible involvement of endogenously expressed separate transcriptional activators (such as MSG1) in Smad4-mediated transcription.
MATERIALS AND METHODS
Yeast One-Hybrid and Two-Hybrid Assays.
Details of the interaction trap yeast two-hybrid system were described by Finley and Brent (19). Human MSG1 was fused to LexA DNA-binding protein followed by a removal of the CR2 transcriptional activating domain (15), generating LexA-MSG1ΔCR2 bait (His+ plasmid). Smad4(C) (amino acids 302–552) was isolated as an MSG1-interacting protein by screening a human melanocyte cDNA library (Trp+ vector) with Leu+ and β-galactosidase (Ura+ plasmid) markers. For LexA-based transcriptional activation assay, Smad4(C) (amino acids 266–552) was fused to LexA. Protein expression of all LexA fusion proteins were confirmed by anti-LexA Western blotting.
The prey insert (Smad4[302–552]) was fused to GAL4DB or to GAL4 transcriptional-activating domain. GAL4-based yeast one-hybrid transactivation assay (15) and two-hybrid assay (20) were performed as described, by using p53 and simian virus 40 large T antigen as positive control interactors.
Mammalian Cell Expression Plasmids.
Expression plasmids for influenza hemagglutinin (HA)-tagged MSG1, HA-MRG1, GAL4DB-fusion MSG1, and GAL4DB-MRG1 were described previously (14, 15). Plasmids for Flag-tagged Smad2 (21), GAL4DB-Smad2 (22), and TGFβ type I receptor kinase [TβRI(T204D)] (23) were provided by J. L. Wrana and L. Attisano (The Hospital for Sick Children, Toronto, Canada); Myc-tagged Xenopus FAST-1 and ARE-Luc (24) were from M. Whitman (Harvard Medical School, Boston). Plasmids for GAL4DB-Smad4 (partial and full-length) were constructed by using the pSG424 vector (25); plasmid for Flag-Smad3 and Flag-Smad4 were constructed on pCMV2-Flag (Kodak) and pcDNA3 (InVitrogen), respectively, by a standard PCR-based method. Deletion mutants of MSG1 or Smad4 were generated by site-directed mutagenesis using a kit (CLONTECH). All new constructs were sequenced.
Mammalian Cell Modified One-Hybrid Assay.
Cells were maintained in DMEM supplemented with 10% fetal calf serum. Expression plasmids were transfected together with a GAL4-dependent chloramphenicol acetyltransferase (CAT) reporter plasmid (pG5CAT, 0.5 μg/well) and a β-galactosidase plasmid (pSVβGal, 0.5 μg/well) by lipofection (4 × 105 cells per 3.5 cm well; 2 μg/well total plasmids with pBSKm carrier) as described previously (15). Reporter gene expression was evaluated by CAT assay 16–24 h after transfection (15) with normalization by β-galactosidase activity.
Reconstitution of Activin Responsive Element (ARE)-Dependent, Smad4-Mediated Transcription in Mammalian Cells.
Reconstitution of the ARE-dependent, Smad4-mediated transcription in mammalian cells originally was described by Liu et al. (2). HepG2 human hepatoblastoma cells were cotransfected with expression plasmids for FAST-1 and HA-MSG1 together with a luciferase reporter plasmid for ARE (26) and a β-galactosidase plasmid by lipofection. Cells were cultured for 24 h after transfection in the presence of 10% fetal calf serum, then treated with TGFβ1 (5 ng/ml) for 16 h in the presence of 0.2% fetal calf serum. Luciferase activity was determined as described previously (6) with normalization by β-galactosidase activity.
RESULTS
MSG1 Functionally Interacts with the C-Terminal Domain of Smad4 in Yeast.
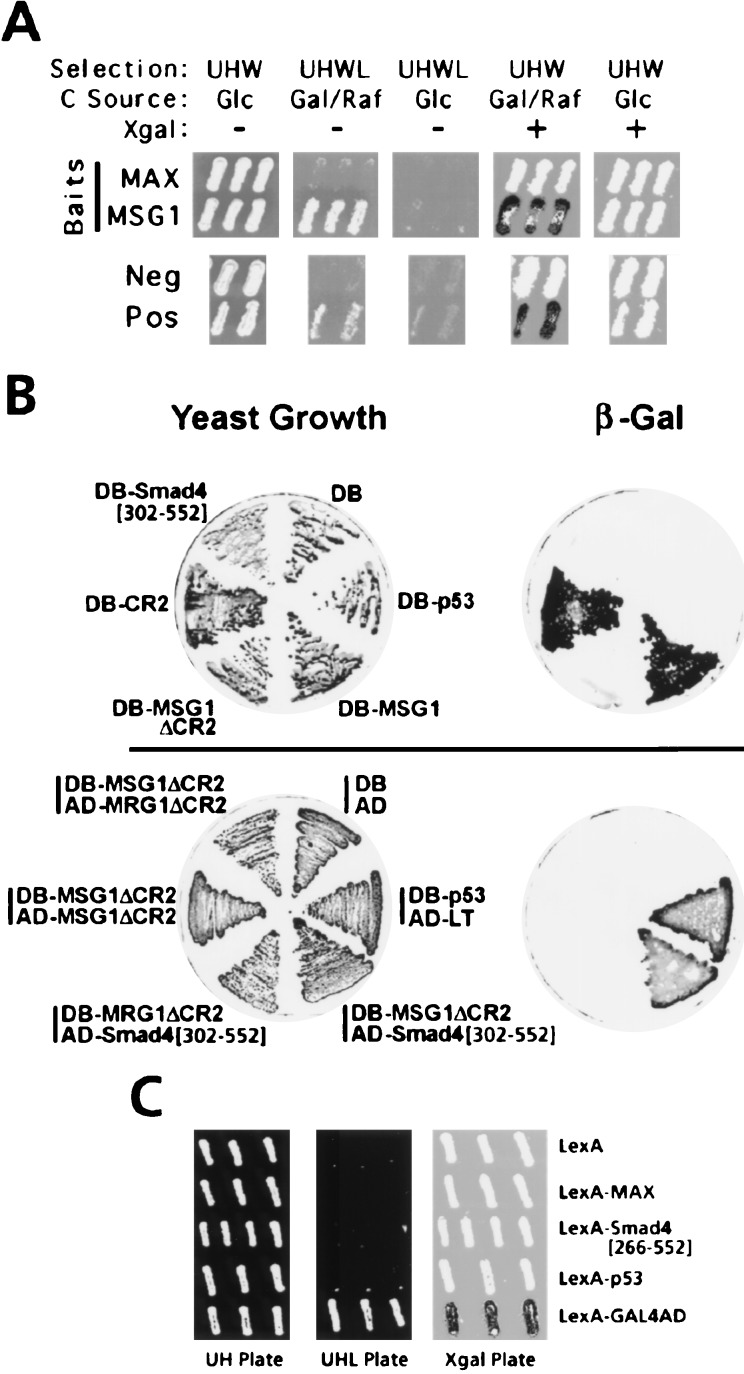
Smad4(C) (amino acids 302–552) was isolated as an MSG1-interacting protein from a human melanocyte cDNA library by yeast two-hybrid screening (Fig. 1A). Smad4(C) has been reported to be responsible for Smad4 functions, including transcriptional transactivation, homotrimer formation, and hetero-oligomerization with the pathway-restricted SMAD proteins (1). The Smad4(C) prey did not interact with an unrelated bait (the Max protein, ref. 27). To eliminate possible involvement of the LexA DNA-binding protein in the observed interaction, the MSG1/Smad4 interaction was further confirmed by an independent yeast two-hybrid assay system (20) based on the GAL4 transcription factor (Fig. 1B, Lower). MRG1, an MSG1-related protein that shares two highly conserved domains (CR1 and CR2) with MSG1 (15), did not interact with Smad4(C). In the absence of the transactivator domain, MSG1 interacted with neither MSG1 itself nor with MRG1.
Figure 1.
Functional interaction and transcriptional-activating activities of MSG1 and Smad4(C) in yeast. (A) Functional interaction of MSG1 and Smad4(C) demonstrated by the interaction-trap yeast two-hybrid assay (19, 27). Smad4(C) prey is expressed on galactose/raffinose (Gal/Raf) plates but not on glucose (Glc) plates (19, 27). Positive (Pos) and negative (Neg) control interactors and the LexA-Max bait (MAX, nonspecific interactor control) are described (27). Selection markers were used for: β-galactosidase reporter plasmid (Ura+, U); LexA-fusion bait plasmids (His+, H); B16 activator-fusion prey plasmids (Trp+, W); and the auxotrophic phenotype resulting from interaction of the bait and prey (Leu+; L). (B) Functional interaction and transactivating activities of MSG1, MRG1, and Smad4(C) analyzed by GAL4-based yeast one-hybrid (Upper) or two-hybrid (Lower) assays (20). DB, GAL4 DNA-binding domain; AD, GAL4 transcriptional activating domain; LT, simian virus 40 large T antigen. Transcription of the β-galactosidase reporter gene by transactivating activity of GAL4DB fusion proteins (Upper) or resulting from protein–protein interaction GAL4DB- and GAL4AD-fusion proteins (Lower) were evaluated by the β-galactosidase replica filter assay (15). (C) Absence of transactivation by Smad4[266–552] in yeast. Transactivating activities of LexA-fusion proteins (His+ selection, H) were evaluated as described (19), by the LexA-dependent Leu+ (L) auxotrophic phenotype (determined on the UHL plate) or transcription of a LexA-dependent β-galactosidase reporter plasmid (Ura+ selection, U) (determined on the Xgal plate). The carbon source of all plates were galactose/raffinose. A fusion protein of LexA and GAL4 transactivating domain (GAL4AD) was used as positive control.
Absence of Detectable Transactivating Activity of Smad4(C) in Yeast.
MSG1 strongly activated transcription in yeast, and the transactivating activity was localized to the acidic CR2 domain (Fig. 1B, Upper), as reported previously (15). On the other hand, Smad4(C) (amino acids 302–552) did not activate transcription in yeast (Fig. 1B, Upper). We sought to determine whether a longer Smad4(C) fragment (amino acids 266–552), which activates transcription in mammalian cells (see Fig. 3A and ref. 4), would activate transcription in yeast; still no activity was detected for LexA-Smad4[266–552] (Fig. 1C) even by the very sensitive Leu+ auxotrophic phenotype assay of the interaction trap system (19). Strong expression of LexA-Smad4[266–552] protein in yeast was confirmed by anti-LexA Western blotting (data not shown).
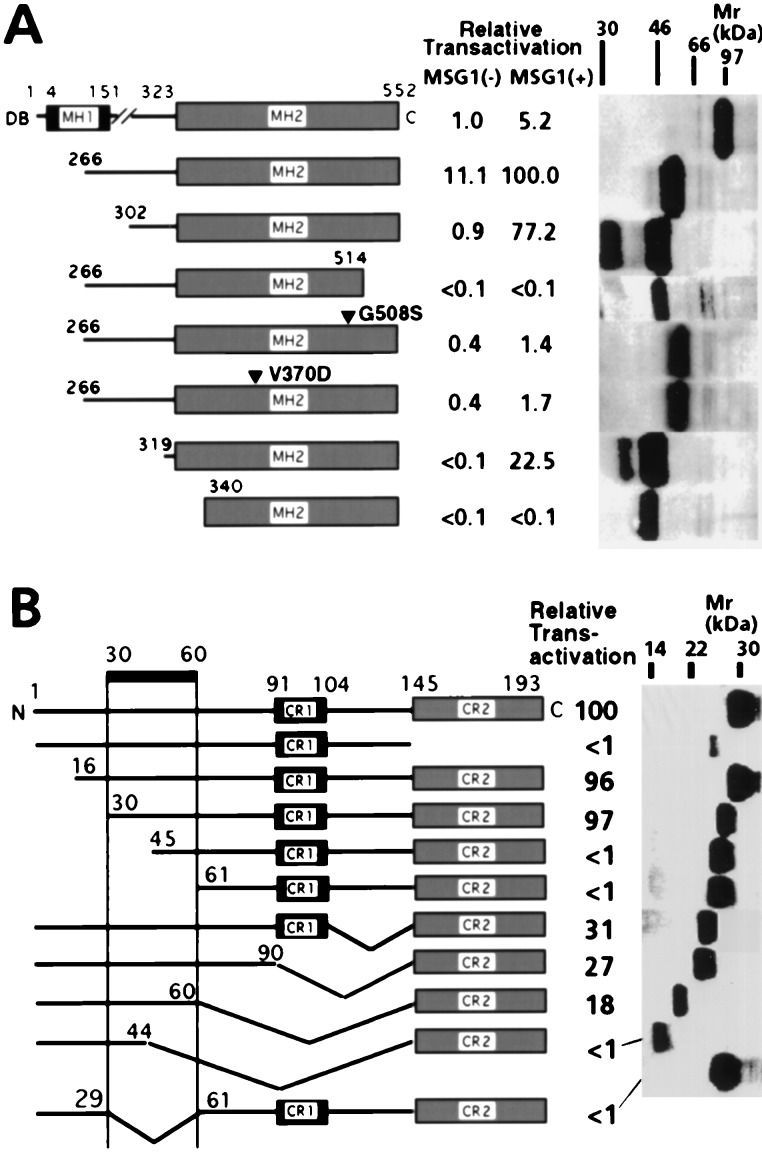
Figure 3.
Effects of mutations on Smad4(C)-mediated transcriptional activation in NIH 3T3 cells. (A) Cells were cotransfected with HA-MSG1 and GAL4DB-Smad4(C) fusion proteins (indicated) together with pG5CAT, followed by incubation in the presence of 10% fetal calf serum for 16 h and CAT assay. Because protein expression of GAL4DB fusions in NIH 3T3 cells were not detectable by Western blotting because of very weak simian virus 40 early promoter, expression of these fusion proteins were evaluated in COS-1 cells by anti-GAL4DB Western blotting. (B) Cells were cotransfected with GAL4DB-Smad4[302–552] and deletion mutants of HA-MSG1 together with pG5CAT, followed by incubation in the presence of 10% fetal calf serum for 16 h and CAT assay. Protein expression of MSG1 deletion mutants in NIH 3T3 cells was analyzed by anti-HA Western blotting.
MSG1 Enhances Smad4(C)-Mediated Transcription in Mammalian Cells.
Fusion proteins of MSG1 and MRG1 with GAL4DB strongly activated transcription in NIH 3T3 cells (Fig. 2A). We previously showed that the acidic CR2 domain of MSG1 is required and sufficient for this transactivating activity (15). A GAL4DB fusion of Smad4(C) (amino acids 302–552) did not show significant transactivating activity in the absence of MSG1, but it strongly activated transcription when MSG1 was cotransfected; this effect depended on the amounts of MSG1 protein expressed in the cells (data not shown). The MSG1-enhanced transcription was not observed for GAL4DB control. MRG1, a nuclear protein that shares the CR2 domain with MSG1 (15), showed minimal effects. HA epitope tagging of MSG1 did not affect MSG1-enhanced transactivation (data not shown).
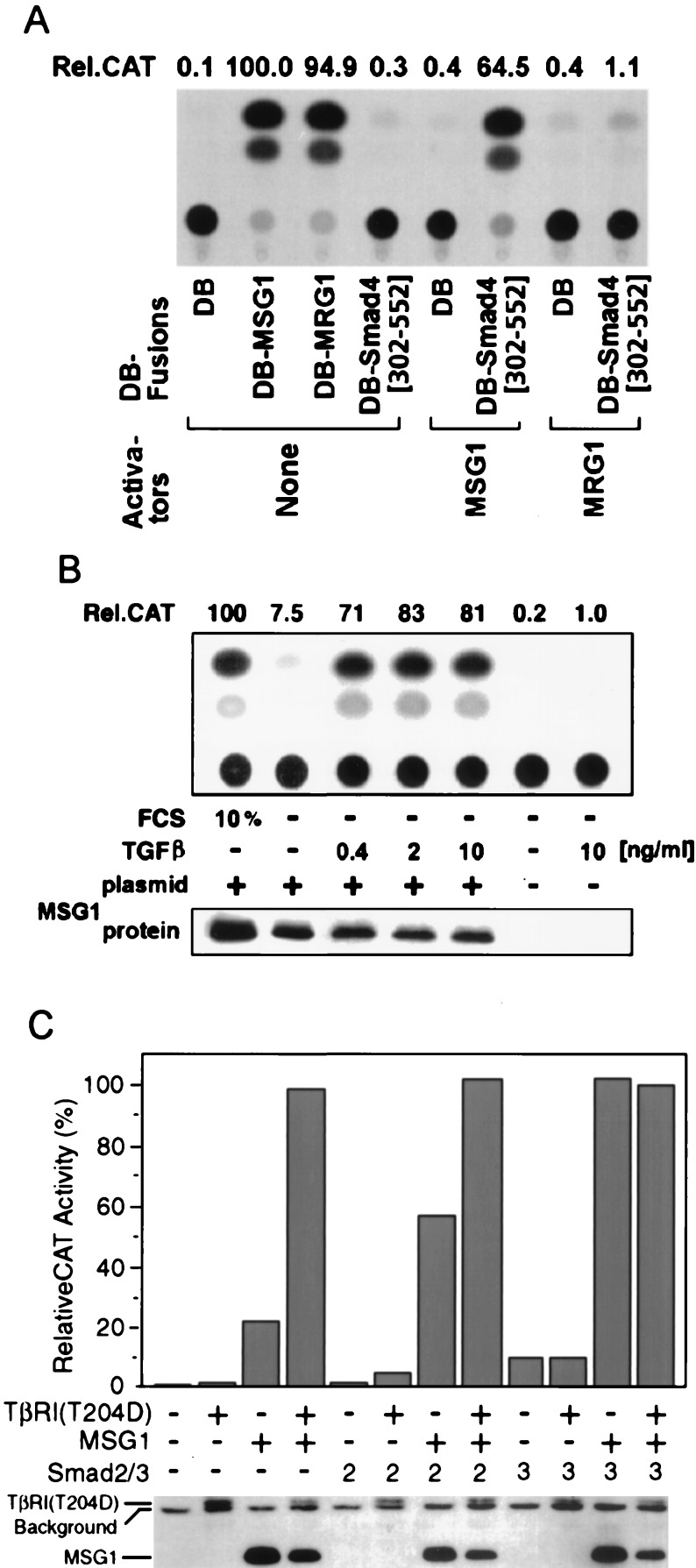
Figure 2.
Effects of MSG1 on transcriptional activation mediated by Smad4 C-terminal domain (amino acids 302–552) in NIH 3T3 cells. (A) Cells were cotransfected with expression plasmids for GAL4 DNA-binding domain (DB) fusion proteins and activators together with a GAL4-dependent CAT reporter plasmid (pG5CAT), followed by incubation in the presence of 10% fetal calf serum for 16 h and determination of relative CAT activity (Rel.CAT). (B) Cells were cotransfected with GAL4DB-Smad4[302–552], HA epitope-tagged MSG1 and pG5CAT, followed by incubation in serum-free medium supplemented with TGFβ1 for 16 h and CAT assay. (C) Cells were cotransfected with GAL4DB-Smad4[302–552] and pG5CAT together with HA-MSG1, HA-TβRI(T204D) activated TGFβ type I receptor kinase, Flag epitope-tagged Smad2, or Flag-Smad3, followed by incubation in serum-free medium for 16 h and CAT assay. Protein expression of MSG1 and TβRI(T204D) (B and C) was evaluated by anti-HA Western blotting.
Enhancement of Smad4(C)-Mediated Transcription by MSG1 Depends on TGFβ Signaling.
Because Smad4 plays pivotal roles in transcriptional activation by TGFβ superfamily cytokines (1), we sought to determine whether the enhancing effect of MSG1 on Smad4(C)-mediated transcription is affected by TGFβ signaling. Because NIH 3T3 cells are responsive to TGFβ (28) and can activate the latent form of TGFβ present in serum (29), they are exposed to background stimulation by TGFβ when cultured in serum-containing medium. When NIH 3T3 cells were cultured in serum-free medium for 16 h, the transcription-enhancing effect of MSG1 was dramatically suppressed, but was restored by addition of low concentrations of TGFβ (Fig. 2B). Two ng/ml of TGFβ led to a 10-fold (10.2 ± 2.1; mean ± SEM, n = 7) enhancement of Smad4(C)-mediated transcription under serum-free condition but only when MSG1 was cotransfected. Without MSG1 cotransfection, even 10 ng/ml of TGFβ did not show significant effects. MSG1 protein expression was comparable in the presence or absence of serum or TGFβ (Fig. 2B). TGFβ did not affect expression of the GAL4DB fusion proteins, because transactivating activity by GAL4DB fusion of VP16 viral transactivator was not affected by TGFβ treatment (data not shown). TGFβ did also not affect transactivating activity by GAL4DB fusion of full-length MSG1 (data not shown), indicating that TGFβ does not influence the intrinsic transactivating activity of MSG1.
We next attempted to determine whether cotransfection of the pathway-restricted SMAD proteins and/or activated TβRI(T204D) affects the transcription-enhancing effect of MSG1. Under serum-free culture conditions, GAL4DB-Smad4(C) alone did not activate transcription effectively, even when TβRI(T204D) was cotransfected (Fig. 2C). In the presence of MSG1, but without TβRI(T204D), transcriptional activation by GAL4DB-Smad4(C) was weak, but it was enhanced about 5-fold by TβRI(T204D). Without cotransfection of MSG1, Smad4(C)-mediated transcription was weak even when Smad2 or Smad3 was cotransfected, in the presence or absence of TβRI(T204D). When MSG1 was cotransfected, further cotransfection of Smad2 or Smad3 enhanced Smad4(C)-mediated transcription strongly even in the absence of TβRI(T204D). The Smad4(C)-mediated transcription observed in the presence of MSG1 and Smad2 cotransfection was further enhanced by TβRI(T204D). Protein expression of MSG1 and TβRI(T204D) was not enhanced by Smad2/3 cotransfection (Fig. 2C, Lower), and expression of Smad2 and Smad3 proteins was not enhanced by MSG1 cotransfection (anti-Flag Western blotting: data not shown). Taken together, these results strongly suggest the essential role of the pathway-restricted SMAD proteins in the effect of MSG1 to enhance Smad4(C)-mediated transcriptional activation.
Mutations of Smad4(C) that Disrupt SMAD Hetero-oligomerization Suppress Both Basal and MSG1-Enhanced Transcriptional Activation.
To further characterize the requirement of the pathway-restricted SMAD proteins in the basal and MSG1-enhanced transcriptional activation mediated by Smad4(C), we examined the effects of defined mutations that impair hetero-oligomerization of Smad4(C) in serum-stimulated NIH 3T3 cells. As has been reported for other cell lines (4, 30), full-length Smad4 activated transcription only marginally when fused N-terminally to GAL4DB, but MSG1 enhanced this weak transactivating activity by about 5-fold (Fig. 3A). GAL4DB fusion of Smad4(C) (amino acids 266–552) activated transcription weakly but significantly even in the absence of MSG1 cotransfection; and this activity was strongly enhanced by MSG1 about 9-fold (Fig. 3A). A shorter fragment of Smad4(C) (amino acids 302–552) did not activate transcription significantly in the absence of MSG1, but again it showed strong activity in the presence of MSG1. The MSG1-enhanced transactivating activity of GAL4DB-Smad4[266–552] was greater than that of GAL4DB-Smad4[302–552].
Two Smad4 mutations (Arg-515 to stop codon and Gly-508 to Ser) have been reported to suppress SMAD hetero-oligomerization and Smad4-mediated transactivation, whereas Smad4 homotrimerization is kept intact (3, 7). When introduced in GAL4DB-Smad4[266–552], both of these mutations resulted in dramatic reduction in the transcriptional activating activity in the presence or absence of MSG1 (Fig. 3A). An additional mutation, Val-370 to Asp, which impairs both homo- and hetero-oligomerization of Smad4 (7), also resulted in loss of transactivating activity. Protein expression levels of the mutants were comparable to that of the wild type (Fig. 3A, Right). These results strongly support the requirement of SMAD hetero-oligomerization for both the basal and MSG1-enhanced transcriptional activation mediated by GAL4DB-Smad4(C).
Because we previously reported the possible existence of a transcriptional-transactivating domain of Smad4 (SAD for Smad4 activation domain) at amino acids 274–321 (6), we tested whether removal of this region affected basal and/or MSG1-enhanced transactivation. Deleting amino acids 302–318 resulted in a decrease in the MSG1-enhanced Smad4(C)-mediated transactivation by about 70% and a total loss of the basal transactivation (Fig. 3A). Further deletion to amino acid 339 resulted in a total loss of transactivation in the presence or absence of MSG1. These results indicate that the region involving SAD is essential for both basal and MSG1-enhanced transcriptional activation mediated by Smad4(C).
Domains of MSG1 Necessary to Enhance Smad4(C)-Mediated Transcriptional Activation.
Deletion mutants of MSG1 were cotransfected with GAL4DB-Smad4[302–552] in serum-stimulated NIH 3T3 cells, and their transcription-enhancing effects were determined (Fig. 3B). Removal of the CR2 transactivating domain resulted in a total loss of transactivation; although protein expression of MSG1 was decreased by this deletion (Fig. 3B, Right), further overexpression of MSG1 lacking the CR2 domain still did not show detectable transactivation (data not shown). By systemic deletion of MSG1, an essential sequence for enhancing Smad4(C)-mediated transcription was localized within the region of amino acids 30–60 (Fig. 3B). Removal of the CR1 domain did not have a significant effect, consistent with the absence of the enhancing effect for MRG1, which shares the CR1 domain with MSG1 (Fig. 2A).
MSG1 Enhances Transcription Mediated by Full-Length Smad4.
Because N-terminal modification of full-length Smad4 by GAL4DB fusion resulted in inactivation of Smad4 (Fig. 3A and refs. 4 and 30), we reconstituted the Smad2/Smad4-mediated TGFβ signaling pathway by using GAL4DB-fused full-length Smad2 and full-length Smad4 (nonfusion) to determine whether MSG1 enhanced transcriptional activation by full-length Smad4 as well. To avoid confusion caused by endogenous Smad4 (2), we used a Smad4-deficient cell line MDA-MB-468 (6, 31) for this purpose. Without providing Smad4 by transfection, GAL4DB-Smad2 did not show any detectable transactivating activity in the presence or absence of MSG1 or TβRI(T204D) (Fig. 4A), consistent with a previous report (2). In the presence of exogenous Smad4, MSG1 enhanced Smad2/4-mediated transcriptional activation by about 70%. Although TβRI(T204D) alone also enhanced Smad2/4-mediated transcriptional activation, further cotransfection of MSG1 resulted in an additional enhancement by about 100%.
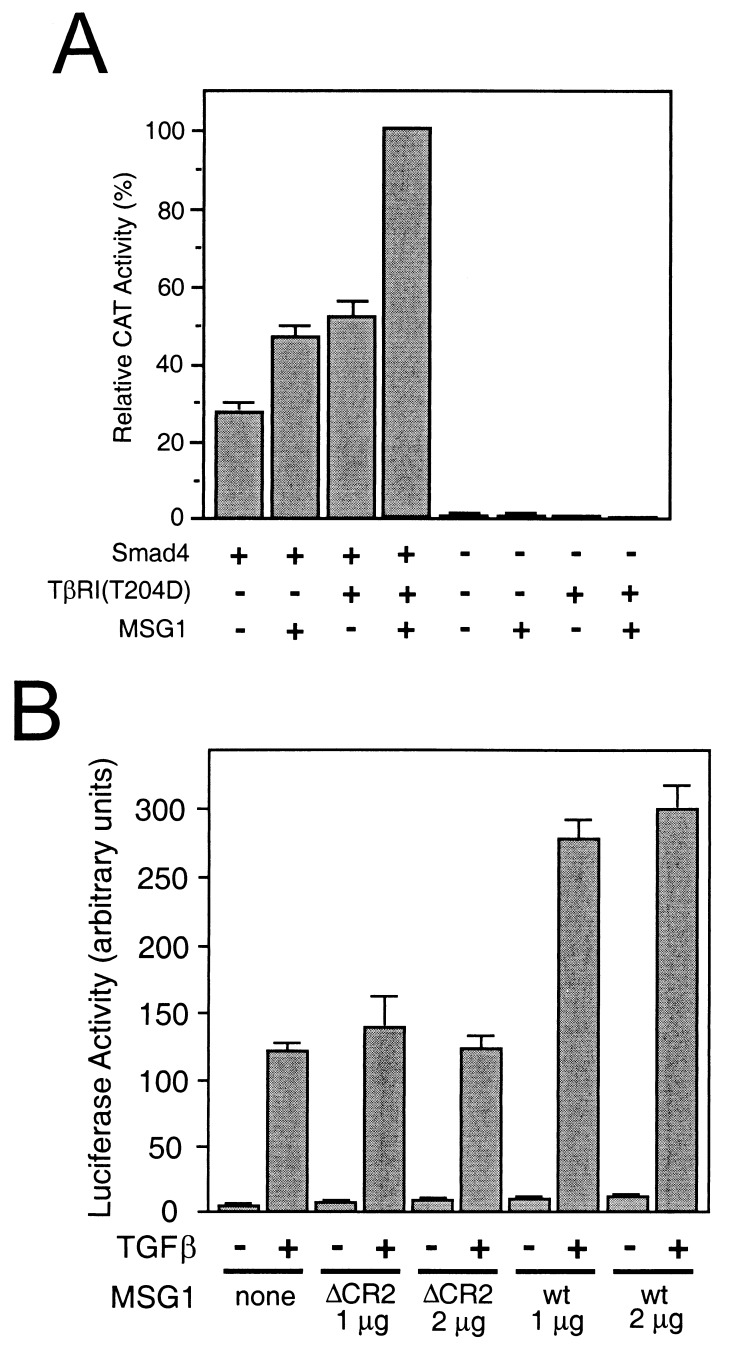
Figure 4.
MSG1 enhances transcription mediated by full-length Smad4 in mammalian cells. (A) MDA-MB-468 cells (Smad4-deficient) were cotransfected with a GAL4DB fusion of full-length Smad2, full-length Smad4 (without fusion), TβRI(T204D), HA-MSG1, together with pG5CAT reporter followed by incubation in the presence of 10% fetal calf serum for 16 h and CAT assay. Results are mean ± SEM of three independent experiments. (B) HepG2 cells were cotransfected with FAST-1, HA-MSG1 [wild type (wt) or a deletion mutant lacking the CR2 transactivating domain (ΔCR2)] together with an ARE-dependent luciferase reporter plasmid. Transfected cells were treated with TGFβ1 (5 ng/ml) for 16 h in the presence of 0.2% fetal calf serum, and the luciferase activity was determined. Results are mean ± SEM of duplicate measurements of triplicate wells from a representative experiment.
We next attempted to determine whether MSG1 enhanced transcription mediated by the FAST-1 DNA-binding protein that binds to the ARE. This transcriptional activation is a physiological example in which a multisubunit transcription factor complex involving SMAD proteins is formed in response to TGFβ family cytokines in Xenopus (26). We reconstituted this FAST-1/ARE-dependent transcription in HepG2 human hepatoblastoma cells and evaluated the effects of MSG1. In this reconstituted system, the SMAD proteins are provided endogenously by the host mammalian cells (2). FAST-1/ARE-mediated transactivation was observed only in the presence of TGFβ (Fig. 4B), and MSG1 enhanced this TGFβ-stimulated transactivation about 2.5-fold. MSG1 lacking the CR2 transactivating domain showed no effect. In the absence of TGFβ signaling, no significant transactivation was observed even when MSG1 was cotransfected. These results suggest that MSG1 is able to enhance transcriptional activation mediated by the SMAD proteins expressed at low endogenous levels in mammalian cells.
DISCUSSION
We have provided evidence that MSG1, a recently reported orphan transcriptional activator, functionally interacts with Smad4(C) (Fig. 1) and enhances transcriptional activating activity of Smad4(C) as well as full-length Smad4 (Figs. 2–4). In mammalian cells, MSG1 enhanced Smad4(C)-mediated transcriptional activation (Fig. 2); and this effect of MSG1 depended on TGFβ signaling (Fig. 2B), augmented by overexpression of the pathway-restricted SMAD proteins (Fig. 2C), and suppressed by Smad4(C) mutations that disrupt SMAD hetero-oligomerization (Fig. 3A). MSG1 enhanced transcription mediated by endogenously expressed SMAD proteins in mammalian cells when SMAD proteins were activated by TGFβ signaling (Fig. 4B). These results suggest that MSG1 may function as a physiological modifier of SMAD-mediated transcriptional activation.
We have shown that the basal transcriptional-activating activity of GAL4DB-Smad4[266–552] detected without MSG1 cotransfection also requires SMAD hetero-oligomerization (Fig. 3A). LexA-fusion Smad4[266–552] did not show any detectable transactivating activity in yeast (Fig. 1C), which does not express endogenous pathway-restricted SMAD proteins. The requirement of SMAD hetero-oligomerization for SMAD-mediated transcriptional activation has been emphasized in previous studies from other laboratories (3, 5, 8, 10, 22, 32); its roles in releasing the autosuppression by Smad4(N) on transcriptional-activating domain located in Smad4(C) (11) as well as in recruiting Smad4 from cytosol to nucleus (2, 12, 13) have been proposed. Our approach using GAL4DB-fusion Smad4(C) mutants has provided evidence that SMAD hetero-oligomerization is still essential for transcriptional transactivating activity located in Smad4(C), even in the absence of autosuppressive Smad4(N) and in the presence of the potent nuclear localization signal of GAL4DB.
Although we cannot exclude the possibility that Smad4 may contain an intrinsic potential transactivating domain that is activated only when SMAD hetero-oligomers are formed, our results suggest an alternative interpretation—namely, Smad4 may require separate, endogenously expressed transcriptional activators for its transactivating activity, at least in part, in mammalian cells. In this case, SMAD hetero-oligomerization might be required for functional interaction between Smad4 and such transactivators. Deletion of the SAD sequence of Smad4, which previously has been proposed as an intrinsic transactivating domain (6), resulted in the simultaneous reduction of both the basal and MSG1-enhanced transcriptional activation mediated by GAL4DB-Smad4 (Fig. 3A). This finding suggests that the SAD sequence actually may be necessary for the functional interaction of separate transactivators (such as MSG1). However, although MSG1 appears to interact functionally with Smad4, biochemical demonstration of their physical interaction has been difficult in our hands, suggesting that the molecular mechanisms of this functional interaction are complex. It should be noted that the spontaneous interaction of MSG1 and Smad4(C) detected by the yeast two-hybrid system (Fig. 1 A and B) does not conflict with the fact that their interaction in mammalian cells depended on TGFβ signaling (Fig. 2B). Because the interacting proteins are strongly overexpressed and potentially modified by endogenous enzymes that are not yet completely characterized, the yeast two-hybrid system can detect any possible protein–protein interactions that actually are regulated (i.e., do not occur spontaneously) in mammalian cells. For example, interaction of Smad2 and Smad4 depends on TGFβ signaling in mammalian cells but occurs spontaneously in a yeast two-hybrid system (8, 10, 22).
MSG1 initially was reported as a melanocyte-specific gene (14, 17). However, extra-melanocytic expression of the Msg1 mRNA transcripts in embryonic tissues recently has been demonstrated (16), and NIH 3T3 cells actually express the Msg1 mRNA transcript at a low level detectable by reverse transcription–PCR but not readily by Northern blotting (data not shown). Considering the very strong transcriptional activating activity of MSG1, which is as high as 73% of the extraordinarily strong transactivating activity of VP16 (15, 33), such a low level of Msg1 expression in NIH 3T3 cells still may be sufficient to contribute to the weak basal transactivation mediated by Smad4[266–552] (Fig. 3A). It is, of course, also possible that there may be other transactivators that functionally interact with Smad4.
Although we do not yet know the physiological role of MSG1, our present study implies that expression levels of MSG1 may affect SMAD-mediated transcription in vivo. We recently have reported that MSG1 expression in human melanocytes is enhanced by endothelin-1 and basic fibroblast growth factor, both of which are physiological melanocyte-acting cytokines (17). The strong expression of the Msg1 mRNA transcripts in B16-F1 melanoma cells (14) was dramatically suppressed by TGFβ treatment (M.H.F., unpublished observation). In addition, a recent study by us has suggested that the intracellular localization of MSG1 also is regulated (17). Such regulation of MSG1 expression and translocation could, in turn, affect the signaling of TGFβ superfamily cytokines. It should be noted that MSG1 did not affect TGFβ-stimulated p3TPLux transactivation in HepG2 cells (R.J.L., unpublished observation). This finding may imply a possible diversity in the mechanisms of transcriptional activation for different target genes of the TGFβ superfamily cytokines.
Finally, although MSG1 was initially characterized as a melanocyte-specific gene (14), based on our present study, the term (S)mad supporting gene appears to be more appropriate.
Acknowledgments
We thank A. S. Zervos for human melanocyte cDNA expression library and help in the interaction trap cloning; J. L. Wrana, L. Attisano, and M. Whitman for providing plasmids; and J. Ryan and C. Eng for technical support and plasmid construction.
ABBREVIATIONS
- ARE
activin responsive element
- CAT
chloramphenicol acetyltransferase
- HA
hemagglutinin
- GAL4DB
GAL4 DNA-binding domain
- Smad4(C)
carboxy-terminal domain of Smad4
- Smad4(N)
amino-terminal domain of Smad4
- TGFβ
transforming growth factor β
- TβRI
TGFβ receptor type I
References
- 1.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 2.Liu F, Pouponnot C, Massagué J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 4.Liu F, Hata A, Baker J C, Doody J, Cárcamo J, Harland R M, Massagué J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 6.de Caestecker M P, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts A B, Lechleider R J. J Biol Chem. 1997;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Hata A, Lo R S, Massagué J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 8.Wu R-Y, Zhang Y, Feng X-H, Derynck R. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 10.Hata A, Lo R S, Wotton D, Lagna G, Massagué J. Nature (London) 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 11.Wrana J, Pawson T. Nature (London) 1997;388:28–29. doi: 10.1038/40290. [DOI] [PubMed] [Google Scholar]
- 12.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J-I, Heldin C-H, Miyazono K, ten Dijke P. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souchelnytskyi S, Tamaki K, Engström U, Wernstedt C, ten Dijke P, Heldin C-H. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 14.Shioda T, Fenner M H, Isselbacher K J. Proc Natl Acad Sci USA. 1996;93:12298–12303. doi: 10.1073/pnas.93.22.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shioda T, Fenner M H, Isselbacher K J. Gene. 1997;204:235–241. doi: 10.1016/s0378-1119(97)00551-9. [DOI] [PubMed] [Google Scholar]
- 16.Dunwoodie S L, Rodriguez T, Beddington R S P. Mech Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Ahmed N U, Fenner M H, Ueda M, Isselbacher K J, Shioda T. Exp Cell Res. 1998;242:478–486. doi: 10.1006/excr.1998.4123. [DOI] [PubMed] [Google Scholar]
- 18.Fenner, M. H., Parrish, J. E., Boyd, Y., Reed, V., MacDonald, M., Nelson, D. L., Isselbacher, K. J. & Shioda, T. (1998) Genomics, in press. [DOI] [PubMed]
- 19.Finley R L, Jr, Brent R. In: DNA Cloning—Expression Systems: A Practical Approach. Glover D, Hames B D, editors. Oxford: Oxford Univ. Press; 1996. pp. 169–203. [Google Scholar]
- 20.Chien C-T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis I L, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 22.Abdollar S, Macías-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana J L. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 23.Wieser R, Wrana J L, Massagué J. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Nature (London) 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 25.Sadowski I, Bell B, Broad P, Hollis M. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 27.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 28.Pironin M, Clement G, Benzakour O, Barritault D, Lawrence D, Vigier P. Int J Cancer. 1992;51:980–988. doi: 10.1002/ijc.2910510624. [DOI] [PubMed] [Google Scholar]
- 29.Pierson B A, Gupta K, Hu W S, Miller J S. Blood. 1996;87:180–189. [PubMed] [Google Scholar]
- 30.Atfi A, Buisine M, Mazars A, Gespach C. J Biol Chem. 1997;272:24731–24734. doi: 10.1074/jbc.272.40.24731. [DOI] [PubMed] [Google Scholar]
- 31.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 32.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X-F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;333:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]