Abstract
RasG is the most abundant Ras protein in growing Dictyostelium cells and the closest relative of mammalian Ras proteins. We have generated null mutants in which expression of RasG is completely abolished. Unexpectedly, RasG − cells are able to grow at nearly wild-type rates. However, they exhibit defective cell movement and a wide range of defects in the control of the actin cytoskeleton, including a loss of cell polarity, absence of normal lamellipodia, formation of unusual small, punctate polymerized actin structures, and a large number of abnormally long filopodia. Despite their lack of polarity and abnormal cytoskeleton, mutant cells perform normal chemotaxis. However, rasG − cells are unable to perform normal cytokinesis, becoming multinucleate when grown in suspension culture. Taken together, these data suggest a principal role for RasG in coordination of cell movement and control of the cytoskeleton.
Ras proteins are a family of small GTPases that control a range of fundamental processes in all known eukaryotic cells. Ras was first discovered in an activated form as the product of a viral oncogene (Shih et al., 1979). It has since been found to be a central regulator of mammalian cell growth and division, differentiation, shape, and motility (for reviews see Sorcher et al., 1993; Valencia and Sander, 1995). Distinct roles for Ras have been found in several organisms. In particular, Ras proteins mediate signaling through receptors such as the Sevenless protein in Drosophila (Fortini et al., 1992), and a Ras homologue (the let-60 gene product) is required for the specification of the vulval cells of Caenorhabditis elegans (Han and Sternberg, 1990). The RAS1 and RAS2 genes of Saccharomyces control cell growth by regulating the activity of adenylyl cyclase (Toda et al., 1985) and may also influence the cytoskeleton through the CAP protein (Wang et al., 1993). Relatives of Ras proteins in the Rac and Rho subfamilies act downstream of Ras; again, they appear to control both growth and motility (Ridley et al., 1992). Rac and Rho seem to control separate aspects of cell architecture, with Rac and its relative Cdc42 controlling actin-rich protrusions (ruffles and filopodia, respectively) and Rho regulating the formation of actomyosin bundles known as stress fibers (Ridley et al., 1992; Nobes and Hall, 1995).
Ras proteins are active when GTP is bound and become inactive by hydrolyzing this GTP to GDP. Two families of proteins regulate Ras activity by controlling the bound nucleotide. Guanine nucleotide exchange factors (GEFs)1 such as CDC25 and Sos activate Ras by allowing GDP to dissociate and be replaced by GTP. GTPase-activating proteins (GAPs), on the other hand, inactivate Ras by binding to the active form and stimulating the hydrolysis of GTP to GDP (Boguski and McCormick, 1993). Both may be controlled by different stimuli. In mammalian cells, the binding of receptor tyrosine kinases to their ligands can cause recruitment of both GEFs and GAPs to the membrane through a family of adaptor proteins such as Grb2 and Shc (Lowenstein et al., 1992; Pelicci et al., 1992); the interaction between Drosophila Sevenless and Boss proteins is transmitted to Ras through the Grb2 homologue Drk (Olivier et al., 1993). In yeast, the signals that regulate Ras activity through CDC25 and IRA1&2 are not yet understood.
The work described in this paper suggests a connection between Ras proteins and cytokinesis. Correct cell division involves two distinct processes: nuclear division (karyokinesis) and then partitioning of the cytoplasm and organelles (cytokinesis; Rappaport, 1986). Karyokinesis is mainly accomplished by the microtubules that form the spindle, whereas cytokinesis is apparently based around actin and myosin II (classical, double-headed myosin). Soon after karyokinesis, a concentration of polymerized actin (F-actin) is visible at the equator of the cell, and a cleavage furrow containing myosin II forms which pinches the daughter cell in two (Fishkind and Wang, 1995). Mutants in several species affected in myosin II function lose the ability to perform cytokinesis properly, despite apparently normal karyokinesis (De Lozanne and Spudich, 1987; Karess et al., 1991). The simple eukaryote Dictyostelium discoideum has recently proved to be an excellent subject for the study of cytokinesis, in particular because the cells possess an alternative method of partitioning cell contents when normal partition cannot take place (De Lozanne and Spudich, 1987). This process, which has been named “traction-mediated cytofission” (Fukui et al., 1990), allows the survival of mutants with strong cytokinesis phenotypes, which would be inviable in other systems. Dictyostelium also offers relatively simple gene disruption by homologous recombination, and is a much-studied target for analysis of cytoskeletal proteins.
The first cytokinesis mutants to be isolated in Dictyostelium, and consequently the most extensively studied mutants, are defective in cytoskeletal proteins, for example, myosin II heavy chain (DeLozanne and Spudich, 1987) and coronin (de Hostos et al., 1993). However, several recent reports have identified two GAP proteins of unknown substrate specificity (Faix and Dittrich, 1996; Lee et al., 1997) and the small GTP-binding protein RacE (Larochelle, 1996) as putative regulators of cytokinesis. Neither GAP proteins nor RacE are likely to be directly involved in the physical separation of the daughter cells; instead they must lie on the signaling pathways controlling the process.
Dictyostelium possesses an unusual, extended family of ras genes (Daniel et al., 1995). Two of the products (RasG and RasD; Reymond et al., 1984; Robbins et al., 1989) are closely related to mammalian Ras proteins (68% and 65% overall identity to human H-ras, respectively), whereas the RasB, RasC, and RasS gene products (Daniel et al., 1993, 1994) are more divergent (though still clearly members of the Ras subfamily). Dictyostelium cells grow unicellularly but aggregate and form a multicellular fruiting body upon starvation. The rasD and rasS genes are only expressed during multicellular development (Reymond et al., 1984; Daniel et al., 1994). RasD, furthermore, is expressed at higher levels in the stalk cell precursors than the prespore cells. The gene products are therefore presumed to have a function specifically connected with aggregation or cell-type differentiation. RasG, RasB, and RasC are expressed during growth (Robbins et al., 1989; Daniel et al., 1993). RasG mRNA expression ceases as soon as multicellular development begins, and RasG protein is lost from the cells during development, suggesting a specific requirement during growth (Khosla et al., 1990, 1996).
Aside from their different patterns of expression, little is known about the functions of Dictyostelium Ras proteins. Overexpression of an activated form of RasD causes aberrant signaling and developmental arrest in aggregates (Reymond et al., 1986). Similar activating mutants of RasG cause a block in aggregation (Khosla et al., 1996) and cytoskeletal changes (Rebstein et al., 1997). Neither of these lines has allowed elucidation of the function of the Ras proteins in normal Dictyostelium cells. In this work we report the disruption of the gene that encodes RasG; disruptants show a range of phenotypes based around the control of the actin cytoskeleton, in particular during cytokinesis. This suggests that one major role of RasG is to control cell architecture, rather than growth or differentiation.
Materials and Methods
Unless otherwise indicated, all chemicals were obtained from Sigma Chemical Company (St. Louis, MO) and all enzymes from New England Biolabs (Beverly, MA).
Cell Strains, Growth, and Transformation
Dictyostelium discoideum AX2 cells were maintained at 22°C in HL-5 medium (Sussman, 1987). For bacterially grown cells, SM plates were inoculated with 105–106 Dictyostelium cells plus 200 μl of a suspension of Klebsiella aerogenes in L-broth. Transformation was performed by a modification of Howard et al. (1988); briefly, cells in log phase growth were mixed with 25 μg of linearized DNA and electroporated at 1.0 or 1.1 mV, 3 μF with a 5 Ω resistance in series. After 10 min incubation on ice, cells were warmed for 15 min in the presence of 2 μl healing solution (100 mM MgCl2, 100 mM CaCl2), and then HL-5 was added. Blasticidin (ICN, Irvine, CA) or G418 (GIBCO BRL, Gaithersburg, MD) at 10 μg/ml final concentration were added 24 h after electroporation. Transformants appearing after 4–5 d were cloned on lawns of Klebsiella growing on SM agar plates.
Molecular Biology
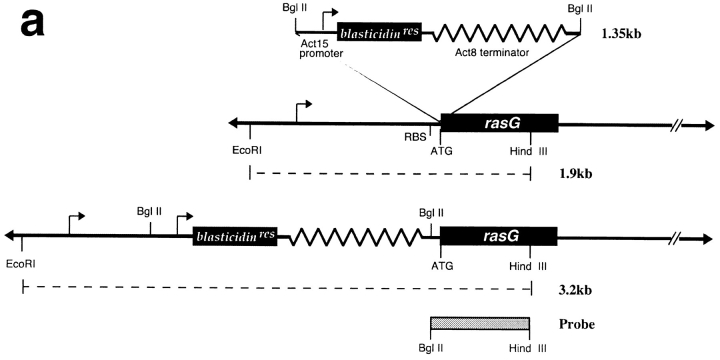
The rasG knockout vector was constructed by inserting a version of the pBsrΔBam marker (Adachi et al., 1994) with BamHI linkers into the single BglII site of pRASG (Robbins et al., 1992). Transformants in which the marker was oriented in the same direction as the rasG gene were selected to ensure the presence of a terminator between the actin promoter and the rasG coding sequence. From the resulting vector a 3.2-kb fragment was digested with EcoRI and HindIII restriction enzymes and used to transform Dictyostelium strain AX2 as described above. To analyze clones, AX2 or rasG − genomic DNA prepared by the method of Sun et al. (1990) was digested with HindIII and EcoRI, blotted onto nylon membrane (Qiagen Inc., Chatsworth, CA), and probed with the [α-32P]dATP labeled BglII–HindIII fragment from the rasG-coding sequence (Robbins et al., 1989) as described in Sambrook et al. (1989).
Western Blotting
rasG − + AX2 cell lysates were separated by SDS-PAGE and blotted onto 0.45 μm PVDF membrane (Amersham Life Science, Pittsburgh, PA) by standard procedures (Sambrook et al., 1989). The membrane was probed with rat α-Ras mAb Y13-259 (Oncogene Science, Mineola, NY) at 10 μg/ml final concentration, or with a polyclonal antibody to RasG as described in Khosla et al. (1994). Secondary HRP-conjugated anti–rat antibody or anti– rabbit antibody (Pierce, Rockford, IL) was used at 1:2,500 dilution. The signal was detected using an enhanced chemiluminescence kit (Amersham Corp., Arlington Heights, IL).
Microscopy
Cells from axenic culture were seeded onto acid-washed coverslips and fixed with 1% glutaraldehyde, 0.1% Triton X-100 in KK2 buffer (20 mM potassium phosphate, pH 6.2, 2 mM MgCl2) for 10 min. Autofluorescence was quenched using 5 mg/ml NaBH4 for 10 min. Actin filaments were stained with 100 nM Rhodamine- or Texas red–conjugated phalloidin (Sigma Chemical Co. and Molecular Probes, Inc., Eugene, OR, respectively) and nuclei with 500 nM Hoechst 33258 or 33342. Cells were observed using an epifluorescence microscope (Axiophot; Zeiss, Inc., Thornwood, NY) or a scanning confocal microscope (MRC1024; Bio Rad, Hercules, CA). Images were recorded with a Hamamatsu CCD camera or on Kodak Ektachrome 400 film, and digitized with an Agfa Arcus II scanner in conjunction with Adobe Photoshop 3.0 software.
Cells were synchronized to observe cytokinesis using aphidicolin (Pedrali-Noy et al., 1987). The medium of axenically growing cells was supplemented with 10 μg/ml aphidicolin (final concentration) for 16 h. The aphidicolin was removed with three washes of fresh axenic medium and the cells allowed to continue through the cell cycle. At the first signs of mitotic activity (usually about 5 h after release) cells were fixed as described above.
Time-lapse video microscopy of cells was used to observe cytokinesis. Cells growing in petri dishes were synchronized as above, and a time-lapse video recorder (Betacam; Sony, Inc., Tokyo, Japan) was used to record phase contrast images collected by a CCD camera (Sony, Inc.) from an inverted microscope (Zeiss, Inc.).
To observe traction-mediated cytofission, rasG − cells were grown for 5 d in shaking flasks and transferred to tissue culture plates. Phase-contrast photomicrographs were taken using an inverted microscope (Zeiss, Inc.) and Kodak T-Max 100 film. Alternatively, electronic images were obtained using a Panasonic video camera and a frame grabber connected to a Macintosh computer running NIH Image 1.60 software.
For the effects of azide, axenically growing cells were seeded onto coverslips and perfused with KK2 buffer for 5 min using a perfusion chamber as described in Devreotes et al. (1987). The buffer was replaced with 0.01% sodium azide in KK2 and an inverted microscope (Zeiss, Inc.) with phase contrast optics used to observe adhering cells.
To measure speed and persistence, cells were followed in a perfusion chamber, and images of several cells were recorded every minute. Images were grabbed as above, and the position of the centroids of several cells calculated using NIH Image 1.60 software and expressed as an x,y coordinate. The coordinates were used to calculate the velocity and persistence of each cell as described below.
Firstly, the length of the path between successive centroids was taken as the instantaneous velocity of each cell. Secondly, the cosine of the angle of the path between centroids was used as a measure of the persistence of movement of a cell. A cell moving in a constant direction will produce a path with angle 0° and therefore a cosine of one, while a cell changing direction at 90° will produce a cosine value of zero.
At least eight cells were followed for at least 20 min, and the velocities and persistences were pooled and used to give mean values.
Chemotaxis Assay
Bacterially growing cells were washed with KK2 buffer to remove bacteria. Cells were seeded onto coverslips and submerged in KK2 buffer. A micropipette filled with 10 μM folic acid in KK2 was placed just above the coverslip and pressurized to expel the folic acid. The assay was performed with the same pipette for both rasG − and wild-type cells.
Results
Disruption of the Dictyostelium rasG Gene
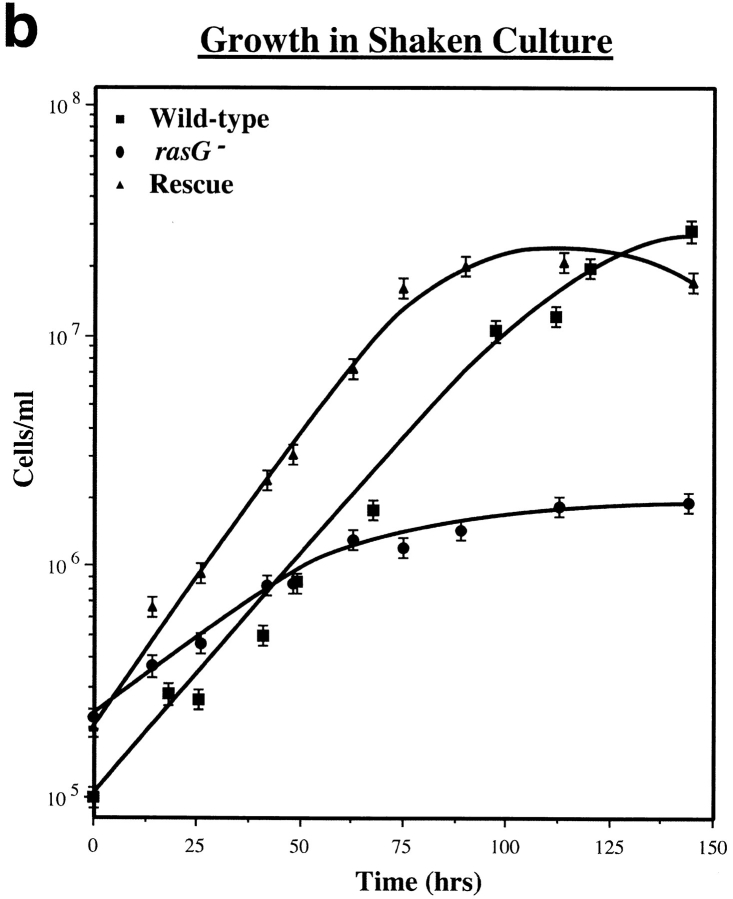
Cells containing a disrupted rasG gene were generated by homologous recombination by the strategy shown in Fig. 1 A. A construct was made containing 1.9 kb of rasG genomic DNA (Robbins et al., 1992), with a bsr marker (Sutoh, 1993) inserted between the promoter and coding sequence of the gene, in the same orientation so the strong act8 terminator blocked any read-through from the rasG or bsr promoters. The construct was transfected into AX2 cells, and transformants were cloned after 7 d of blasticidin selection. 2 independent clones, out of 74 examined, were found to contain a disruption in rasG (Fig. 1 B), with the rest apparently containing nonhomologous integration of the vector. Both independent lines were found to behave similarly, so one (IR15) was used for all the work described here. When Western blots are probed with the broad-spectrum Ras antibody Y13-259 (Furth et al., 1982), disrupted cells show approximately half the wild-type level of Ras proteins (Fig. 1 C, left). Y13-259 detects the diverged RasB and RasC proteins, albeit less well than RasG or RasD. The Ras protein seen in IR15 therefore presumably derives from the rasB and rasC genes (which are expressed at lower levels than rasG in wild-type cells) or rasD (which is barely expressed in growing wild-type cells). No RasG protein is found in disruptants when a specific RasG antibody is used (Fig. 1 C, right).
Figure 1.
Disruption of the rasG gene. (a) Schematic representation of the cloning strategy employed to disrupt the rasG gene. A 1.7-kb fragment encoding the cDNA for the blasticidin resistance gene (bsr) driven by the constitutive actin15 promoter was inserted by homologous recombination into the rasG promoter between the promoter and the ATG start codon. A probe from the rasG coding sequence (shaded bar) was used to detect correct disruptants by Southern blotting of genomic DNA. The expected bands in the parental strain and disruptants are indicated by dotted lines. (b) Southern blot of rasG − and wild-type parental genomic DNA. Nuclear DNA from strains IR15 (rasG −) and AX2 (wt) was digested with EcoRI and HindIII, separated on an 0.8% agarose gel, blotted onto nylon, and probed with the rasG coding sequence (see above). The 1.9-kb parental band and 3.2-kb rasG − disrupted band are marked. (C) Western blot of rasG − and AX2 wild-type cells. Whole cell lysates were separated by PAGE using a 15% acrylamide gel, blotted onto PVDF, and probed with the general Ras antibody Y13-259 (left) and a RasG specific antibody (right). Y13-259 recognizes several different Dictyostelium Ras proteins with varying efficiency.
Growth of rasG− Cells
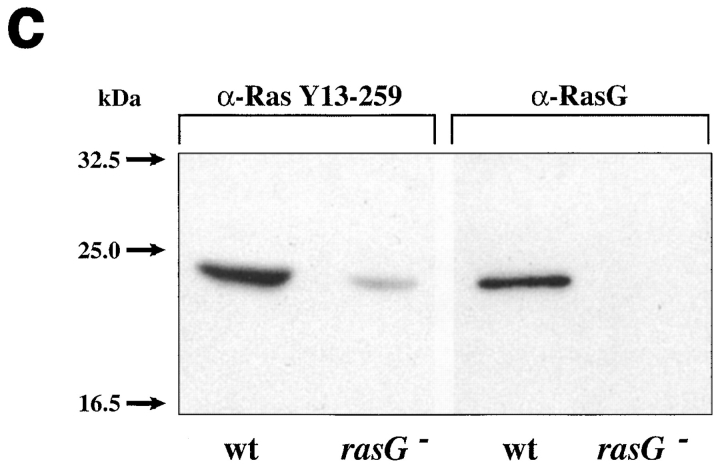
Unexpectedly, rasG appears to be dispensable for growth under most conditions. As shown in Fig. 2 A, rasG − cells in liquid medium grow only slightly more slowly than wild-type (14.5 vs 13.4 h doubling time). rasG − cells also grow somewhat more slowly than wild-type on bacteria, forming colonies that are a little smaller than AX2 (data not shown).
Figure 2.
Growth of wild-type and rasG − cells. (a) Growth on surfaces. Wild-type (▪), rasG − (•), and rasG − cells rescued by expression of rasG (▴) were seeded in Petri plates (105 cells/ plate) with 10 ml axenic medium. At intervals the cells were detached from the surface, and the number of cells in a small aliquot was counted. (b) Growth in suspension culture. Wild-type (▪), rasG − (•), and rasG − cells rescued by expression of rasG (▴) were transferred from Petri plates into axenic medium at time zero and counted at intervals thereafter.
When cells are grown in shaken suspension, however, a different result is seen (Fig. 2 B). rasG − cells double far more slowly than wild-type (27 vs 15 h doubling time), and cease growing at a considerably lower cell density (Fig. 2 B). This weak growth in suspension culture, despite nearly normal growth on surfaces, is reminiscent of the phenotype of mhcA − cells (De Lozanne and Spudich, 1987), which do not produce myosin II, and therefore cannot perform cytokinesis. The role of rasG in cytokinesis was examined further, as described below.
Retransfection of rasG − cells with a genomic copy of the rasG gene and promoter reverses both growth phenotypes, allowing a normal doubling time whether cells are grown on plates, in shaken suspension (Fig. 2, A and B), or on bacteria (data not shown). The mutant phenotypes are therefore solely due to a lack of RasG.
Aberrant Morphology and Adhesion in rasG Mutants
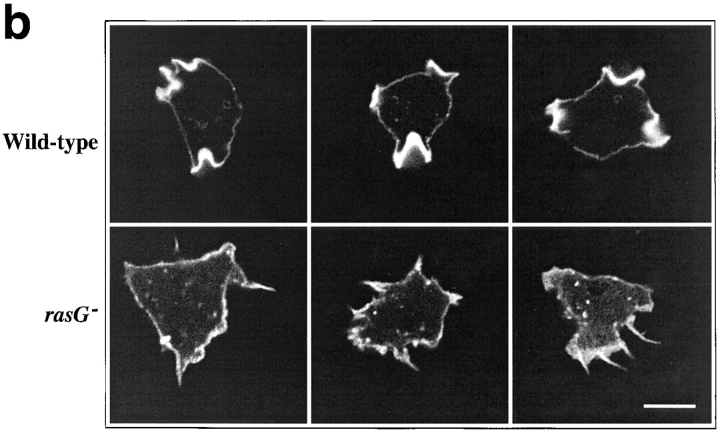
Observed by phase contrast microscopy, rasG − cells differ from wild-type in several respects. Cells grown on bacteria (which behave more consistently than axenically grown cells) and then allowed to migrate on a glass coverslip (Fig. 3 A) are far more flattened than wild-type and adhere more tightly. They are also less polar; most wild-type cells possess a clear orientation, whereas rasG − cells are nearly all circular (Fig. 3 A). When the motion of cells was recorded by video microscopy, rasG − cells travelled considerably slower than wild-type (64% of the mean speed; Table I). Two aspects of the motion of rasG − cells are surprising. The defect in polarization of rasG − cells would suggest that they are unable to maintain a specific direction, yet they move with nearly the same persistence (a measure of the cell's ability to move in a constant direction) as wild-type (Table I). Secondly, wild-type cells alternate between polarized and rounded morphologies, with nearly all movement taking place in the polar phase. rasG − cells seem to move nearly as fast when rounded as when polar, which explains how they are able to move at as much as 64% of wild-type speeds despite the majority being apparently unpolarized.
Figure 3.
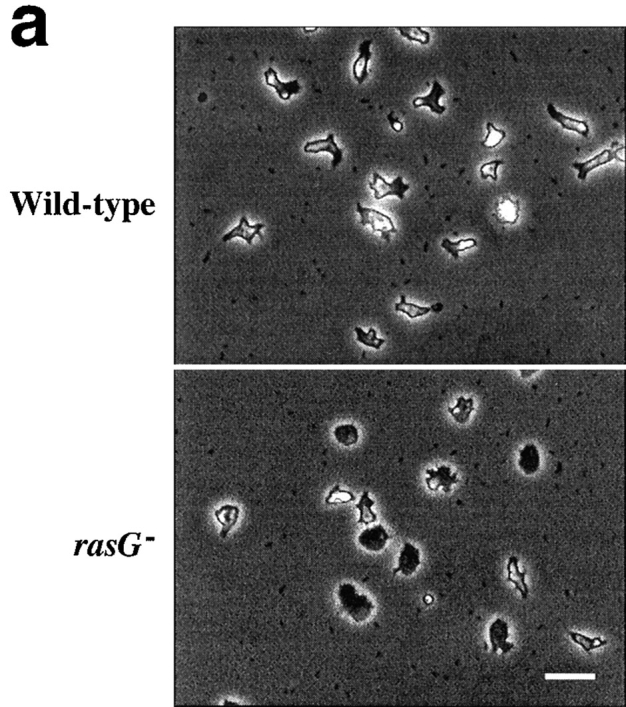
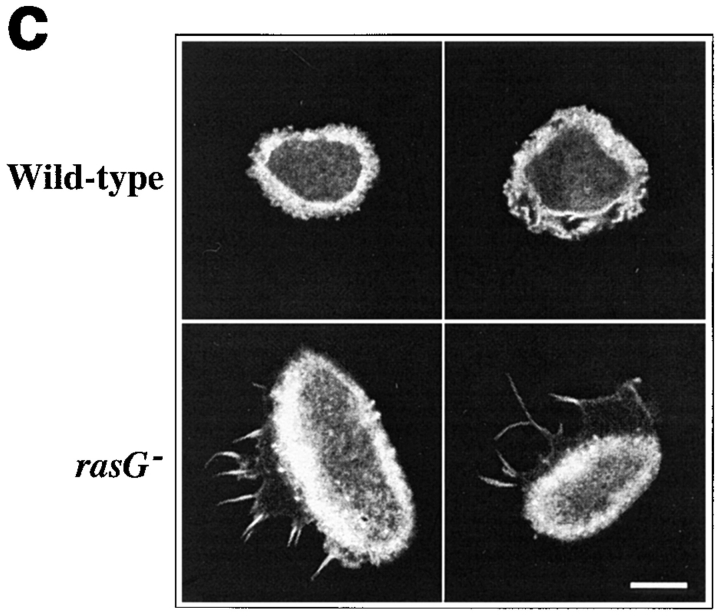
Morphology of wild-type and rasG − cells. (a) Phase-contrast micrographs of cells adhering to glass. Wild-type cells (top) show a rounded and polarized morphology, while rasG − cells (bottom) are more flattened and nonpolar. (b and c) Scanning confocal micrographs of cells stained with rhodamine–phalloidin to visualize F-actin. (b) Moving wild-type cells (top) and rasG − cells (bottom). (c) Stationary wild-type (top) and rasG − cells (bottom). Bar: (a) 20 μm; (b and c) 10 μm.
Table I.
Speed and Persistence Measurements for Wild-type and rasG− Cells
| Wild-type (AX2) | rasG − (IR15) | |||
|---|---|---|---|---|
| Mean speed (μm/min) | 4.5 ± 2.6 | 2.9 ± 2.4 | ||
| Persistence | 0.46 | 0.44 |
Cells were examined in a perfusion chamber as described in Materials and Methods. Images were recorded every minute, and the distance moved by the centroids of several cells was used to calculate the mean cell speed. The persistence, which represents the constancy of direction of each cell, was calculated as the cosine of the angle between consecutive paths.
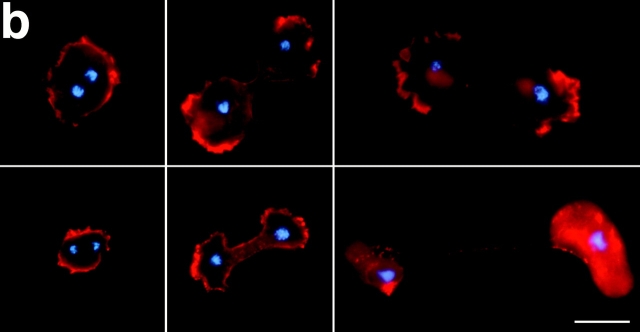
The distribution of F-actin is also aberrant in axenically grown rasG − cells. Staining with fluorescent phalloidin (Fig. 3 B) reveals two major differences between rasG − and wild-type. Firstly, the lamellipodia seen at the leading edges of polarized wild-type cells are replaced by large numbers of elongated filopodia. While similar filopodia are seen in a proportion of wild-type cells (∼10–15%; data not shown), they are present in nearly all rasG − cells, frequently in large numbers, often reaching considerably greater lengths than in wild-type cells (Fig. 3 B). Again, nearly all rasG − cells show no obvious polarity; the filopodia appear to be spread randomly around the perimeter of most cells. Wild-type cells in the rounded phase frequently show a continuous, broad cortex of F-actin, with no actin-rich protrusions; equivalent rasG − cells exhibit a similar cortex, but usually also possess long filopodia (Fig. 3 C). Various highly unusual morphologies are also common in rasG − cells, in particular crescent and hour glass shapes (Fig. 4). In these cells the cortex, rather than being continuous, is divided into discrete F-actin–rich lobes, as if the different ends of the cell were trying to move in opposite directions. The nuclei are usually found between the lobes of F-actin, and the two halves of the cells pulling in opposite directions can squeeze the nuclei into cylindrical shapes.
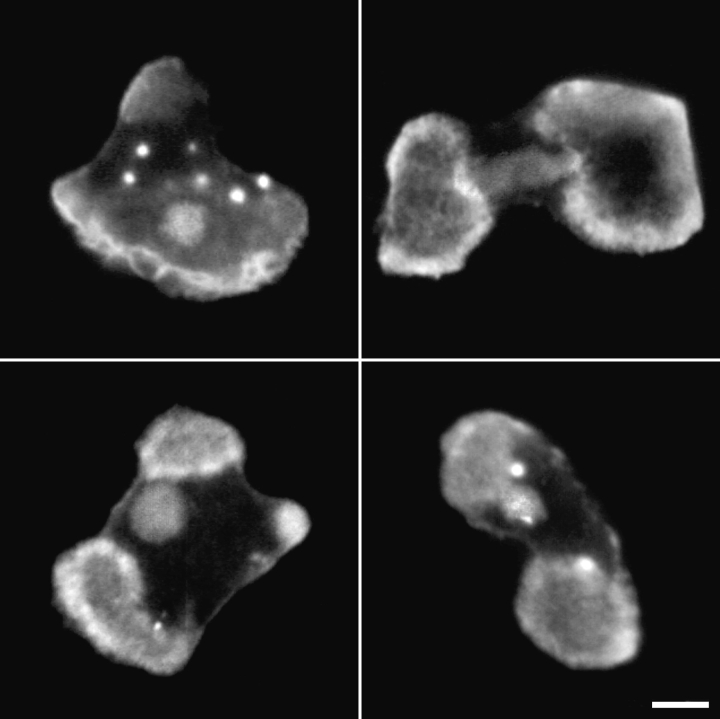
Figure 4.
Epifluorescence micrographs of stationary rasG − cells stained with Texas red– phalloidin to visualize F-actin and Hoechst 33342 to visualize the nuclei. The cells adopt abnormal morphologies and adhere to the glass through large pads of F-actin. Small, intensely stained masses of F-actin are also present in some cells. Bar, 5 μm.
A second unusual feature of rasG − cells is the abundance of small, punctate F-actin structures within the cell body. These are conspicuously present in the majority of cells (Figs. 3, B and C, and 4; particularly clear actin structures are also visible in Fig. 6 A). Similar structures are seen in a small proportion of wild-type cells, but in lower numbers (1–2 per cell; data not shown). Three-dimensional reconstruction of confocal Z-series indicates that the structures are roughly spherical and are localized to the cortex of the base and the upper surface of the cells (data not shown; a QuickTime video showing an example of a three-dimensional reconstruction is available on the World Wide Web at http://www.ucl.ac.uk/∼dmcbrob/movies.html). The precise nature of these structures is as yet unknown. Mutant and wild-type cells contain similar quantities of F-actin per cell (data not shown); the differences appear to reflect a failure of organization, rather than an inability to polymerize actin.
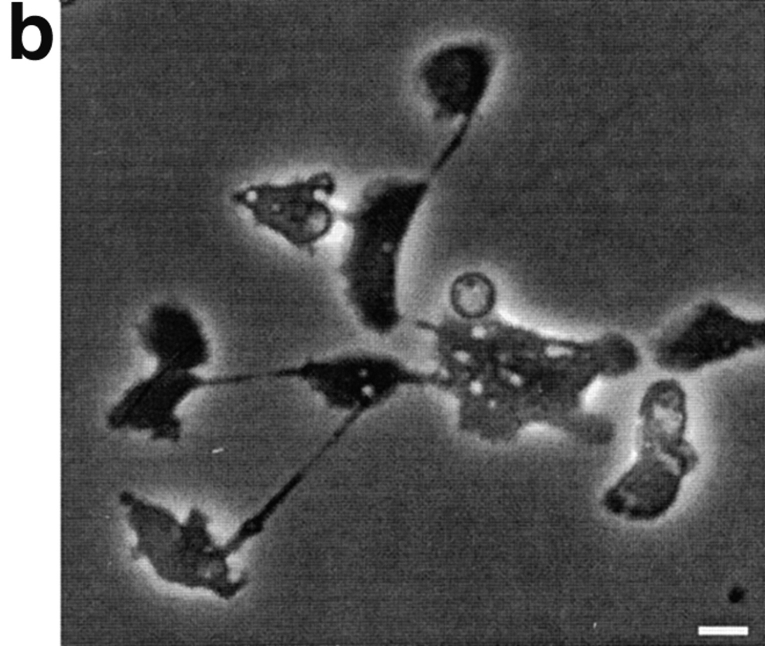
Figure 6.
Defective cytokinesis in rasG − cells. (a) Epifluorescent micrograph of a multinucleate rasG − cell that has been growing in shaken culture for 5 d. F-actin is visualized with rhodamine-conjugated phalloidin, nuclei with Hoechst 33258. (b) Phase contrast micrograph of a multinucleate rasG − cell undergoing traction-mediated cytofission, 10 min after plating. Bars, 20 μm.
Normal Chemotaxis in rasG− Cells
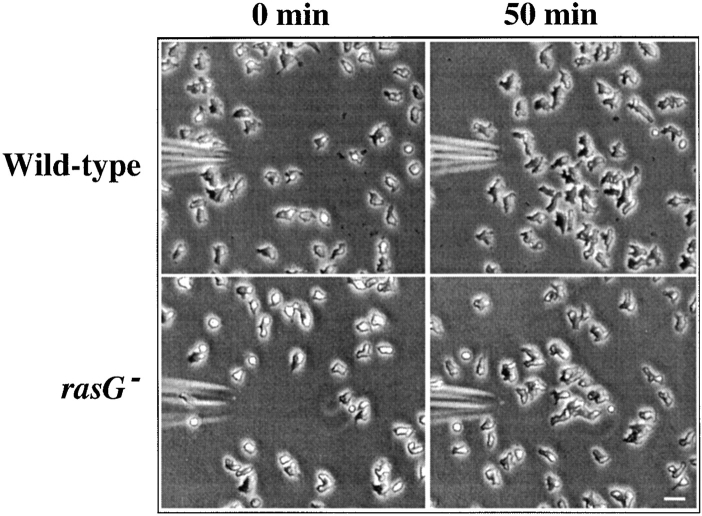
Chemotaxis up chemical gradients involves both cell polarity and changes in the actin cytoskeleton. Since both the cell polarity and cytoskeleton of the rasG − cells are aberrant (Fig. 3), and in the light of the observation that the disruption of the Aimless RasGEF demonstrates a requirement for a ras pathway for Dictyostelium chemotaxis (Insall et al., 1996), we measured chemotaxis of rasG − cells towards folic acid, which is used by growing cells to detect bacteria. As documented in Fig. 5, rasG − cells are almost equal to wild-type in their ability to migrate towards a micropipette filled with folic acid. Considering their impaired cytoskeletal structure and slow movement, they perform chemotaxis surprisingly successfully.
Figure 5.
Chemotaxis in wild-type and rasG − cells. Phase contrast micrographs of wild-type cells (top) and rasG − cells (bottom) migrating towards a micropipette filled with 10 μM folic acid. Cells are shown 0 (left) and 50 min (right) after presentation of the pipette. Bar, 20 μm.
Cytokinesis and Cell Fission
The poor growth of rasG − cells in shaken suspension suggests a defect in cytokinesis. When cells are transferred from growth on petri plates, where they maintain a wild-type number of nuclei (data not shown), to shaken culture, they rapidly become multinucleate (Fig. 6 A). Within 3 d of shaken growth, the majority of cells has several nuclei, and many have >10. Multinucleate cells the size of the one shown in Fig. 6 A are common, and considerably larger examples are sometimes observed. After 6 or 7 d of growth in shaken suspension, cells become so large that they lyse, presumably from the shear forces generated by shaking. When multinucleate cells are allowed to adhere to a surface such as a glass coverslip, they rapidly tear themselves into mononucleate fragments. The single cell shown in the process of splitting in Fig. 6 B had divided into at least 11 daughters within 30 min of plating on a surface. This process, which has been named traction-mediated cytofission (Fukui et al., 1990), appears not to greatly harm the cells, and is the mechanism which allows Dictyostelium cytokinesis mutants to divide at nearly normal rates on a surface (a QuickTime video showing a rasG − cell splitting by traction-mediated cytofission after growth in suspension culture can be viewed at the World Wide Web site http://www.ucl.ac.uk/∼dmcbrob/movies.html).
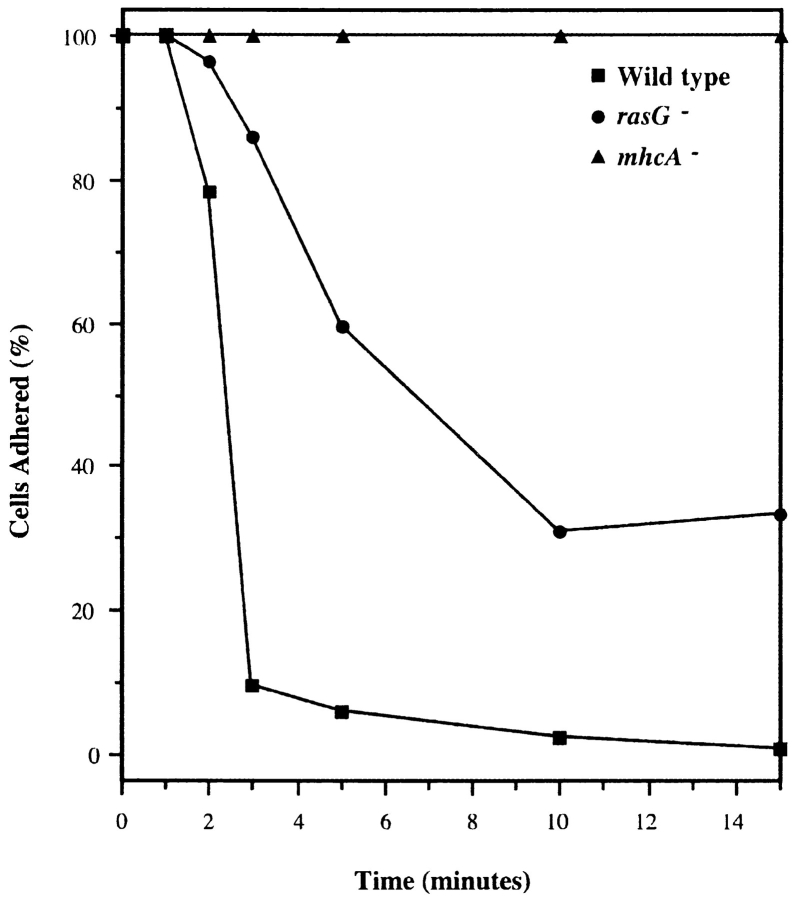
Nearly all the cytokinesis mutants that have been previously described in Dictyostelium have defects in the structural proteins of the cytoskeleton. The most comprehensively studied mutants lack myosin II function (De Lozanne and Spudich, 1987; Knecht and Loomis, 1987; Chen et al., 1995). These cells are unable to complete cytokinesis because of an inability to generate the contractile ring which pinches the daughter cells apart. The phenotype of rasG disruptants is not caused by a lack of myosin II activity. Treatment of cells with sodium azide causes a rapid depletion of cellular ATP levels, which causes myosin II to bind irreversibly to actin (Patterson and Spudich, 1995). When wild-type cells in a perfusion chamber are treated with 0.01% sodium azide, they round up and are washed off the substratum. mhcA − cells, which lack the heavy chain of myosin II, remain flattened and stuck down. Azide treatment caused nearly normal rounding and loss of rasG − cells (Fig. 7). The small number of mutant cells that did not wash away had rounded up in the same way as wild-type cells but were still adhering to the glass coverslip. This seems to be caused by the aberrant adhesion of rasG − cells rather than any lack of myosin II function.
Figure 7.
Normal myosin function in rasG − cells. Cells adhering to glass coverslips were perfused with buffer containing 0.01% azide. Cells lacking myosin II (mhcA −, ▴) remain flattened and adherent, while both wild-type cells (▪) and rasG − cells (•) round up and detach from the glass.
We wished to discern whether rasG − cells were unable to perform cytokinesis because of a mechanical defect preventing the physical separation of the daughter cells, like that in mhcA − cells, or because of a problem with the initiation of cytokinesis due to the loss of a signal communicating the completion of nuclear division to the cytoplasm. To answer these questions, synchronized cells in mitosis were required, but we found existing methods of synchronizing Dictyostelium cells unsatisfactory. By releasing cells whose cell cycle progression was blocked by aphidicolin (an inhibitor of DNA polymerase; Pedrali-Noy et al., 1980), we observed good synchrony. 50% of cells passed through mitosis ∼4 h after release. When synchronized wild-type cells are observed by time-lapse video microscopy, cytokinesis is observed as a rounding of the cell followed by a rapid pinching movement, taking ∼5 min to complete (Fig. 8 A, top rows). The initial stages of cytokinesis in rasG − cells appear indistinguishable from those in wild-type cells (Fig. 8 A, lower rows). However, the daughter cells appear unable to separate completely and remain attached by a thin bridge of cytoplasm, which is eventually resolved by traction between the daughter cells (Fig. 8 A).
Figure 8.
Cytokinesis in wild-type and rasG − cells. (a) Video frames showing cytokinesis in wild-type and rasG − cells. Cells were observed by time-lapse videomicroscopy after synchronization by aphidicolin. Representative cytokineses from each strain are shown. (b) Synchronized wild-type and rasG − cells were stained with Texas red-conjugated phalloidin and Hoechst 33342 to visualize F-actin and nuclei, respectively. RasG − cells show a characteristic failure to separate after formation of the cleavage furrow. Bar, 10 μm.
Wild-type cells that are fixed during cytokinesis and stained for F-actin and chromosomes show a characteristic binucleate hour-glass shape with the chromosomes condensed throughout the process (Fig. 8 B, top rows). Again, with fixed cells the initial stages of cytokinesis in rasG − cells appear identical to those in wild-type cells (Fig. 8 B, lower rows), but the bridge of cytoplasm that results from the failure of the rasG − daughter cells to separate is shown clearly. The chromosomes of the two daughters decondense (Fig. 8 B, lower right image) indicating that the cells can remain attached for some time. In contrast, wild-type cells show condensed nuclei even as the daughters separate completely (Fig. 8 B, upper right image). Staining with an anti-tubulin antibody indicates that the microtubules also return to an interphase morphology before complete separation in the rasG − cells but remain in a mitotic morphology throughout cytokinesis in wild-type cells (data not shown).
The failure of rasG − cells late in cytokinesis suggests that they have a mechanical defect. RasG does not appear to be required for the spatial or temporal specification of the cleavage furrow. As demonstrated by Fig. 7, myosin II activity is present in rasG − cells, but RasG may be required for the correct regulation of myosin II or other cytoskeletal proteins during cytokinesis. Alternatively, RasG may be required for the correct regulation of actin polymerization during cytokinesis with the result that an aberrant F-actin framework is established in the cleavage furrow that prevents the necessary contractile forces being generated by myosin II and other cytoskeletal proteins.
Discussion
The most obvious phenotype of rasG − mutant cells is, unexpectedly, a defect in cytokinesis. Cells also exhibit several other cytoskeletal defects, including alterations in cell polarity, morphology, motility, and F-actin structures. The slightly reduced growth rate of mutants is most probably due to the defects in motility and cytokinesis, although we cannot rule out other minor difficulties. Clearly rasG is not essential for growth. These results could be interpreted in two ways. Firstly, RasG may be concerned solely with the regulation of motility, while differentiation and growth control are mediated through other Ras proteins, or not by a Ras at all. Alternatively, RasG could be important for the control of a wide range of cellular processes, just like its mammalian homologues, but the phenotype of rasG disruption can be moderated by partial functional redundancy. If the latter is true, RasG might normally be central to regulation of growth but be substituted by RasB, RasC, or ectopically expressed RasD in rasG − cells. RasD is highly homologous to RasG: 100% identical in the effector and effector-proximal domains and 82% identical over the entire length (Daniel et al., 1995), and therefore might be expected to substitute for some of the functions of RasG if expressed during growth. The low recovery of homologous recombinants compared to random integrants might suggest a central role for RasG, but it is notoriously hard to predict the efficiency of homologous integration. On the other hand, the phenotypes observed after overexpression of mutant forms of RasG are mainly concerned with cell morphology. Cells containing an activated RasG appear flattened, while those transformed with inhibitory mutants are polarized (Rebstein et al., 1997). Whichever explanation is true, cytokinesis and actin organization appear to be the only processes that cannot survive rasG disruption.
Ras and Actin
The role for Ras proteins in motility has been observed in a range of species, though until recently it has been overshadowed by the importance of growth control. Ras activation leads to enhanced motility of endothelial cells (Fox et al., 1994) and apparently mediates the chemotactic response of fibroblasts to PDGF (Kundra et al., 1995). When the genes encoding RasGAP and/or NF1 are disrupted in mice, one of the causes of embryonic lethality is malformation of blood vessels due to hypermotile endothelial cells (Henkemeyer et al., 1995). Likewise, Saccharomyces CDC42, which controls actin polymerization during bud formation, is regulated through Ras (Mosch et al., 1996).
In Dictyostelium rasG − cells, all of the phenotypes we observe are connected with the control of the actin cytoskeleton. The lamellipodia at the leading edge of wild-type cells, and the filopodia found in rasG − cells, are both transitory structures made of freshly polymerized actin (Hall et al., 1988), and the cleavage furrow is also actomyosin rich (Fukui and Inoue, 1991). The absolute quantity of F-actin in rasG − cells is normal, though the part played by the unusual punctate actin structures is unknown.
A range of experiments has shown that small GTPases such as Rac and Cdc42 proteins are controlled by Ras pathways (Nobes and Hall, 1995). Dictyostelium contains a large number of rac homologues (Bush et al., 1993), so the different effects of RasG on the cytoskeleton and motility could be transmitted through several Rac proteins with separate roles. The specific role of the racE gene in cytokinesis (Larochelle et al., 1996), described below, is a possible example. Another known effector of Ras proteins is p110 PI 3-kinase (Rodriguez-Viciana et al., 1994), which may constitute the direct connection between Ras and Rac proteins. Three different p110 homologues have recently been cloned in Dictyostelium (Zhou et al., 1995), and it is significant that mutants also show defects in the control of the cytoskeleton. We look forward to observing the effects of rasG disruption on PI 3-kinase and Rac activity.
Ras and Cytokinesis
Ras proteins in other organisms have been associated with both control of the cytoskeleton and cell proliferation but not usually with cytokinesis. However, since in other systems Ras is essential for cell growth, and therefore nuclear division, its role in the division of the cytoplasm thereafter has been difficult to study. Despite this, there are several reports connecting Ras activity with cytokinesis. Schizosaccharomyces byr4, a suppressor of ras1 mutations (Song et al., 1996), appears to encode a regulated inhibitor of cytokinesis. Furthermore, activating Ras mutants increase the rate of cytokinesis in mammalian cells (Ng et al., 1992). There is also an increasing amount of evidence that the small GTPases Rac, Rho, and Cdc42 are centrally involved in cytokinesis (Dutartre et al., 1996), and Ras appears to act as an upstream regulator of their activities. It is therefore likely that the central role of RasG in Dictyostelium cytokinesis is mirrored in other organisms.
There are several likely candidates for proteins downstream of RasG. Interestingly, the disruption of one GAP gene (Lee et al., 1997) and overexpression of the other (Faix and Dittrich, 1996) both result in deficiencies in cytokinesis, as does overexpression of activating RasG mutants (Rebstein et al., 1997). This may reflect a need for limited, local activation of RasG during cytokinesis. RasG activation at an inappropriate place or time could be just as harmful to efficient cytokinesis as the complete lack of RasG in disrupted cells. The protein encoded by the racE gene is required for cytokinesis (Larochelle et al., 1996); unlike rasG − cells, however, racE − mutants appear normal in all other respects, including the formation of lamellipodia, adhesion, and rate of motility. Again, since Rac proteins in other systems can be activated by Ras, a direct connection between RasG and RacE seems possible. Similarly, cells containing concurrent deletions in two PI 3-kinase genes (pik1 and pik2) lose the ability to grow in shaken culture (Zhou et al., 1995), despite nearly normal growth on surfaces, which suggests that they have similar defects in cytokinesis to rasG − cells. Alternatively, the defect in cytokinesis in rasG − cells may be indirect, caused by the general disruption of the actin cytoskeleton rather than the loss of a specific signaling pathway.
Like the ras genes themselves, the biochemical pathways between Ras and the cytoskeleton are apparently conserved between Dictyostelium and mammalian cells. This again suggests that cytokinesis in mammalian cells may turn out to be affected by Ras as it is in Dictyostelium discoideum. The fascinating prospect is that Ras proteins through their involvement in the signaling pathways that commit cells to division and by regulating the cytoskeleton during cytokinesis are major players at the start and at the end of the cell cycle.
Acknowledgments
We thank Dr. David Drechsel for helpful discussions and Drs. Jeff Williams and Adrian Harwood for constructive comments on the text.
Footnotes
1. Abbreviations used in this paper: GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factors.
The work described in this paper was supported by a Wellcome Trust Career Development Fellowship 043754 to R.H. Insall, by a Medical Research Council Career Development Fellowship to L.M. Machesky, and by a grant from the Medical Research Council of Canada to G. Weeks. R.I. Tuxworth is supported by the Medical Research Council graduate program from the Institute of Molecular Cell Biology, University College London.
Please address all correspondence to Robert Insall, MRC Laboratory for Molecular Cell Biology, University College London, Gower Street, London WC1E 6BT, UK. Tel.: (44) 171 380 7270; Fax: (44) 171 380 7805; E-mail: dmcbrob@ucl.ac.uk
References
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideumcytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem Biophys Res Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature (Lond) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bush J, Franek K, Cardelli J. Cloning and characterization of seven novel Dictyostelium discoideumRac-related genes belonging to the Rho family of GTPases. Gene. 1993;136:61–68. doi: 10.1016/0378-1119(93)90448-c. [DOI] [PubMed] [Google Scholar]
- Chen TLL, Kowalczyk PA, Ho GY, Chisholm RL. Targeted disruption of the Dictyosteliummyosin essential light chain gene produces cells defective in cytokinesis and morphogenesis. J Cell Sci. 1995;108:3207–3218. doi: 10.1242/jcs.108.10.3207. [DOI] [PubMed] [Google Scholar]
- Daniel J, Spiegelman GB, Weeks G. Characterization of a third ras gene, rasB, that is expressed throughout the growth and development of Dictyostelium discoideum. . Oncogene. 1993;8:1041–1047. [PubMed] [Google Scholar]
- Daniel J, Bush J, Cardelli J, Spiegelman GB, Weeks G. Isolation of two novel ras genes in Dictyostelium discoideum: evidence for a complex, developmentally regulated ras gene subfamily. Oncogene. 1994;9:501–508. [PubMed] [Google Scholar]
- Daniel, J., G. Weeks, and G. Spiegelmann. 1995. Dictyostelium ras genes. In Guidebook to the Small GTPases. M. Zerial and L. Huber, editors. Oxford University Press, Oxford, UK. 100–104.
- de Hostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyosteliummutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyosteliummyosin heavy chain gene by homologous recombination. Science (Wash DC) 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: a focus on leucocytes and Dictyostelium. . Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- Devreotes P, Fontana D, Klein P, Sherring J, Theibert A. Transmembrane signaling in Dictyostelium. . Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Dutartre H, Davoust J, Gorvel J-P, Chavrier P. Cytokinesis arrest and redistribution of actin-cytoskeleton regulatory components in cells expressing the Rho GTPase CDC42S. J Cell Sci. 1996;109:367–377. doi: 10.1242/jcs.109.2.367. [DOI] [PubMed] [Google Scholar]
- Faix J, Dittrich W. DGap1, a homolog of Ras-GTPase activating proteins that controls growth, cytokinesis and development in Dictyostelium discoideum. . FEBS (Fed Eur Soc Biochem) Lett. 1996;394:251–257. doi: 10.1016/0014-5793(96)00963-5. [DOI] [PubMed] [Google Scholar]
- Fishkind D, Wang Y. New horizons for cytokinesis. Curr Opin Cell Biol. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Fortini M, Simon M, Rubin G. Signalling by the sevenless protein tyrosine kinase is mimicked by ras1 activation. Nature (Lond) 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- Fox P, Sa G, Dobrowolski S, Stacey D. The regulation of endothelial cell motility by p21 ras. Oncogene. 1994;9:3519–3526. [PubMed] [Google Scholar]
- Fukui Y, Inoue S. Cell division in Dictyosteliumwith special emphasis on actomyosin organization in cytokinesis. Cell Motil Cytoskel. 1991;18:41–54. doi: 10.1002/cm.970180105. [DOI] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyosteliummyosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth M, Davis L, Fleurdelys B, Scolnick E. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras family. J Virol. 1982;43:294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AL, Schlein A, Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J Cell Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- Han M, Sternberg P. Let-60, a gene which specifies cell fates during vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M, Rossi D, Holmyard D, Puri M, Mbamalu G, Harpal K, Shih T, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase activating protein. Nature (Lond) 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- Howard PK, Ahern KG, Firtel RA. Establishment of a transient expression system for Dictyostelium discoideum. . Nucl Acids Res. 1988;16:2613–2623. doi: 10.1093/nar/16.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R, Borleis J, Devreotes P. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. . Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- Karess R, Chang X-J, Edwards K, Kulkarni S, Aguilera I, Kiehart D. The regulatory chain of non-muscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. . Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Khosla M, Robbins SM, Spiegelman GB, Weeks G. Regulation of DdrasG gene expression during Dictyosteliumdevelopment. Mol Cell Biol. 1990;10:918–922. doi: 10.1128/mcb.10.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla M, Spiegelman GB, Weeks G, Sands TW, Virdy KJ, Cotter DA. RasG protein accumulation occurs just prior to amoebae emergence during spore germination in Dictyostelium discoideum. . FEMS (Fed Eur Microbiol Soc) Microbiol Lett. 1994;117:293–298. doi: 10.1111/j.1574-6968.1994.tb06782.x. [DOI] [PubMed] [Google Scholar]
- Khosla M, Spiegelman GB, Weeks G. Overexpression of an activated RasG gene during growth blocks the initiation of Dictyosteliumdevelopment. Mol Cell Biol. 1996;16:4156–4162. doi: 10.1128/mcb.16.8.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. . Science (Wash DC) 1987;236:1081–1085. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Kundra A, Anand-Apte B, Feig L, Zetter B. The chemotactic response to PDGF-BB: evidence of a role for Ras. J Cell Biol. 1995;130:725–731. doi: 10.1083/jcb.130.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle D, Vithalani K, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133:1321–1329. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Escalante R, Firtel RA. A Ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium. . Development. 1997;124:983–996. doi: 10.1242/dev.124.5.983. [DOI] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signalling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/ Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G, Boylan J, Zimmer SG, Sisken JE. Cytokinesis is more rapid in Ha-T24-ras transfected rat embryo fibroblasts than in non-transfected control cells. Cell Motil Cytoskeleton. 1992;21:159–166. doi: 10.1002/cm.970210209. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Olivier J, Raabe T, Henkemeyer M, Dickson B, Mbamalu G, Margolis B, Schlessinger J, Hafen E, Pawson T. A DrosophilaSH2-SH3 adaptor protein implicated in coupling the Sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange. Cell. 1993;73:179–191. doi: 10.1016/0092-8674(93)90170-u. [DOI] [PubMed] [Google Scholar]
- Patterson B, Spudich JA. A novel positive selection for identifying cold-sensitive myosin II mutants in Dictyostelium. . Genetics. 1995;140:505–515. doi: 10.1093/genetics/140.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrali-Noy G, Spadari S, Miller-Faures A, Miller AO, Kruppa J, Koch G. Synchronization of HeLa cell cultures by inhibition of DNA polymerase alpha with aphidicolin. Nucleic Acids Res. 1980;8:377–387. doi: 10.1093/nar/8.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci P. A novel transforming protein (shc) with a SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- Rappaport R. Repeated furrow formation from a single mitotic apparatus in cylindrical sand dollar eggs. J Exp Zool. 1985;234:167–171. doi: 10.1002/jez.1402340120. [DOI] [PubMed] [Google Scholar]
- Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cyt. 1986;105:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- Rebstein, P.J., M. Khosla, G.B. Spiegelman, and G. Weeks. 1997. Pleiotropic effects of the overexpression of an activated of rasG gene in Dictyostelium. J. Cell Science. In press.
- Reymond CD, Gomer RH, Mehdy MC, Firtel RA. Developmental regulation of a Dictyosteliumgene encoding a protein homologous to mammalian Ras protein. Cell. 1984;39:141–148. doi: 10.1016/0092-8674(84)90199-5. [DOI] [PubMed] [Google Scholar]
- Reymond CD, Gomer RH, Nellen W, Theibert A, Devreotes P, Firtel R. Phenotypic changes induced by a mutated Ras gene during the development of Dictyosteliumtransformants. Nature (Lond) 1986;323:340–343. doi: 10.1038/323340a0. [DOI] [PubMed] [Google Scholar]
- Ridley A, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Robbins SM, Williams JG, Jermyn KA, Spiegelman GB, Weeks G. Growing and developing Dictyosteliumcells express different ras genes. Proc Natl Acad Sci USA. 1989;86:938–942. doi: 10.1073/pnas.86.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins SM, Williams JG, Spiegelman GB, Weeks G. Cloning and characterization of the Dictyostelium discoideumrasG genomic sequences. Biochim Biophys Acta. 1992;1130:85–89. doi: 10.1016/0167-4781(92)90467-e. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne P, Dhand R, Vanhaesenbrock B, Gout I, Fry M, Waterfield M, Downward J. Phosphatidyl-3-OH kinase as a direct target of Ras. Nature (Lond) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. In Molecular Cloning: A Laboratory Manual. 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- Shih TY, Weeks MO, Young HA, Scolnick EM. Identification of a sarcoma virus-coded phosphoprotein in nonproducer cells transformed by Kirsten or Harvey murine sarcoma virus. Virology. 1979;96:64–79. doi: 10.1016/0042-6822(79)90173-9. [DOI] [PubMed] [Google Scholar]
- Song K, Mach K, Chen C, Reynolds T, Albright C. A novel suppressor of RAS1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorcher, S., A. Schonthal, A. Alberts, J. Meinkoth, and J. Feramisco. 1993. The ras superfamily of GTPases. In The ras superfamily of GTPases. J. Lacal and F. McCormick, editors. CRC Press, Boca Raton, FL. 85–101.
- Sun TJ, Van Haastert PJM, Devreotes PN. Surface cAMP receptors mediate multiple responses during development in Dictyostelium, evidenced by antisense mutagenesis. J Cell Biol. 1990;110:1549–1554. doi: 10.1083/jcb.110.5.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyosteliumunder controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideumwith a new selectable marker Bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Valencia, A., and C. Sander. 1995. The ras superfamily. In Guidebook to the Small GTPases. M. Zerial and L. Huber, editors. Oxford University Press, Oxford, UK. 12–19.
- Wang J, Suzuki N, Nishida Y, Kataoka T. Analysis of the 70-kilodalton cyclase-associated protein (CAP) by using mutants of yeast adenylyl cyclase defective in CAP binding. Mol Cell Biol. 1993;13:4087–4097. doi: 10.1128/mcb.13.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KM, Takegawa K, Emr SD, Firtel RA. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: Biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol Cell Biol. 1995;15:5645–5656. doi: 10.1128/mcb.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]