Abstract
In Xenopus egg extracts, spindles assembled around sperm nuclei contain a centrosome at each pole, while those assembled around chromatin beads do not. Poles can also form in the absence of chromatin, after addition of a microtubule stabilizing agent to extracts. Using this system, we have asked (a) how are spindle poles formed, and (b) how does the nucleation and organization of microtubules by centrosomes influence spindle assembly? We have found that poles are morphologically similar regardless of their origin. In all cases, microtubule organization into poles requires minus end–directed translocation of microtubules by cytoplasmic dynein, which tethers centrosomes to spindle poles. However, in the absence of pole formation, microtubules are still sorted into an antiparallel array around mitotic chromatin. Therefore, other activities in addition to dynein must contribute to the polarized orientation of microtubules in spindles. When centrosomes are present, they provide dominant sites for pole formation. Thus, in Xenopus egg extracts, centrosomes are not necessarily required for spindle assembly but can regulate the organization of microtubules into a bipolar array.
During cell division, the correct organization of microtubules in bipolar spindles is necessary to distribute chromosomes to the daughter cells. The slow growing or minus ends of the microtubules are focused at each pole, while the plus ends interact with the chromosomes in the center of the spindle (Telzer and Haimo, 1981; McIntosh and Euteneuer, 1984). Current concepts of spindle assembly are based primarily on mitotic spindles of animal cells, which contain centrosomes. Centrosomes are thought to be instrumental for organization of the spindle poles and for determining both microtubule polarity and the spindle axis. In the prevailing model, termed “Search and Capture,” dynamic microtubules growing from two focal points, the centrosomes, are captured and stabilized by chromosomes, generating a bipolar array (Kirschner and Mitchison, 1986). However, while centrosomes are required for spindle assembly in some systems (Sluder and Rieder, 1985; Rieder and Alexander, 1990; Zhang and Nicklas, 1995a ,b), in other systems they appear to be dispensable (Steffen et al., 1986; Heald et al., 1996). Furthermore, centrosomes are not present in higher plant cells and in female meiosis of most animal species (Bajer and Mole, 1982; Gard, 1992; Theurkauf and Hawley, 1992; Albertson and Thomson, 1993; Lambert and Lloyd, 1994). In the absence of centrosomes, bipolar spindle assembly seems to occur through the self-organization of microtubules around mitotic chromatin (McKim and Hawley, 1995; Heald et al., 1996; Waters and Salmon, 1997). The observation of apparently different spindle assembly pathways raises several questions: Do different types of spindles share common mechanisms of organization? How do centrosomes influence spindle assembly? In the absence of centrosomes, what aspects of microtubule self-organization promote spindle bipolarity?
To begin to address these questions, we have used Xenopus egg extracts, which can be used to reconstitute different types of spindle assembly. Spindle assembly around Xenopus sperm nuclei is directed by centrosomes (Sawin and Mitchison, 1991). Like other meiotic systems (Bastmeyer et al., 1986; Steffen et al., 1986), Xenopus extracts also support spindle assembly around chromatin in the absence of centrosomes through the movement and sorting of randomly nucleated microtubules into a bipolar structure (Heald et al., 1996). In this process, the microtubule-based motor cytoplasmic dynein forms spindle poles by cross-linking and sliding microtubule minus ends together. Increasing evidence suggests that the function of dynein in spindle assembly depends on its interaction with other proteins, including dynactin, a dynein-binding complex, and NuMA1 (nuclear protein that associates with the mitotic apparatus) (Merdes et al., 1996; Echeverri et al., 1996; Gaglio et al., 1996). In this paper, we demonstrate that both in the presence and absence of centrosomes, spindle pole assembly occurs by a common dynein-dependent mechanism. We show that when centrosomes are present, they are tethered to spindle poles by dynein. In the absence of dynein function, microtubules are still sorted into an antiparallel array, indicating that other aspects of microtubule self-assembly independent of pole formation promote spindle bipolarity around mitotic chromatin. Since centrosomes are dispensable for pole formation in this system, what is their function? We show here that if only one centrosome is present, it acts as a dominant site for microtubule nucleation and focal organization, resulting in a monopolar spindle. Therefore, although centrosomes are not required in this system, they can influence spindle pole formation and bipolarity.
Materials and Methods
Preparation of Extracts, DMSO Asters, and Spindles
10,000-g cytoplasmic extracts of unfertilized Xenopus eggs arrested in metaphase of meiosis II by CSF activity were prepared fresh as described (Murray, 1991). FITC-labeled tubulin prepared from calf brain tubulin was added to 0.2 mg/ml (Hyman, 1991). DNA beads and chromatin bead spindles were prepared as described (Heald et al., 1996). DMSO asters were assembled by the addition of 5% DMSO and a 30-min incubation at 20°C (Sawin and Mitchison, 1994). Mitotic spindles were assembled around demembranated sperm nuclei by either the half spindle or the interphase to mitotic pathway as described (Sawin and Mitchison, 1991).
Immunofluorescence and Video Experiments
For immunofluorescence of sperm DNA and chromatin bead spindles, 20-μl reactions were diluted with 1 ml 30% glycerol, 1% Triton X-100 in BRB80 (80 mM Pipes, 2 mM MgCl2, 1 mM EGTA) and spun onto coverslips in modified corex tubes as described (Mitchison and Kirschner, 1984). For DMSO aster reactions and for visualizing dissociating centrosomes, 15% glycerol was used instead of 30%. For samples to be processed for dynein heavy chain immunofluorescence, 4% formaldehyde was included. After spinning, coverslips were fixed in methanol at −20°C for 5 min and then blocked in 3% BSA for 10 min at room temperature. Primary antibodies used were raised against Xenopus proteins and affinity purified. Polyclonal anti-NuMA antibodies were provided by A. Merdes (University of California, San Diego, CA). Polyclonal anti–γ tubulin antibodies were raised to a COOH-terminal peptide by T. Ashford (European Molecular Biology Laboratory, Heidelberg, Germany), and anti– dynein heavy chain antibodies raised to a conserved sequence in the motor domain (Vaisberg et al., 1993) were provided by S. Reinsch (European Molecular Biology Laboratory). Rhodamine-conjugated secondary antibodies were used, and in some cases DNA was stained with propidium iodide. Rhodamine-labeled seeds were prepared and video microscopy was performed as described (Heald et al., 1996). Seed movement data was acquired using NIH Image. Seed distance from the center of DMSO asters was measured at 5-s intervals.
Dynein Inhibition
The monoclonal IgM anti–dynein intermediate chain antibody (mAb 70.1) and control IgM anti–mouse IgG ascites were obtained from Sigma Chemical Co. (St. Louis, MO) and dialyzed against 50 mM potassium glutamate, 0.5 mM MgCl2, then concentrated to 20 mg/ml, flash frozen, and stored in small aliquots at −80°C. For spindle pole disruption experiments, antibodies were diluted 1:10 in assembly reactions, which were then diluted and spun after 5 or 10 min as described above. To block pole assembly, mAb 70.1 was added 1:10 to extracts before spindle assembly.
Preparation of EM Samples
Chromatin bead spindles in 200 μl of extract were magnetically retrieved at 20°C. The extract was removed, and the spindles were gently resuspended in 100 μl of hooking solution containing 1.5 mg/ml calf brain tubulin, 0.5 M Pipes, pH 6.9, 1.5 mM MgCl2, 1 mM EGTA, 1 mM GTP, and 2.5% DMSO. After a 5–7-min incubation at 20°C, spindles were again magnetically retrieved, and the hooking solution was removed. 1 ml of 25% glycerol, 0.2% glutaraldehyde in BRB80 was added, and then more glutaraldehyde was added immediately afterward, bringing the final concentration to 2%. After 30 min at 20°C, the spindles were pelleted by centrifugation and washed several times with PBS. Samples were then dehydrated, imbedded in epon and sectioned, and observed with an electron microscope (Carl Zeiss, Inc., Thornwood, NY). For data acquisition, hook handedness was assessed in 50–100 microtubules in each section, and three sections were analyzed for each condition.
Results
Pole Assembly Proceeds by a Common Mechanism with or without Centrosomes
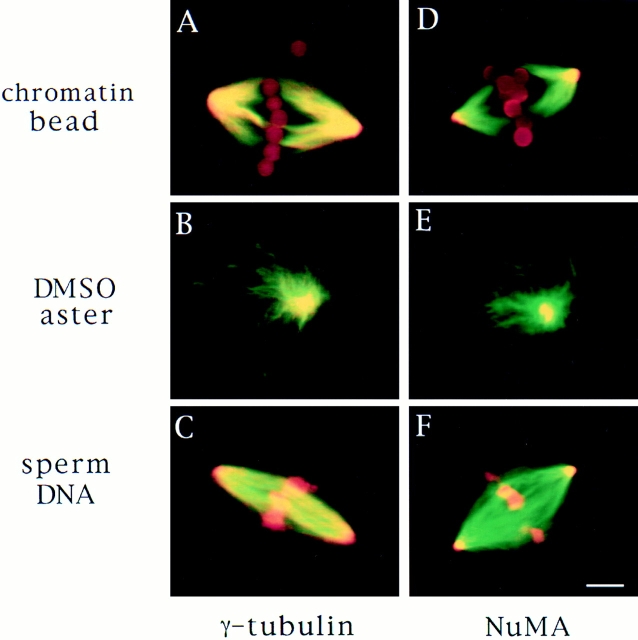
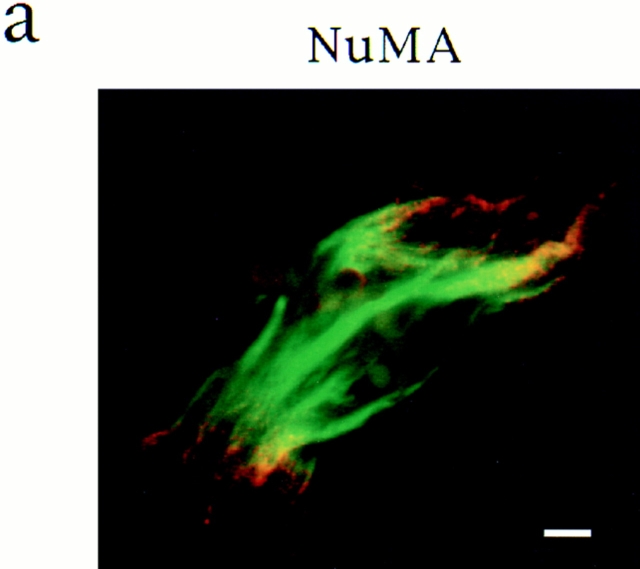
To compare the mechanism of spindle pole assembly in the presence and absence of centrosomes, we examined how spindle poles formed in Xenopus egg extracts under three different conditions: around sperm nuclei that contain centrosomes (Sawin and Mitchison, 1991), around chromatin beads in the absence of centrosomes (Heald et al., 1996), and in self-assembled mitotic asters, which form in the absence of centrosomes and chromatin but in the presence of DMSO, a microtubule stabilizing agent (Sawin and Mitchison, 1994). We first compared the structure of the three different types of poles by immunofluorescence microscopy by examining the distribution of two different proteins usually found associated with microtubule minus ends in polarized arrays, γ tubulin (Lajoie et al., 1994; Stearns and Kirschner, 1994; Debec et al., 1995; Li and Joshi, 1995) and NuMA (Maekawa et al., 1991; Gaglio et al., 1995; Merdes et al., 1996). These proteins were localized similarly in all three cases (Fig. 1). γ Tubulin was enriched at poles but was also present throughout the microtubules. NuMA was highly focused in the center of the poles. Thus, by morphological criteria, centrosomal and noncentrosomal poles appear to be quite similar.
Figure 1.
Spindle pole antigens are similarly localized on poles whether or not centrosomes are present. Microtubules are green, antigens and DNA are red, and overlap is yellow. γ Tubulin (A–C) and NuMA (D–F) immunofluorescence of chromatin bead spindles (A and D) and DMSO asters (B and E) with self-assembled poles and sperm DNA spindles (C and F) with a centrosome at each pole. Bar, 5 μm.
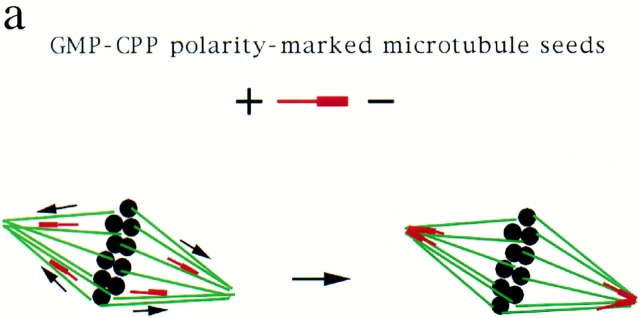
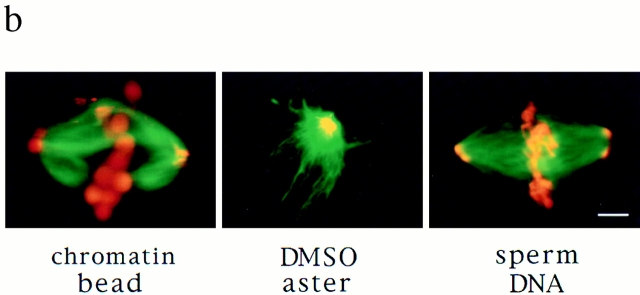
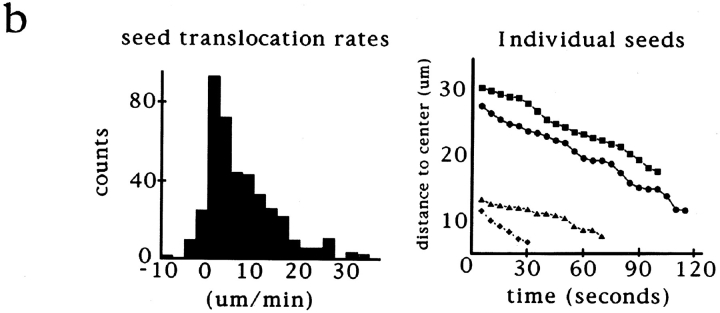
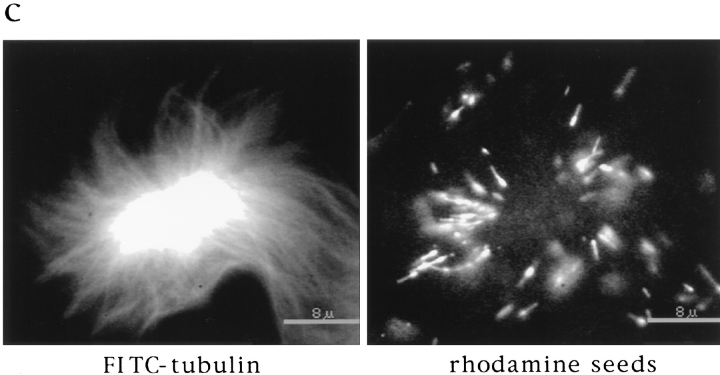
While the visual similarity of these microtubule arrays was striking, it did not indicate whether pole assembly occurred by a common mechanism. To characterize how poles form, we took advantage of an assay developed to follow microtubule movements during spindle assembly. We have shown previously that stable polarity-marked microtubule “seeds,” containing a brightly labeled minus end and a dimly labeled plus end, act as markers for the movement of endogenous microtubules during spindle assembly around chromatin beads (Heald et al., 1996). Seeds added to extracts bind to and translocate along microtubules towards poles with their minus ends leading (Fig. 2 a). We compared seed motility on the three types of polarized microtubule arrays (Fig. 2 b). Just as on chromatin bead spindles, microtubule seeds translocated poleward in DMSO asters, forming bright foci at the poles. A complete analysis of seed movement on DMSO asters is shown in Fig. 3. Poleward seed movement was saltatory, with an average speed of 6 μm/min and a peak velocity of 30 μm/min. Polarity-marked seeds moved poleward with their brightly labeled minus ends leading. Since both chromatin bead and DMSO aster pole structures are generated by microtubule self-organization, these results were not surprising. Importantly, however, seeds also translocated poleward on sperm DNA spindles containing poles derived from centrosomes (Fig. 2 b). In all three cases, seed movement was qualitatively and quantitatively similar (data not shown). Therefore, minus end–directed microtubule movement is a general property of poles, whether or not centrosomes are present.
Figure 2.
Stable microtubule seeds translocate to and accumulate at mitotic poles in the presence and absence of centrosomes. Seeds are red, microtubules are green, and overlap is yellow. (a) Diagram of seed experiment: polarity-marked microtubules polymerized from pure tubulin with a nonhydrolyzable GTP analogue, GMP-CPP, were added to extracts containing spindles. Seeds bound to spindle microtubules and moved poleward, where they accumulated. (b) Seeds accumulate at poles of chromatin bead spindles, DMSO asters, and sperm DNA spindles that contain centrosomes. Bar, 5 μm.
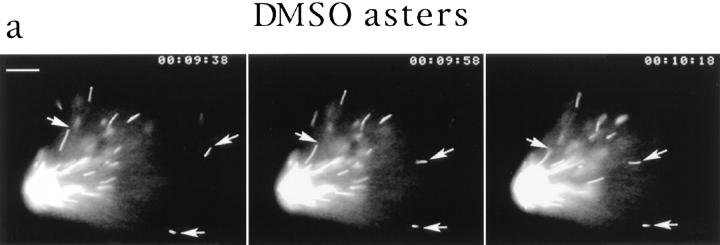
Figure 3.
Video analysis of seed movement on DMSO asters. (a) Successive video frames at 20-s intervals showing that individual seeds move poleward. (b) Rates of seed movement over 5-s intervals and distances of individual seeds from the pole over time. (c) Polarity-marked microtubule seeds are oriented with their minus ends directed toward the center of the aster and its focus of microtubule minus ends. Bars: (a) 5 μm; (c) 8 μm.
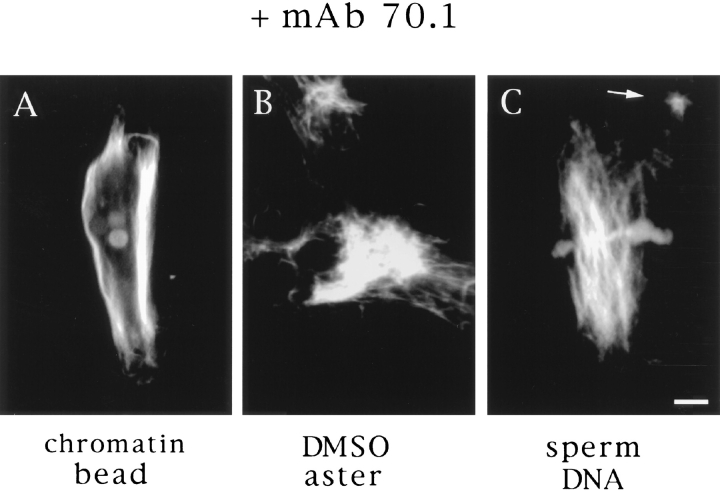
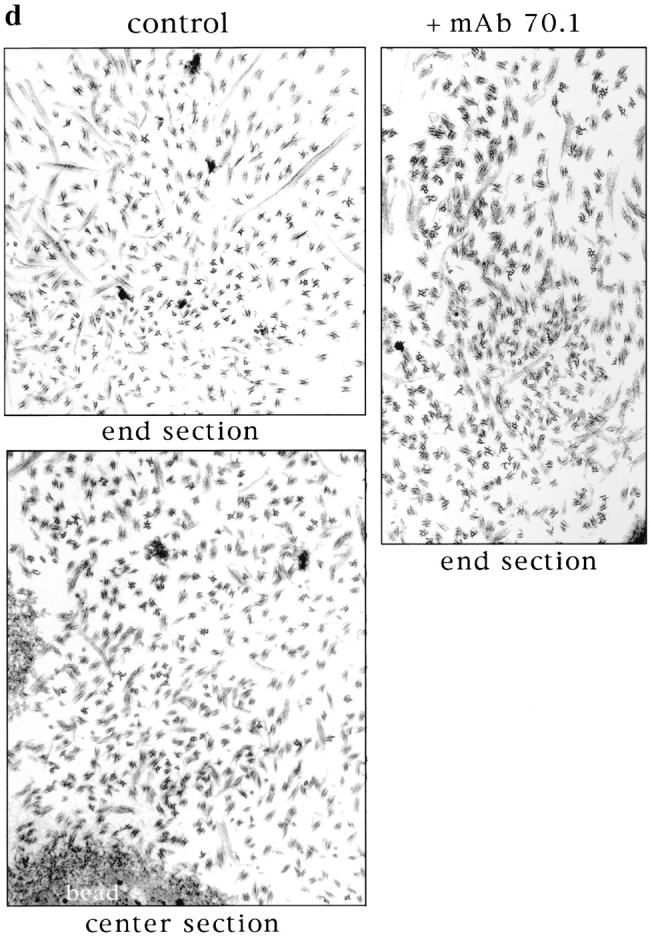
The similar behavior of stable microtubules added to spindles and asters suggested that this movement was due to a common mechanism. We have shown that seed movement on chromatin bead spindles is dependent on cytoplasmic dynein. Inhibition of dynein with vanadate or by addition of a monoclonal antibody to the dynein intermediate chain (mAb 70.1) not only blocked seed translocation but caused dissolution of the poles (Heald et al., 1996; and Fig. 4 A). We wanted to test whether dynein was also involved in the organization of DMSO asters and in spindle poles containing centrosomes. The addition of mAb 70.1 to DMSO asters caused disruption of aster integrity (Fig. 4 B), confirming a requirement for dynein in spontaneous aster assembly shown previously (Verde et al., 1991; Gaglio et al., 1996). Furthermore, addition of mAb 70.1 also disrupted sperm DNA spindle poles containing centrosomes, causing them to splay outwards and become disorganized within 10 min (Fig. 4 C). In all cases, addition of antibody prevented seed movement towards poles (data not shown). When added before initiation of spindle assembly reactions, mAb 70.1 blocked pole formation, yielding similar structures with frayed, unfocused ends (Heald et al., 1996; see Fig. 8 a, D). Therefore, these results indicate that dynein-dependent translocation of microtubules is required for pole formation and maintenance both in the presence and absence of centrosomes.
Figure 4.
Disruption of poles by antidynein antibodies. The monoclonal antibody (mAb 70.1) that recognizes the intermediate chain of cytoplasmic dynein was added to extracts containing chromatin bead spindles (A), DMSO asters (B), or sperm DNA spindles (C) with centrosomes. Pole structures were disrupted within 10 min. Bar, 5 μm.
Figure 8.
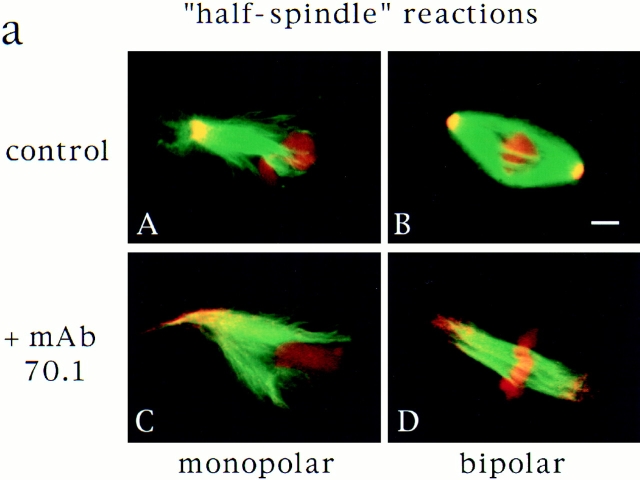
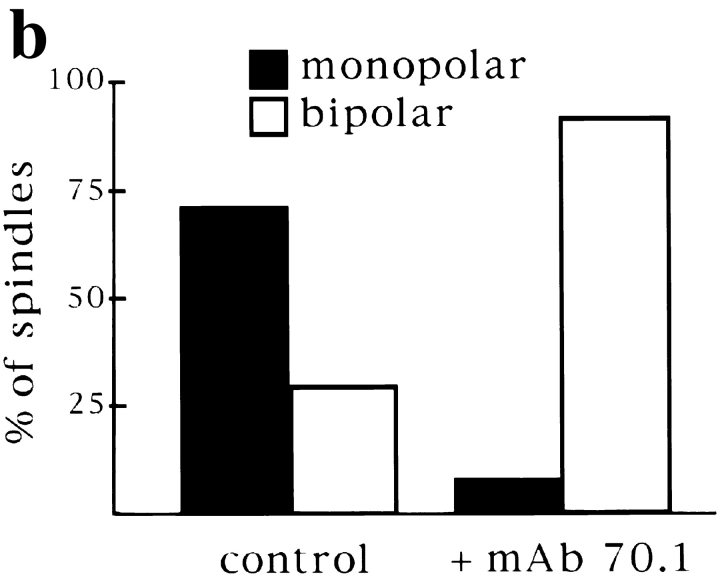
Centrosomes are dominant sites of pole assembly. (a) Sperm half spindle reactions in the presence of control antibodies (A and B) or mAb 70.1 (C and D) were evaluated for microtubule polarity by NuMA immunofluorescence. (b) Quantification of monopolar and bipolar structures under each condition. At least 300 spindles were counted for each condition in two separate experiments. Bar, 5 μm.
We wanted to understand how mAb 70.1 was disrupting dynein activity. Microtubule gliding assays performed with purified dynein revealed that mAb 70.1 did not block motility of the motor (data not shown). We therefore examined whether dynein localization was disrupted by addition of mAb 70.1 to extracts. A polyclonal antibody to the dynein heavy chain was used for immunofluorescent analysis of spindles before and after mAb 70.1 addition (Fig. 5). In agreement with published reports, the dynein heavy chain was localized to the poles of spindles, and faint punctate staining was also visible on chromosomes, probably corresponding to kinetochores (Pfarr et al., 1990; Steuer et al., 1990). Upon addition of mAb 70.1, all spindle staining with the heavy chain antibody disappeared within 5 min. Therefore, mAb 70.1 seems to inhibit dynein by preventing localization and/or accumulation of the motor on spindle microtubules, perhaps by disruption of the interaction of dynein with other proteins, such as the dynactin complex or NuMA.
Figure 5.
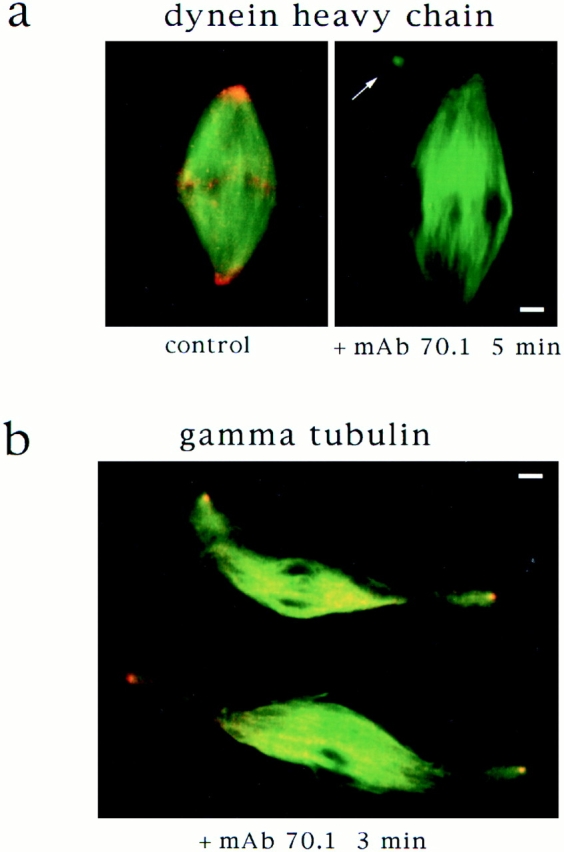
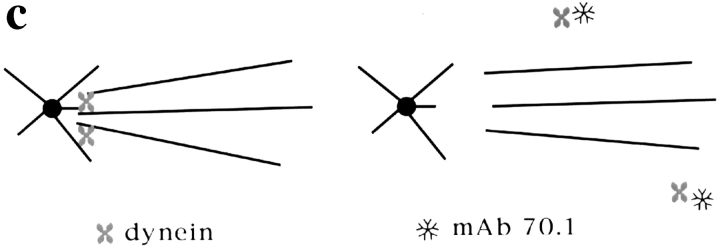
Dynein tethers centrosomes to spindle poles. (a) Cytoplasmic dynein is eluted from spindles by addition of mAb 70.1. Immunofluorescent localization of dynein heavy chain to spindle poles disappears within 5 min after mAb 70.1 addition. Microtubules are green, dynein heavy chain is red, and overlap is yellow. (b) Centrosomes are released from sperm DNA spindle poles 3 min after addition of mAb 70.1. Immunofluorescent localization of γ tubulin on sperm centriolar structures that are dissociating from spindle poles. (c) Model of how dynein tethers spindle microtubules to centrosomal microtubules and how this is disrupted by mAb 70.1. Bars, 5 μm.
Centrosomes Are Tethered to Spindle Poles by Dynein
The disruption of sperm DNA spindle poles by mAb 70.1 showed that centrosomes are not sufficient to maintain the cohesion of poles in the absence of dynein activity. We wanted to know what happened to the centrosomes after dynein inhibition. It has been proposed that centrosomes are tethered to spindle poles by the action of a dynein complex that cross-links spindle microtubules to centrosomal microtubules (Karsenti, 1991; Fig. 5 c). If this were true, then dynein disruption should release centrosomes from spindles. We therefore examined sperm DNA spindles after dynein inhibition and often noted small asters near spindle poles (Figs. 4 and 5 a, arrows). These structures looked very similar to isolated centrosomes and were not found upon mAb 70.1 addition to chromatin bead spindles that lack centrosomes (not shown). To determine whether these structures were dissociated centrosomes, we localized centrosomes shortly after mAb 70.1 addition by immunofluorescent analysis of γ tubulin (Fig. 5 b). Centriolar-like structures were often visible, tenuously linked to spindle microtubules. These experiments indicate that centrosomes are discrete entities independent of spindle poles, tethered to the spindle by cytoplasmic dynein activity (Fig. 5 c).
Sorting of Microtubules into an Antiparallel Array Does Not Require Pole Formation
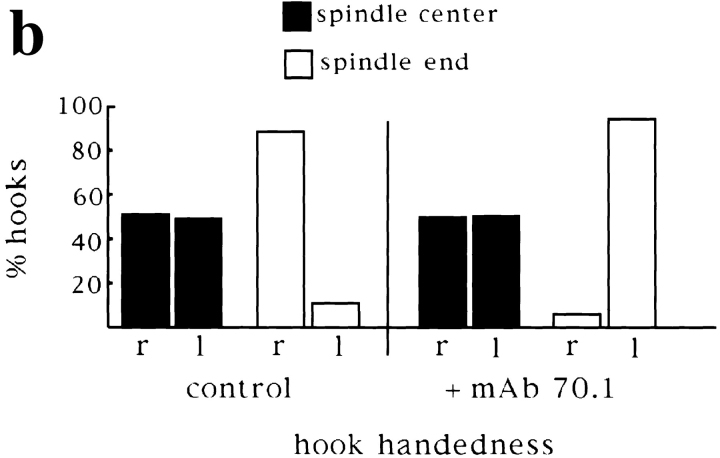
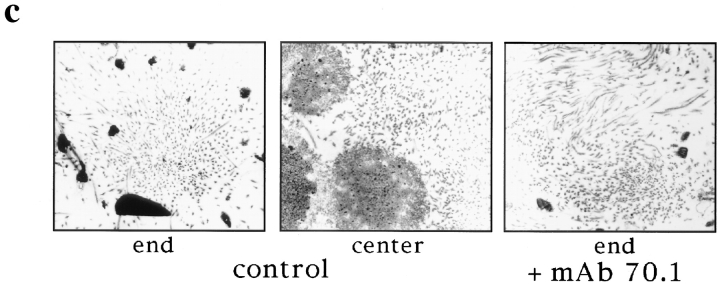
Our results thus far indicate that cytoplasmic dynein plays a central role in spindle organization. We wondered to what extent dynein function and pole assembly contributed to the polarized organization of microtubules in spindles, with microtubule minus ends at the poles and antiparallel microtubule interactions in the center of the spindle. We have shown previously that in the presence of mAb 70.1, a parallel array of microtubules formed around chromatin beads with frayed, unfocused ends, and the beads located in the center of the array (Heald et al., 1996; Fig. 6 a). We wanted to know whether microtubules in such bundles were randomly oriented or sorted into an antiparallel array around chromatin, with plus ends at the chromatin and minus ends at spindle termini. To address this question, we assessed microtubule polarity in spindles assembled in the presence or absence of mAb 70.1 using two independent approaches. First, we examined the localization of NuMA, a protein normally associated with the minus ends of microtubules in polarized arrays (Fig. 1) (Maekawa et al., 1991). We found that NuMA was still localized to the two frayed unfocused ends of the spindle formed in the presence of mAb 70.1, albeit more diffusely (Fig. 6 a). This result indicated that microtubule minus ends were still sorted away from chromatin in the absence of pole formation. Second, we determined directly whether microtubule polarity was uniform or not by the hooking technique. In this technique, microtubules are incubated with pure tubulin under conditions that promote addition of hooked protofilament appendages to the microtubule walls (Heidemann and McIntosh, 1980). The polarity of individual microtubules can then be determined by examination of hook handedness in serial sections by electron microscopy (Euteneuer and McIntosh, 1981; Euteneuer et al., 1982). Here we analyzed single sections to determine hook handedness in chromatin bead spindles assembled in the presence of control antibodies or mAb 70.1 (Fig. 6). Under both conditions, sections that were cut through beads, likely to correspond to the spindle centers, contained microtubules with approximately equal proportions of right- and left-handed hooks, indicating that microtubules were of mixed polarity (Fig. 6, b–d). In control spindles, sections through focused microtubule bundles, corresponding to poles, contained hooks of which 90% were the same handedness, indicating that microtubules were of almost uniform polarity (Fig. 6, b–d). Although spindles assembled in the presence of mAb 70.1 did not contain poles, microtubule bundles, likely to be close to spindle termini, contained microtubules of >95% uniform hook handedness. Therefore, both NuMA staining and hook analysis show that microtubules are sorted into antiparallel arrays around chromatin beads. This sorting is independent of dynein activity and pole formation, indicating that other microtubule-based motors are contributing to the antiparallel organization of microtubules in spindles.
Figure 6.
Microtubules are sorted into antiparallel arrays around chromatin in the absence of pole formation. (a) Immunofluorescent localization of NuMA to the frayed ends of chromatin bead spindles that have formed in the absence of dynein activity. (b and c) Hooking analysis. (b) Quantification of hook handedness in control and poleless spindles. Percentage of right- and left-handed hooks in sections through spindle centers containing chromatin beads, and spindle ends containing microtubule poles (control), or bundles (+ mAb 70.1). (c) Low magnification micrographs (5-μm width) are shown to give an overall impression of microtubule organization seen in sections through spindle ends containing microtubule poles or bundles and through spindle centers containing beads. (d) Higher magnification micrographs (2–2.5-μm width) show hooks on cross sectioned individual microtubules. In the presence of control antibodies, a section through a pole contains right-handed (clockwise) hooks, while a section containing beads contains both right- and left-handed hooks. In the presence of mAb 70.1, a section through a microtubule bundle likely to be at the spindle end is shown that contains almost exclusively left-handed hooks. Note: Hook handedness does not give any information about the polarity of the microtubules in these sections but indicates the degree to which microtubule polarity is uniform. 50–100 microtubules were evaluated in each section, and three sections were evaluated for each condition. Bar, 5 μm.
Centrosomes Regulate Bipolarity by Providing Focal and Dominant Pole Assembly Sites
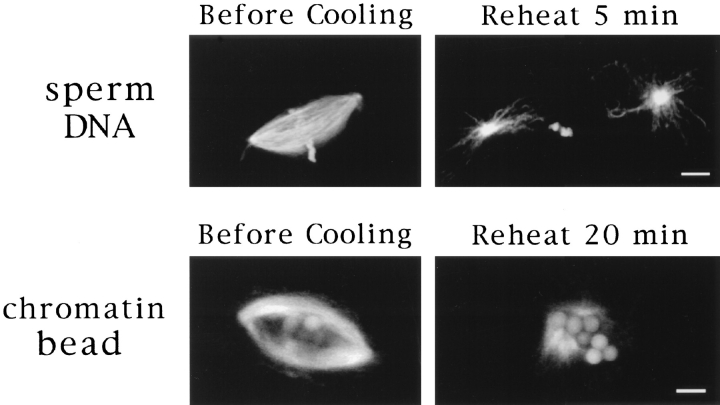
We have shown that pole formation occurs by a general dynein-dependent mechanism in Xenopus egg extracts whether or not centrosomes are present, creating morphologically indistinguishable structures (Fig. 1). These results raise the fundamental question of the role of centrosomes in spindle assembly, and more specifically in the establishment of spindle bipolarity. One potential explanation for our results is that pole assembly mechanisms seem to be equivalent because microtubule organizing centers, similar to centrosomes, are created at poles during microtubule self-organization. To address this issue, we examined the fate of spindle poles after microtubule depolymerization. If chromatin bead spindle pole formation creates stable microtubule organizing structures, we reasoned that this organization should persist after pole disassembly. To test this, we cooled spindle reactions containing either sperm DNA spindles or chromatin bead spindles on ice to cause complete microtubule depolymerization. The reactions were then incubated at 20°C for various times and examined for microtubule regrowth (Fig. 7). In extracts containing sperm nuclei, microtubule asters were visible within 5 min after reheating. Two asters were often found near condensed sperm chromatin, apparently growing from the centrosomes that were at the spindle poles, no longer closely associated with sperm chromatin. In contrast, chromatin bead reactions contained no asters or microtubules 5 min after reheating. Detectable microtubule growth did not occur until ∼15 min later, when microtubules began to grow close to the chromatin beads. This nucleation phenotype corresponds to the first step in spindle assembly in the absence of centrosomes (Heald et al., 1996). Thus, poles assembled around chromatin beads do not become permanent microtubule organizing centers, whereas centrosomes retain this property.
Figure 7.
Centrosomes provide permanent microtubule organizing sites. Sperm DNA and chromatin bead spindles before and 5 or 20 min after incubation on ice to depolymerize microtubules. No microtubules were visible on chromatin beads 5 min after cooling. Bar, 5 μm.
Although the self-assembly of spindle poles does occur in some systems, centrosomes, when present, seem to determine the number of poles formed (Mazia et al., 1981; Bajer, 1982). This phenomenon has also been observed in Xenopus egg extracts, in which monopolar spindles form in the presence of a single centrosome, or when centrosome separation is blocked (Sawin and Mitchison, 1991; Boleti et al., 1996). However, we have shown that in the absence of centrosomes, predominantly bipolar spindles formed around chromatin beads (Heald et al., 1996). These observations suggest that the presence of one centrosome, by providing a dominant focal microtubule nucleation site, could overrule the natural tendency of microtubules to self-organize into a bipolar array around chromosomes. To test centrosome dominance directly, we compared spindle assembly reactions in the presence and absence of a single centrosome. Xenopus “half spindle” reactions mimic the single centrosome reaction because a sperm nucleus containing a single centrosome directs assembly of a monopolar spindle when added directly to mitotic extracts (Fig. 8 a; Sawin and Mitchison, 1991). To remove the centrosome, we added mAb 70.1, reasoning that dynein inhibition would cause release of the centrosome from the microtubule array, preventing its influence. We then assessed the polarity of microtubule arrays formed by immunofluorescent localization of NuMA (Fig. 8). In the control reaction, as expected, more than 75% of the microtubule arrays formed after 45 min were monopolar, with NuMA localized to the single spindle pole. Approximately 25% of the structures observed were bipolar, as a result of half spindle fusion, as previously described (Sawin and Mitchison, 1991). In contrast, ∼90% of the microtubule structures formed in the presence of mAb 70.1 were sorted into an antiparallel array around chromatin with NuMA localized at each end. NuMA staining was no longer focused because spindle poles did not form in the absence of dynein activity. However, these structures were bipolar in the sense that microtubule minus ends were at the two spindle termini. 10% of the structures had a monopolar distribution of NuMA with sperm chromatin asymmetrically located. These results indicate that bipolarity is the favored self-assembly state of microtubules around chromatin. However, the presence of a single centrosome influences microtubule organization, leading to monopolarity. Centrosomes therefore create dominant sites for pole formation.
Discussion
The Role of a Dynein Complex in Pole Formation
In this paper we show that in Xenopus egg extracts, mitotic poles formed from DMSO-stabilized microtubules, around chromatin beads or around sperm chromatin, are generated by the same mechanism; the minus end–directed movement of microtubules across one another, driven by dynein (Verde et al., 1991; Gaglio et al., 1996; Heald et al., 1996). There is an increasing body of evidence supporting a general requirement for dynein in spindle assembly in vivo (Pfarr et al., 1990; Steuer et al., 1990; Vaisberg et al., 1993). Both genetic and biochemical evidence suggest that dynein function is dependent on its interaction with dynactin, a dynein-binding complex (for reviews see Schroer, 1994; Schroer et al., 1996). Here, we show that an antibody directed against the dynein intermediate chain (mAb 70.1) prevents the accumulation of dynein heavy chain on spindle microtubules. Because the dynein intermediate chain interacts with the dynactin subunit p150-glued (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995), mAb 70.1 may be dissociating dynein from spindles by disrupting its interaction with dynactin. Interestingly, overexpression of dynamitin (a component of dynactin) in somatic cells also prevents proper targeting of dynein to spindles and disrupts the poles (Echeverri et al., 1996). Both dynein and dynactin are also required for taxol aster formation in HeLa cell extracts (Gaglio et al., 1996), further supporting the generality of this mechanism.
Another protein known to be required for microtubule organization into poles is NuMA (Kallajoki et al., 1991, 1992, 1993; Yang and Snyder, 1992; Compton and Cleveland, 1993; Compton and Luo, 1995; Gaglio et al., 1995). Recently, experiments in Xenopus egg extracts have shown that dynein and NuMA interact and that depletion of NuMA results in a spindle pole defect very similar to that of dynein inhibition (Merdes et al., 1996). A model consistent with these results is that NuMA is transported to microtubule minus ends in a complex with dynein, where NuMA could help cross-link microtubules and give cohesion to the spindle pole (Merdes et al., 1996). However, our results indicate that at least some NuMA was still enriched at spindle ends in the absence of dynein activity (Fig. 6 a). Furthermore, we have found that inhibition of NuMA blocks movement of microtubule seeds on spindle arrays (Heald, R., and A. Merdes, unpublished data). These data support the idea that, at least in Xenopus, a complex of dynein and NuMA is essential for spindle pole formation. Our results indicate that NuMA is targeted to microtubule minus ends by another mechanism in addition to its association with dynein, perhaps in association with another minus end–directed motor, as proposed (Gaglio et al., 1996), or NuMA could attain a polarized distribution independently of motor activity (Maekawa et al., 1991). Taken together, the data emerging from several laboratories indicate that dynein functions in a multimeric complex with NuMA and dynactin. However, the molecular mechanism by which these proteins interact to form spindle poles is not yet understood.
Microtubule Sorting
In the process of spindle assembly around chromatin beads, randomly nucleated microtubules are organized into a bipolar array with microtubule minus ends at poles and plus ends at chromatin (Heald et al., 1996). We have referred to this process as microtubule sorting and proposed that microtubule-based motors could function as sorting devices by recognizing microtubule polarity and moving microtubules relative to one another and to chromatin. One possibility is that sorting occurs concomitantly with spindle pole assembly, as dynein collects microtubule minus ends into arrays of uniform polarity. Here we have shown by two independent means that microtubule sorting occurs in the absence of dynein activity and pole formation, resulting in an antiparallel array of microtubules around chromatin. While NuMA staining indicated that microtubule minus ends were enriched at the frayed spindle ends, hooking analysis revealed that microtubule bundles were of uniform polarity. Therefore, other motors besides dynein must contribute to microtubule sorting during spindle assembly. One possibility is that plus end–directed motors associated with chromatin sort microtubules by moving towards plus ends, thereby pushing microtubule minus ends away from chromatin (Vernos et al., 1995). Alternatively, or concurrently, spindle tetrameric plus end–directed motors of the BimC family (Walczak and Mitchison, 1996) or motors shown to promote antiparallel microtubule sliding such as MKLP1 (Nislow et al., 1992) could also fulfill this role. Since polarity-marked seed motility appears to be driven predominantly by dynein, other assays need to be developed to visualize the activities of other motors in spindles.
The Nature of a Spindle Pole
The precise nature of spindle poles and centrosomes and the relationship between them has long been a source of confusion and controversy in cell biology (see Mazia, 1984). One reason for the confusion is that centrosomes and mitotic poles are structurally similar, each containing a focus of microtubule minus ends. However, several observations suggest that centrosomes and mitotic poles are functionally different. First, several proteins, including NuMA (Merdes et al., 1996) and the dynein/dynactin complex (Gaglio et al., 1996; this work), are required for pole formation but not for microtubule nucleation by centrosomes. Second, we have shown that after microtubule depolymerization, centrosomes persist as microtubule organizing centers, while self-assembled poles do not (Fig. 7). Thus, focal nucleation and motor-dependent organization are two functionally different mechanisms for generating a focus of microtubule minus ends.
A concept emerging from these studies is that the organization of microtubules into a spindle pole is a process that is superimposed on the centrosomal aster (Karsenti, 1991; Gaglio et al., 1996). Several observations support a dynamic association of spindle microtubules with centrosomal microtubules at poles. First, poleward microtubule flux requires that microtubule minus ends are free to depolymerize (Mitchison and Sawin, 1990). Second, the centrosome and spindle pole appear to organize separate populations of microtubules. These two populations are easily recognized in spindles assembled around sperm nuclei in Xenopus egg extracts because there are two sources of microtubules, chromatin and centrosomes, that can be separated by dynein inhibition (Fig. 5 b). In other meiotic or embryonic systems, such as Lepidoptera and Drosophila, the centrosomes are physically located a significant distance away from the spindle poles (Wolf and Bastmeyer, 1991). These two different microtubule populations seem to exist in somatic systems as well. Serial sectioning of somatic spindles revealed that most spindle microtubules ended a significant distance away from the centrosomes (Mastronarde et al., 1993). In some centrosome-dependent systems, loss or removal of centrosomes does not damage spindle integrity once the spindle is in metaphase or in anaphase (Mitchison and Salmon, 1992; Murray et al., 1996; Nicklas et al., 1989). How are two populations of microtubules created in such systems, in which chromatin-induced nucleation of microtubules does not occur? Spindle microtubules could be generated by release of microtubules from centrosomes. This has been shown to occur in Xenopus egg extracts (Belmont et al., 1990) and somatic cells in interphase (McBeath and Fujiwara, 1990). Thus, centrosomes can provide a source of microtubules and not themselves be poles.
The Role of Centrosomes
Since centrosomes are not always required to form spindle poles, what is their role? We and others have shown that a single centrosome can create a dominant site for pole assembly, enforcing monopolarity (Mazia et al., 1981; Bajer, 1982; Sawin and Mitchison, 1991; Fig. 8). In Xenopus egg extracts, an explanation for this observation is that by functioning as an efficient microtubule nucleator, a centrosome generates microtubules before they appear to grow around chromatin (Fig. 7). Once microtubules begin to grow around chromatin, they may be preferentially shuttled by dynein towards the centrosome, which constitutes a preformed focal point of microtubule minus ends, before they can be sorted into an antiparallel array. Therefore, centrosomes could be dominant for kinetic reasons, suppressing aspects of microtubule self-organization and thereby directing the sites of spindle pole formation. We propose that the critical function of centrosomes is to determine pole assembly sites. Centrosome positioning then provides a mechanism for determining the orientation of the cleavage plane, which is important in many cell types and during development (White and Strome, 1996).
Why are centrosomes required in some systems and not in others? One distinguishing feature of early embryonic systems such as Xenopus results from the presence of stored components in eggs that may be limiting in somatic cells. Therefore, centrosomes may be required in most systems because of the lack of stored microtubule nucleating material in the cytoplasm, which is present only on centrosomes, preventing microtubule growth around chromosomes (discussed in Karsenti et al., 1996). Therefore, the difference between meiotic and mitotic spindle assembly may reflect differences in microtubule nucleation rather than different principles of spindle organization.
Acknowledgments
We thank T. Ashford, A. Merdes, and S. Reinsch for providing antibodies, and S. Andersen, M. Glotzer, C. Gonzalez, and S. Reinsch for comments on the manuscript.
Footnotes
1. Abbreviation used in this paper: NuMA, nuclear protein that associates with mitotic apparatus.
Please address all correspondence to Dr. Rebecca Heald, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69117 Heidelberg, Germany. Tel.: 49-6221-387324. Fax: 49-6221-387306. E-mail:Heald@EMBL-Heidelberg.de
As of October 1997, Dr. Heald's address will be Department of Molecular and Cell Biology, University of California, Berkeley, 311 Life Sciences Addition, Berkeley, CA 94720.
References
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Bajer AS. Functional autonomy of monopolar spindle and evidence for oscillatory movement in mitosis. J Cell Biol. 1982;93:33–48. doi: 10.1083/jcb.93.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer AS, Mole BJ. Asters, poles, and transport properties within spindlelike microtubule arrays. Cold Spring Harbor Symp Quant Biol. 1982;1:263–283. doi: 10.1101/sqb.1982.046.01.029. [DOI] [PubMed] [Google Scholar]
- Bastmeyer M, Steffen W, Fuge H. Immunostaining of spindle components in tipulid spermatocytes using a serum against pericentriolar material. Eur J Cell Biol. 1986;42:305–310. [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Boleti H, Karsenti E, Vernos I. Xklp2, a novel Xenopuscentrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- Compton DA, Cleveland DW. NuMA is required for the proper completion of mitosis. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci. 1995;108:621–633. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- Debec A, Detraves C, Montmory C, Geraud G, Wright M. Polar organization of γ-tubulin in acentriolar mitotic spindles of Drosophila melanogastercells. J Cell Sci. 1995;108:2645–2653. doi: 10.1242/jcs.108.7.2645. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Jackson WT, McIntosh JR. Polarity of spindle microtubules in Haemanthus endosperm. J Cell Biol. 1982;94:644–653. doi: 10.1083/jcb.94.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL. Microtubule organization during maturation of Xenopusoocytes: assembly and rotation of the meiotic spindles. Dev Biol. 1992;151:516–530. doi: 10.1016/0012-1606(92)90190-r. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopusegg extracts. Nature (Lond) 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heidemann SR, McIntosh JR. Visualization of the structural polarity of microtubules. Nature (Lond) 1980;286:517–519. doi: 10.1038/286517a0. [DOI] [PubMed] [Google Scholar]
- Hyman, A.A. 1991. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J. Cell Sci. 14(Suppl.): 125–127. [DOI] [PubMed]
- Kallajoki M, Weber K, Osborn M. A 210 kDa nuclear matrix protein is a functional part of the mitotic spindle; a microinjection study using SPN monoclonal antibodies. EMBO (Eur Mol Biol Organ) J. 1991;10:3351–3362. doi: 10.1002/j.1460-2075.1991.tb04899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallajoki M, Weber K, Osborn M. Ability to organize microtubules in taxol-treated mitotic PtK2 cells goes with the SPN antigen and not with the centrosome. J Cell Sci. 1992;102:91–102. doi: 10.1242/jcs.102.1.91. [DOI] [PubMed] [Google Scholar]
- Kallajoki M, Harborth J, Weber K, Osborn M. Microinjection of a monoclonal antibody against SPN antigen, now identified by peptide sequences as the NuMA protein, induces micronuclei in PtK2 cells. J Cell Sci. 1993;104:139–150. doi: 10.1242/jcs.104.1.139. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Karsenti E. Mitotic spindle morphogenesis in animal cells. Semin Cell Biol. 1991;2:251–260. [PubMed] [Google Scholar]
- Karsenti E, Boleti H, Vernos I. The role of microtubule-based motors in centrosome movements and spindle pole organization during mitosis. Semin Cell Biol. 1996;7:367–378. [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Lajoie MI, Tollon Y, Detraves C, Julian M, Moisand A, Gueth HC, Debec A, Salles PI, Puget A, Mazarguil H. Recruitment of antigenic γ-tubulin during mitosis in animal cells: presence of γ-tubulin in the mitotic spindle. J Cell Sci. 1994;107:2825–2837. doi: 10.1242/jcs.107.10.2825. [DOI] [PubMed] [Google Scholar]
- Lambert, A.M., and C.W. Lloyd. 1994. The higher plant microtubule cycle. In Microtubules. J.S. Hyams and C.W. Lloyd, editors. Wiley-Liss, New York. 325–342.
- Li Q, Joshi HC. γ-Tubulin is a minus end-specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Leslie R, Kuriyama R. Identification of a minus end-specific microtubule-associated protein located at the mitotic poles in cultured mammalian cells. Eur J Cell Biol. 1991;54:255–267. [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazia D. Centrosomes and mitotic poles. Exp Cell Res. 1984;153:1–15. doi: 10.1016/0014-4827(84)90442-7. [DOI] [PubMed] [Google Scholar]
- Mazia D, Paweletz N, Sluder G, Finze EM. Cooperation of kinetochores and pole in the establishment of monopolar mitotic apparatus. Proc Natl Acad Sci USA. 1981;78:377–381. doi: 10.1073/pnas.78.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath E, Fujiwara K. Microtubule detachment from the microtubule-organizing center as a key event in the complete turnover of microtubules in cells. Eur J Cell Biol. 1990;52:1–16. [PubMed] [Google Scholar]
- McIntosh JR, Euteneuer U. Tubulin hooks as probes for microtubule polarity: an analysis of the method and an evaluation of data on microtubule polarity in the mitotic spindle. J Cell Biol. 1984;98:525–533. doi: 10.1083/jcb.98.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Hawley RS. Chromosomal control of meiotic cell division. Science (Wash DC) 1995;270:1595–1601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Microtubule assembly nucleated by isolated centrosomes. Nature (Lond) 1984;312:232–236. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Sawin KE. Tubulin flux in the mitotic spindle: where does it come from, where is it going? . Cell Motil Cytoskel. 1990;16:93–98. doi: 10.1002/cm.970160202. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Desai AB, Salmon ED. Real time observation of anaphase in vitro. Proc Natl Acad Sci USA. 1996;93:12327–12332. doi: 10.1073/pnas.93.22.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Lee GM, Rieder CL, Rupp G. Mechanically cut mitotic spindles: clean cuts and stable microtubules. J Cell Sci. 1989;94:415–423. doi: 10.1242/jcs.94.3.415. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature (Lond) 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature (Lond) 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Mitotic spindle assembly by two different pathways in vitro. J Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ. Microtubule flux in mitosis is independent of chromosomes, centrosomes, and antiparallel microtubules. Mol Biol Cell. 1994;5:217–226. doi: 10.1091/mbc.5.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. New insights into the interaction of cytoplasmic dynein with the actin-related protein, Arp1. J Cell Biol. 1994;127:1–4. doi: 10.1083/jcb.127.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA, Bingham JB, Gill SR. Actin related protein 1 and cytoplasmic dynein-based motility—what's the connection? . Trends Cell Biol. 1996;6:212–215. doi: 10.1016/0962-8924(96)20014-5. [DOI] [PubMed] [Google Scholar]
- Sluder G, Rieder CL. Experimental separation of pronuclei in fertilized sea urchin eggs: chromosomes do not organize a spindle in the absence of centrosomes. J Cell Biol. 1985;100:897–903. doi: 10.1083/jcb.100.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Steffen W, Fuge H, Dietz R, Bastmeyer M, Muller G. Aster-free spindle poles in insect spermatocytes: evidence for chromosome-induced spindle formation? . J Cell Biol. 1986;102:1679–1687. doi: 10.1083/jcb.102.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature (Lond) 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Telzer BR, Haimo LT. Decoration of spindle microtubules with dynein: evidence for uniform polarity. J Cell Biol. 1981;89:373–378. doi: 10.1083/jcb.89.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophilafemales: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued . J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopuseggs: requirement for phosphorylated factors and cytoplasmic dynein. J Cell Biol. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopuskinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ. Kinesin-related proteins at mitotic spindle poles: function and regulation. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]
- Waters JC, Salmon ED. Pathways of spindle assembly. Curr Opin Cell Biol. 1997;9:37–43. doi: 10.1016/s0955-0674(97)80149-4. [DOI] [PubMed] [Google Scholar]
- White J, Strome S. Cleavage plane specification in C. elegans: how to divide the spoils. Cell. 1996;84:195–198. doi: 10.1016/s0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Wolf KW, Bastmeyer M. Cytology of Lepidoptera. V. The microtubule cytoskeleton in eupyrene spermatocytes of Ephestia kuehniella (Pyralidae), Inachis io (Nymphalidae), and Orgyia antiqua (Lymantriidae) . Eur J Cell Biol. 1991;55:225–237. [PubMed] [Google Scholar]
- Yang CH, Snyder M. The nuclear-mitotic apparatus protein is important in the establishment and maintenance of the bipolar mitotic spindle apparatus. Mol Biol Cell. 1992;3:1259–1267. doi: 10.1091/mbc.3.11.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. Chromosomes initiate spindle assembly upon experimental dissolution of the nuclear envelope in grasshopper spermatocytes. J Cell Biol. 1995a;131:1125–1131. doi: 10.1083/jcb.131.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. The impact of chromosomes and centrosomes on spindle assembly as observed in living cells. J Cell Biol. 1995b;129:1287–1300. doi: 10.1083/jcb.129.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]