Abstract
Proper orientation of the mitotic spindle is critical for successful cell division in budding yeast. To investigate the mechanism of spindle orientation, we used a green fluorescent protein (GFP)–tubulin fusion protein to observe microtubules in living yeast cells. GFP–tubulin is incorporated into microtubules, allowing visualization of both cytoplasmic and spindle microtubules, and does not interfere with normal microtubule function. Microtubules in yeast cells exhibit dynamic instability, although they grow and shrink more slowly than microtubules in animal cells. The dynamic properties of yeast microtubules are modulated during the cell cycle. The behavior of cytoplasmic microtubules revealed distinct interactions with the cell cortex that result in associated spindle movement and orientation. Dynein-mutant cells had defects in these cortical interactions, resulting in misoriented spindles. In addition, microtubule dynamics were altered in the absence of dynein. These results indicate that microtubules and dynein interact to produce dynamic cortical interactions, and that these interactions result in the force driving spindle orientation.
M ovement and positioning of organelles within cells occurs through interaction with the cytoskeleton. Microtubules play a particularly important role in this process; microtubules are able to span large distances in the cell, microtubule motor proteins can move organelles in both directions on the polar microtubule polymer, and microtubules themselves are oriented by the microtubule organizing center in animal cells and fungi. In addition to interacting with organelles, microtubules must also interact with determinants of cellular polarity so that the microtubules are in the right place at the right time. Although much is known about the in vitro properties of microtubules and their associated proteins, little is known about the interactions between microtubules and cellular factors in polarized cells, and how these interactions result in directed intracellular movement.
One example of a microtubule-dependent process that responds to cellular polarity is spindle orientation during mitosis. In many organisms, spindle orientation determines the division plane, with cytokinesis occurring at the position of the metaphase plate. In some cases spindle orientation is controlled to give a determined division pattern. For example, in several of the early embryonic divisions of Caenorhabditis elegans, the centrosomes rotate 90° about the nucleus in one of two daughter cells, resulting in a change of division plane that is essential for proper development (Hyman and White, 1987). During mammalian neurogenesis, spindle orientation and the subsequent division plane predict the developmental fate of the daughter cells from the division (Chenn and McConnell, 1995). A related situation exists in the budding yeast Saccharomyces cerevisiae, where the mitotic spindle must be oriented relative to a predetermined division plane. After bud emergence and growth, the nucleus migrates to the mother–bud junction, or neck, and the spindle is oriented such that it is positioned through the neck as it elongates during anaphase (Byers, 1981). Yeast cells undergo cytokinesis at the neck regardless of the position of the mitotic spindle; thus misorientation of the spindle results not in developmental defects as in the animal cell examples above, but in failure to segregate chromosomes to one of the two cells from the division.
Genetic analysis in S. cerevisiae has shown that cytoplasmic microtubules, actin, and components of the dynein/ dynactin complex are important in spindle orientation (Huffaker et al., 1988; Palmer et al., 1992; Sullivan and Huffaker, 1992; Eshel et al., 1993; Li et al., 1993; Clark and Meyer, 1994; Muhua et al., 1994). Mutations affecting any of these components result in a higher percentage of binucleate and anucleate cells because of the failure of the spindle to elongate through the neck. Cytoplasmic dynein is a multisubunit, minus end–directed microtubule motor protein that has been implicated in vesicle transport (Schroer et al., 1989) and mitotic spindle function (Vaisberg et al., 1993) in animal cells. Dynein interacts with dynactin, a large protein complex, which acts to stimulate dynein-dependent movement (Gill et al., 1991; Schroer and Sheetz, 1991). Yeast spindle orientation during mitosis involves characteristic spindle oscillations across the neck region, which are absent in dynein mutants (Yeh et al., 1995). While yeast dynein heavy chain mutants are viable, the defect in spindle orientation is exacerbated at low temperatures (Eshel et al., 1993; Li et al., 1993). This phenotype is consistent with that found for dynein and dynactin mutations in Aspergillus and Neurospora where this complex is required for proper positioning of nuclei within hyphal cells (Plamann et al., 1994; Xiang et al., 1994, 1995; Tinsley et al., 1996). Localization studies in yeast show that dynein is present at the spindle pole, on the cytoplasmic microtubules, and, less certainly, at the cell cortex (Yeh et al., 1995). This complex pattern of localization has made it difficult to understand the mechanism of force transduction and how it is coordinated with cell polarity.
Microtubules in vitro display a behavior termed dynamic instability; individual microtubules in a population can be either growing or shrinking in length, with transitions between these two phases occurring in a stochastic manner (Mitchison and Kirschner, 1984). In animal cells it has been possible to microinject fluorescently labeled tubulin and to observe microtubule behavior in living cells. Animal cell microtubules in vivo also display dynamic instability (Schulze and Kirschner, 1987, 1988; Cassimeris et al., 1988; Sammak and Borisy, 1988), although microtubule dynamics are tempered in vivo by interaction with other components of the cell and regulated by the cell cycle state (Salmon et al., 1984; Saxton et al., 1984; Belmont et al., 1990; Gliksman et al., 1992; Verde et al., 1992). It is likely that selective modulation of dynamics allows the microtubule cytoskeleton to change in response to differing needs within the cell (Kirschner and Mitchison, 1986).
EM and immunofluoresence experiments on fixed cells show that the microtubule cytoskeleton of S. cerevisiae is much simpler than that of animal cells, providing an excellent opportunity to study a specific process such as directed organelle movement. The yeast microtubule organizing center, termed the spindle pole body, is embedded in the nuclear envelope and nucleates microtubules from both the cytoplasmic and nuclear faces (Byers and Goetsch, 1975). The intranuclear mitotic spindle is a bundle of microtubules that is seen as a bar by immunofluorescence (Kilmartin and Adams, 1984), but can be resolved into ∼40 microtubules per haploid cell by EM (Winey et al., 1995). In contrast, immunofluorescence and EM reveal only a few cytoplasmic microtubules. These appear to be single microtubules, often with their distal end close to the edge of the cell (Kilmartin and Adams, 1984). The genetic evidence for involvement of cytoplasmic microtubules in spindle orientation, and their proximity to the cell cortex, led to a model in which the cytoplasmic microtubules interact with a cortical attachment site through which they exert force on the nucleus (Snyder et al., 1991; Hyman and Stearns, 1992), similar to what is observed for spindle rotation in C. elegans embryos (Hyman, 1989).
Previous studies have characterized aspects of spindle orientation and elongation in living yeast cells (Palmer et al., 1989; Koning et al., 1993; Kahana et al., 1995; Yeh et al., 1995). Although the effects of microtubules were clearly shown in these studies, microtubules themselves could not be observed. In this study, we have characterized microtubules in living yeast cells using a green fluorescent protein (GFP)1–tubulin fusion protein that is incorporated into microtubules. The small size and simple microtubule cytoskeleton of yeast cells have allowed us to visualize cytoplasmic microtubules during the process of spindle orientation in wild-type and dynein-mutant cells. These experiments reveal that microtubule interactions with the cell cortex are associated with movement of the spindle and show that these interactions are dynein dependent.
Materials and Methods
Strains and Media
Yeast strains used in this study are listed in Table I. Yeast extract/peptone, synthetic dextrose, and sporulation media were as described (Sherman et al., 1986). Strains carrying galactose-inducible constructs were grown on synthetic media (−Ura) containing 2% galactose, and in some experiments either 0.5 or 1% glucose was added. Yeast molecular genetic methods were as described (Stearns et al., 1990). Benomyl, 98.6% pure, was a generous gift from E.I. duPont de Nemours and Co., Inc. (Wilmington, DE), and was kept as a 10 mg/ml stock in DMSO at −20°C. Benomyl was added to media to final concentrations of 1, 2.5, 5, 10, and 15 μg/ml.
Table I.
Yeast Strains
| Name | Genotype | Source | ||
|---|---|---|---|---|
| TPS507 | MATa/α ADE2/ade2 his3-Δ200/his3- Δ200 leu2-3,112/leu2-3,112 lys2- 801/lys2-801 ura3-52/ura3-52 | This laboratory | ||
| TSY410 | TPS 507 (pTS403) | This study | ||
| TSY423 | TPS507 (pTS414) | This study | ||
| TSY425 | TPS507 (pTS417) | This study | ||
| TSY510 | TPS507 (pTS331) | This study | ||
| TSY511 | TPS507 (pTS337) | This study | ||
| TSY524 | MATa/α his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 ura3-52/ ura3-52 dyn1::HIS3/dynl::HIS3 | This study | ||
| TSY526 | TSY524 (pTS417) | This study | ||
| DBY2375 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 tub3::TRP1 | D. Botstein | ||
| TSY682 | DBY2375 (pTS417) | This study | ||
| TSY683 | DBY2375 (pTS414) | This study | ||
| TSY684 | DBY2375 (pTS403) | This study | ||
| TSY685 | DBY2375 (pTS337) | This study | ||
| TSY686 | DBY2375 (pTS331) | This study |
GFP–Tubulin Constructs
The vector plasmid pTS210, a YCp50-based construct, contained the GAL1/GAL10 promoter, a short polylinker (BamHI, HindIII, XbaI), and the ACT1 transcriptional terminator (Marschall et al., 1996). pTS331 was similar to pTS210, but was constructed from pRS316 (Sikorski and Hieter, 1989), while pTS337 had the TUB3 gene cloned into the BamHI and XbaI sites of pTS331. Two GFP plasmids were constructed so that genes could be inserted either upstream or downstream of GFP. Primers homologous to GFP sequences, containing added restriction sites, were used to PCR amplify wild-type GFP from pGFP10.1 (Chalfie et al., 1994). PCR-amplified GFP, containing restriction sites XbaI and NheI on either end, was cloned into the XbaI site of pTS210, creating pTS395 that contained the polylinker upstream of GFP and was used to make COOH-terminal GFP fusions. pTS408 was constructed by PCR amplifying GFP with BamHI and BglII sites and by inserting this into the BamHI site of pTS210, creating a plasmid with the polylinker downstream of GFP to create NH2-terminal GFP fusions.
COOH-terminal GFP fusions were made after PCR amplification of either TUB1 or TUB3 containing added restriction sites (BamHI and SpeI) on either end. The TUB1 or TUB3 PCR product was cloned into the BamHI and XbaI site of pTS395, creating plasmids pTS414 and pTS403, respectively. pTS417, an NH2-terminal GFP fusion to TUB3, was constructed by inserting TUB3 into the BamHI and SalI sites of pTS408. All GFP constructs were transformed into the diploid yeast strain, TPS507, and were induced on galactose-containing synthetic −Ura media for all subsequent experiments.
Dynein Disruption
A dyn1::HIS3 disruption deletion vector was generously provided by M. Andrew Hoyt ( Johns Hopkins University, Baltimore, MD) (Geiser et al., 1997). The HIS3 deletion removes 1701 bp from an EcoRV to SacI site within the 3.5-kb R2 fragment as shown in Fig. 2 of Eshel et al., 1993. The EcoRV site is not shown and is upstream of the two HindIII sites used to make the URA3 disruption deletion. The vector was digested with EcoRI and transformed into the diploid strain, TPS507. Heterozygotes containing the dynein disruption integrated at the DYN1 locus were sporulated and dissected to obtain haploids of the opposite mating type. Haploids containing dyn1::HIS3 were mated, and the resulting diploid strain, TSY524, was transformed with GFP-TUB3 plasmid, pTS417, generating strain TSY526. A smaller deletion of sequences contained within the HIS3 disruption showed a similar phenotype to a larger 7-kb deletion allele, and thus probably acts as a null allele (Li et al., 1993). Both strains TSY524 and TSY526 showed phenotypes consistent with previously published results.
Figure 2.
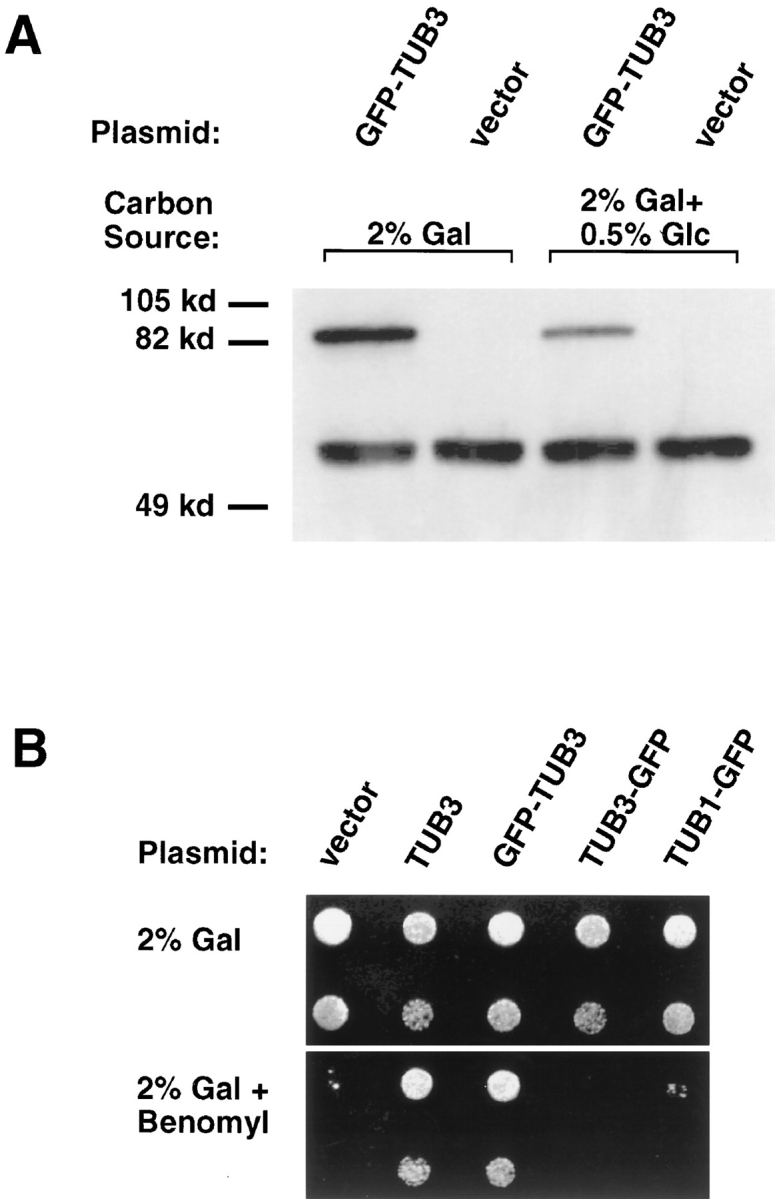
GFP–TUB3 expression and incorporation into functional microtubules. (A) Western blot of crude yeast protein extracts probed with anti–α-tubulin antibody. Strain TSY425, containing the GFP–TUB3 plasmid, and strain TPS510, containing the vector plasmid, were grown in the presence of either 2% galactose, or 2% galactose + 0.5% glucose as the carbon source. Both strains show endogenous total α-tubulin (TUB1p, TUB3p) running as a doublet at 56 kD, while the GFP–TUB3 fusion protein runs at 83 kD. Similar results were obtained with a COOH-terminal fusion protein, TUB3–GFP (data not shown). (B) GFP– TUB3 expression rescues the benomyl supersensitivity of a tub3:: TRP1 haploid strain (DBY2375). (Top) Growth of tub3::TRP1 cells transformed with plasmids as noted above, and a 1:10 dilution of cells (second row), when grown with 2% galactose. (Bottom) Growth of the same strains, and 1:10 dilutions, in the presence of 5 μg/ml benomyl. All cells were incubated at 30°C. tub3:: TRP1 containing either vector alone, TUB3–GFP, or TUB1–GFP remain benomyl supersensitive, while both TUB3 and GFP– TUB3 rescue the benomyl supersensitivity.
Western Blots
Protein samples for Western analysis were prepared by disrupting yeast cells with glass beads as described (Kaiser et al., 1987). Yeast cells were grown overnight in galactose-containing synthetic −Ura media and checked for fluorescence before disruption. Equal amounts of protein were loaded and separated on a 10% acrylamide gel. Extracts were run in two parallel sets, and one set was transferred to nitrocellulose (Protran; Schleicher & Schuell, Keene, NH) while the other set was Coomassie stained. Ponceau S staining of the nitrocellulose blot confirmed equal loading of the lanes. A monoclonal anti–α-tubulin antibody, TAT-1 (Woods et al., 1989), was used at 1:50, followed by an HRP-conjugated anti–mouse antibody at 1:5,000 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Immunoreactive proteins were visualized with chemiluminescent reagents (Dupont–New England Nuclear, Wilmington, DE).
Fluorescence Microscopy and Image Analysis
Immunofluorescence was performed as described (Pringle et al., 1989) with the following modifications. To preserve GFP fluorescence and visualize microtubules, cells were fixed by adding formaldehyde to a final concentration of 3.7% and by incubating at room temperature for 30 min, followed by methanol/acetone treatment. An anti-tubulin primary antibody, YOL1/34 (Kilmartin et al., 1982), was detected with Texas red–conjugated donkey anti–rat antibodies (Jackson ImmunoResearch Laboratories, Inc.).
To visualize GFP in living cells, strains containing GFP plasmids under the control of the GAL1 promoter were grown overnight at 30°C in galactose-containing media. 5 μl of cell culture was spotted directly onto slides, and slides with coverslips were briefly blotted to minimize cells from drifting during imaging. Fluorescent imaging was done in the laboratory of Dr. James Spudich (Department of Biochemistry, Stanford University, CA). Images of living cells were obtained at room temperature using an inverted fluorescence microscope (Axiovert; Carl Zeiss, Inc., Thornwood, NY), equipped with a HiQ fluorescein filter set (Chroma Optics, Brattleboro, VT) and a Planapochromat ×100/1.4 objective lens used in conjunction with ×2.5 optovar. A cooled charge-coupled device (CCD) camera (Princeton Research Instruments, Inc., Princeton, NJ) was attached via a basement port on the microscope, and was used to acquire time-lapse images. Images were acquired using 100-ms exposures every 2 or 10 s, and time-lapse images were generally acquired up to 10 min.
Time-lapse images were obtained and analyzed using MetaMorph imaging software (Universal Imaging, West Chester, PA). To follow single microtubules during time-lapse imaging, the focus was manually adjusted throughout the imaging to keep the microtubule and spindle pole body in focus. Microtubule lengths were tracked by tracing lines over microtubules throughout time-lapse sequences. Tracing of each microtubule was repeated two to three times, and the average pixel length was then converted to microns. Microtubules tended to be straight; in no cases were acutely bent microtubule ends observed. Only microtubules that remained in focus along the entire length were used for subsequent analysis. Dynamic analysis was performed using StatView software (Abacus Concepts, Berkeley, CA), and phases of growth, shrinkage, and pausing were determined by eye from life-history graphs obtained for individual microtubules. Linear regression analysis was used to obtain rates of microtubule growth and shrinkage, and only rates having R2 values >0.7 and P values ⩽0.05 (95% confidence level) were used for analysis. Statistical analysis (unpaired t test; P ⩽ 0.05) was used to compare rates obtained from both wild-type and dynein-mutant cells. Catastrophe and rescue frequencies were calculated by dividing the total number of events by the total time that all microtubules spent growing or shrinking, respectively. Survival analysis was used to obtain values for expected catastrophe and rescue events, which were then compared with the observed values using the χ2 test. Microtubule ends were determined to be at the cortex if they were within three pixels (0.22 μm) of the edge of the cell.
Results
GFP–α-Tubulin Is Incorporated into Microtubules In Vivo
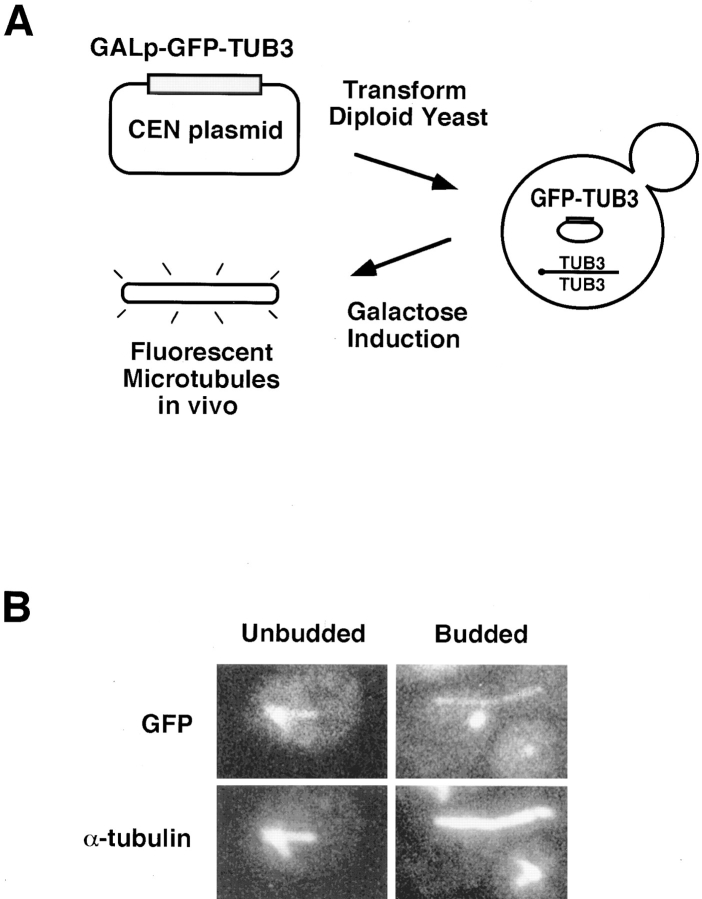
To visualize microtubules in living yeast cells, fusions were constructed between the yeast α-tubulin genes and the Aequorea victoria GFP (Prasher et al., 1992; Chalfie et al., 1994; Stearns, 1995). Yeast has four tubulin genes: two α-tubulin (TUB1 and TUB3), one β-tubulin (TUB2), and one γ-tubulin (TUB4). The TUB1 and TUB2 genes are essential for viability, whereas strains deleted for TUB3 are viable but supersensitive to the microtubule-destabilizing drug, benomyl (Neff et al., 1983; Schatz et al., 1986). The α-tubulin genes were chosen for fusion to GFP because yeast cells are relatively insensitive to the level of expression of α-tubulin, whereas overexpression of β-tubulin is lethal (Burke et al., 1989; Katz et al., 1990; Weinstein and Solomon, 1990). TUB1 and TUB3 encode functionally identical α-tubulin proteins (Schatz et al., 1986). Fusions between GFP and the amino and carboxy terminus of TUB3 (GFP–TUB3 and TUB3–GFP, respectively) were constructed and placed under the control of the inducible GAL1 promoter (Fig. 1 A and Materials and Methods).
Figure 1.
GFP–TUB3 produces fusion protein that incorporates into microtubules. (A) Schematic showing the process by which fluorescent microtubules are generated in vivo. The CEN plasmid carrying GFP–TUB3, pTS417, is transformed into the diploid yeast strain, TPS507, and the resulting strain, TSY425, is then grown in the presence of 2% galactose as the carbon source. After galactose induction, fluorescent microtubules are formed in vivo. (B) Colocalization of GFP–TUB3 with α-tubulin in microtubules. After growth of TSY425 in 2% galactose, cells were fixed and stained for α-tubulin. Fluorescence of rhodamine- labeled α-tubulin was compared with GFP fluorescence; both α-tubulin and GFP colocalized to microtubule structures in unbudded and budded cells. Fluorescent cytoplasmic microtubules and the spindle pole body are seen in unbudded cells (left), whereas a fluorescent mitotic spindle is seen in the budded cell shown (right). The right panel also contains an unbudded cell in which GFP fluorescence of the spindle pole body, but not the microtubules, can be seen.
Cells expressing each of the fusion proteins were examined by fluorescence microscopy; in each case, fluorescent microtubule structures similar to those seen in fixed cells were observed. To demonstrate that GFP–α-tubulin was incorporated into all microtubules in yeast, cells expressing GFP–α-tubulin were fixed and stained with antibody against α-tubulin (Fig. 1 B). Although the fluorescence of GFP–α-tubulin was reduced by the fixation, there was an exact correspondence between GFP- and antibody-labeled microtubules in cells showing GFP fluorescence (Fig. 1 B). In addition, there were occasional bright patches of GFP fluorescence in the cytoplasm that did not appear to be associated with the microtubule cytoskeleton, and did not stain with anti-tubulin antibody. The presence of these patches did not correlate with any specific microtubule behavior.
Cells expressing GFP–α-tubulin grew at rates indistinguishable from cells expressing wild-type α-tubulin from the same promoter, or from cells with vector alone, at normal growth temperatures. The amount of fusion protein produced was approximately equivalent to the total amount of endogenous TUB1 and TUB3 α-tubulin protein (Fig. 2 A, lanes 1 and 2). Expression of either GFP–α-tubulin or α-tubulin alone did have subtle effects on cell growth; such cells were cold sensitive and supersensitive to benomyl (data not shown). Because of the concern that the modest overexpression of α-tubulin observed was affecting microtubule behavior, expression from the inducible promoter was reduced by altering the carbon source. Under these conditions, expression was approximately half that of full induction (Fig. 2 A, lanes 3 and 4), and microtubule dynamics, as assayed below, were similar to those observed under full induction conditions (data not shown). Thus, the ratio of GFP–α-tubulin to α-tubulin does not have a significant effect on microtubule behavior. GFP–α-tubulin constructs were also tested for their ability to complement an α-tubulin mutation. GFP–TUB3 complemented the benomyl supersensitivity of a tub3 null mutant, whereas TUB3–GFP did not complement the mutant phenotype (Fig. 2 B). The failure of TUB3–GFP to complement the null mutant was not specific to TUB3, as a carboxy-terminal fusion of GFP to TUB1, TUB1–GFP, also failed to complement the tub3 null mutant (Fig. 2 B). The complementing GFP–TUB3 construct was used for all subsequent experiments.
Microtubule Organization in Living Yeast Cells
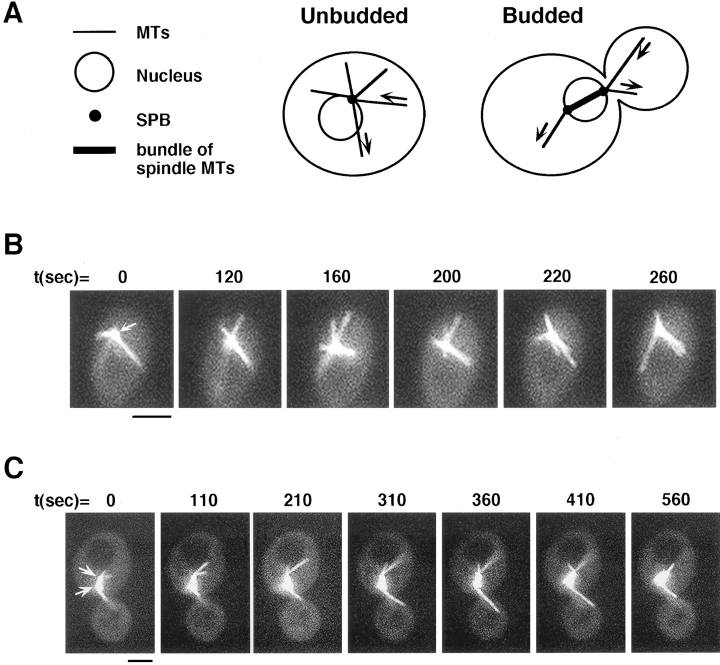
GFP-labeled microtubules were visualized in living yeast cells using fluorescence microscopy and a cooled CCD camera. Time-lapse sequences were used to analyze the behavior of microtubules throughout the cell cycle. Unbudded cells had an average of three, and as many as six, GFP-labeled cytoplasmic microtubules emanating from a single focus (Fig. 3 B). This focus represents either an unduplicated spindle pole body, or duplicated but unseparated spindle pole bodies. The spindle pole body also incorporated GFP–tubulin and could be identified as a bright fluorescent dot, similar to what is observed in fixed cells by anti-tubulin immunofluorescence.
Figure 3.
Yeast microtubules in living yeast cells as visualized by GFP fluorescence. (A) Schematic diagram showing relevant structures in unbudded and budded cells. Arrows on the microtubules denote the dynamic growing and shrinking behavior of microtubules. During mitosis (right), the spindle pole bodies duplicate, and a mitotic spindle is formed, consisting of a bundle of microtubules. Note: although the nuclei are drawn in the schematic, they cannot be seen in fluorescent images of yeast cells. (B) Cytoplasmic microtubules and the spindle pole body are visualized during time-lapse images of a single unbudded cell. The spindle pole body, at the center of the microtubule array, is marked by an arrow. The time, in seconds, is noted above each frame. A faint vacuole, which has lower background staining, can be seen below the spindle pole body. Up to six cytoplasmic microtubules grow and shrink, and come in contact with the cortex during the time sequence. (C) Cytoplasmic microtubules, spindle pole bodies, and a short spindle are visualized in a budded cell. The two spindle pole bodies at each end of the spindle are marked by arrows. Growing and shrinking of cytoplasmic microtubules are shown over the time course, during which the spindle twists slightly out of focus. Vacuoles in both the mother and bud can also be seen. Bars, 2 μm.
In budded cells, both the mitotic spindle microtubules and the cytoplasmic microtubules were labeled (Fig. 3 C). Budded cells had an average of three cytoplasmic microtubules per cell, with a maximum of three per single spindle pole body. As in fixed cells, the mitotic spindle appeared as a bundle of microtubules. The fluorescence intensity of cytoplasmic microtubules was compared to that of mitotic spindles; given the number of microtubules per spindle (Winey et al., 1995), the ratio of intensities was consistent with the observed cytoplasmic microtubules being single microtubules, rather than bundles of microtubules (data not shown). The fluorescence intensity of short spindles was greater than that of elongated spindles, consistent with the known reduction in microtubule numbers that accompanies elongation (Winey et al., 1995). In some cells with elongated spindles, there was a zone of increased fluorescence in the center of the spindle (Fig. 4 C), possibly representing overlapping microtubules. The kinetics of spindle elongation and the oscillatory behavior of the spindle were consistent with previous studies (Kahana et al., 1995; Yeh et al., 1995) (data not shown).
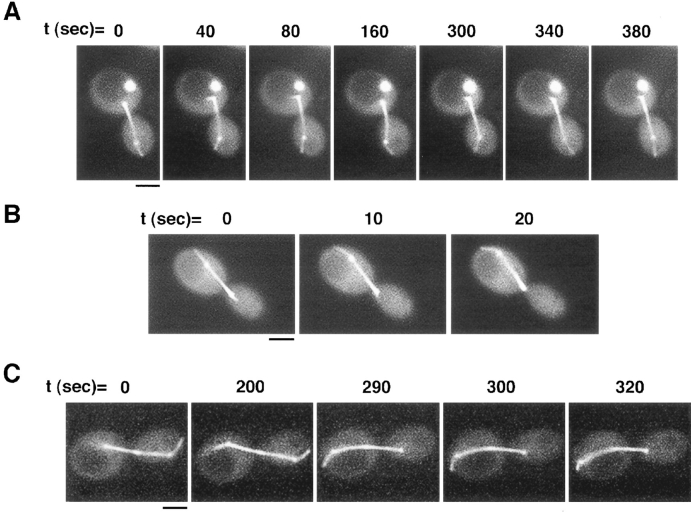
Figure 4.
Cytoplasmic microtubules interact dynamically with the cell cortex and result in associated movements of the spindle during mitosis. (A) The cytoplasmic microtubule sweeps back and forth along the bud cortex over the course of the time sequence. A short, faint cytoplasmic microtubule can also be seen in the mother cell, as well as the two spindle pole bodies and long spindle that spans the bud neck. In this cell, a bright spot of GFP staining, not corresponding to any microtubule structure, is seen in the mother cell. (B) Sweeping of a cytoplasmic microtubule along the cortex of the mother cell results in the spindle being pulled further into the mother cell. Note how the end of the spindle and the spindle pole body in the bud cell are pulled up into the neck by the third frame. (C) Shrinking of a microtubule at the cortex results in spindle movement toward the direction of the cortical attachment. A cytoplasmic microtubule appears in focus by t = 290 s and remains attached at the mother cortex while shrinking. This shrinking is coupled to a movement of the spindle toward the cortex. By the last frame the left spindle pole body is close to the cortex, and only a short cytoplasmic microtubule remains between them. Bars, 2 μm.
Cytoplasmic Microtubule Behavior
The sole function of cytoplasmic microtubules in vegetatively growing yeast cells is to localize and orient the mitotic spindle (Huffaker et al., 1988). To determine how cytoplasmic microtubules perform this function, we characterized their spatial and temporal regulation by time-lapse microscopy. As in animal cells, cytoplasmic microtubules in yeast exhibited dynamic instability, typically either growing or shrinking independently of one another. Pausing, in which a microtubule did not change in length over a given time interval, was also occasionally observed. Microtubules grew throughout the cytoplasm and did not seem to be restricted to particular areas of the cell. All microtubules had one end at the spindle pole body, and the spindle pole body itself was very mobile. Spindle pole body movement was particularly evident in unbudded cells that had a single focus of microtubules and no spindle.
The distal ends of cytoplasmic microtubules often grew until they encountered the cell cortex (Figs. 3 and 4), with at least one such interaction occurring in 85% of cells observed (Table II, Total cortex interactions). On average, microtubule ends spent 40–45% of the time in contact with the cortex during the time-lapse sequences examined (Table II). Interestingly, most movements of the spindle were associated with such microtubule–cortex interactions. Interactions of the cytoplasmic microtubules with the cortex fell into four classes, described in Table II as the percentage of cells in which a microtubule showed a particular behavior. The analysis is not shown per microtubule, as it was difficult to follow conclusively individual microtubules moving throughout the cell over a 10-min period. In class 1 (observed in 71% of cells), a microtubule grew until it came in contact with or “hit” the cell cortex, and then immediately shortened. In class 2 (observed in 33% of cells), a microtubule contacted the cortex and continued to grow, resulting in either increased distance between the cortex and spindle pole body, or in curvature of the microtubule end along the cortex. These types of encounters with the cell cortex occurred with almost equal frequency in unbudded and budded cells, as can be seen by the percentage of cells in which microtubules either hit or grew against the cortex in wild-type cells (Table II).
Table II.
Microtubule Interactions with the Cell Cortex
| Parameter | Wild type | Dynein mutant | ||
|---|---|---|---|---|
| Percentage of time at cortex (n = MTs) | 42.7 (n = 39) | 83.0 (n = 36) | ||
| Unbudded* | 44.5 (n = 24) | 92.8 (n = 10) | ||
| Budded | 41.1 (n = 15) | 79.3 (n = 26) | ||
| Maximum length of MT (μm) | 2.0 | 4.0 | ||
| Unbudded | 1.9 | 2.3 | ||
| Budded | 2.0 | 4.6 | ||
| Total cells analyzed | 52 | 57 | ||
| Unbudded | 40 | 24 | ||
| Budded | 12 | 33 | ||
| Total cortex interactions (%) | 84.6 | 77.2 | ||
| Unbudded | 85.0 | 79.2 | ||
| Budded | 83.3 | 75.8 | ||
| Hit cortex (%) | 71.2 | 68.4 | ||
| Unbudded | 72.5 | 70.8 | ||
| Budded | 66.7 | 66.7 | ||
| Grow at cortex (%) | 32.7 | 19.3 | ||
| Unbudded | 35.0 | 16.7 | ||
| Budded | 25.0 | 21.2 | ||
| Sweep on cortex (%) | 32.7 | 33.3 | ||
| Unbudded | 25.0 | 33.3 | ||
| Budded | 58.3 | 33.3 | ||
| Shrink at cortex (%) | 5.8 | − | ||
| Unbudded | 2.5 | − | ||
| Budded | 16.7 | − | ||
| Total nondynamic cells (%) | 1.9 | 14.0 | ||
| Unbudded | − | 12.5 | ||
| Budded | 8.3 | 15.2 |
Data from unbudded cells and cells containing a very small bud but no discernible spindle were combined for this and all subsequent measurements.
In contrast with the first two types of interactions at the cell cortex, the third and fourth classes occurred predominantly in budded cells. Most often, these interactions between a cytoplasmic microtubule and the cortex resulted in associated movements of the spindle. In class 3 (observed in 33% of cells), a microtubule grew to the cortex and then slid back and forth several times, seeming to “sweep” the cortex (Fig. 4 A). Although sweeping occurred more often in budded cells than in unbudded cells (58% vs 25%), the sweeping behavior in budded cells was equally prevalent at both the mother and bud cortical regions. In the majority of cases (71%), sweeping of the cytoplasmic microtubule along the cortex was associated with movement of the spindle toward the cortex (Fig. 4 B). The fourth class of interactions at the cell cortex (observed in 6% of cells) involved the shrinking of a microtubule while its end remained in association with the cortex (Fig. 4 C). Although this type of interaction did not occur often, in all cases it resulted in a striking coupling of spindle movement to the shortening of a cytoplasmic microtubule. The consequence of a microtubule shrinking while remaining at the cortex was that the spindle pole body and spindle moved toward the microtubule–cortex attachment.
Dynamics of Cytoplasmic Microtubules
Rates of microtubule growth and shrinkage in living yeast cells were determined by examining changes in microtubule length that occurred during time-lapse recordings (Fig. 5 A and Materials and Methods). Transitions between growth and shrinkage of microtubules were also examined, and catastrophe and rescue frequencies were determined (Materials and Methods). Cytoplasmic microtubules grew on average at a rate of 0.491 μm/min and shrank on average at a rate of 1.350 μm/min (Table III). Interestingly, a significant difference was seen in shrinking rates between unbudded cells and budded mitotic cells; shrinking occurred in unbudded cells at a rate of 1.784 μm/ min, as compared with a rate of 0.820 μm/min in budded cells (Table III). In contrast, no significant difference was seen in microtubule growth rates, indicating that more microtubule turnover occurs in unbudded cells. Small-budded cells with duplicated spindle pole bodies but no spindle exhibited microtubule dynamics similar to that of unbudded cells (data not shown).
Figure 5.
Linear regression analysis of microtubule polymerization and depolymerization rates. (A) Two examples of life-history graphs of wild-type cytoplasmic microtubules, plotting the microtubule length vs time. In the first example, a microtubule in a budded cell shrinks and then undergoes a rescue event. After the next growth phase, a catastrophe event occurs, and the microtubule continues shrinking until the end of the time sequence analyzed. In the second example, one catastrophe occurs between the growing and shrinking phases of a microtubule in a budded cell. The microtubule analyzed in the second graph is shown in Fig. 3 C and is present in the bud from t = 210 to t = 560 s. Rates, as determined by linear regression, are shown over the corresponding growth or shrinkage phases. (B) Examples of life-history graphs of cytoplasmic microtubules in dynein-mutant cells. Note the overall longer length of the microtubules as compared with wild-type microtubules. Both examples are microtubules from budded cells.
Table III.
Dynamic Parameters of Microtubules
| Wild type | Dynein mutant | |||||||
|---|---|---|---|---|---|---|---|---|
| Rate | Number of events | Rate* | Number of events | |||||
| Total | ||||||||
| Grow (μm/min) | 0.491 ± 0.036 | 21 | 0.331* ± 0.032 | 27 | ||||
| Shrink (μm/min) | 1.350 ± 0.159 | 20 | 0.386* ± 0.052 | 25 | ||||
| Catastrophe (s−1) | 0.006 | 15 | 0.003‡ | 15 | ||||
| Rescue (s−1) | 0.002 | 2 | 0.002 | 7 | ||||
| Unbudded | ||||||||
| Grow (μm/min) | 0.514 ± 0.047 | 14 | 0.420 ± 0.074 | 7 | ||||
| Shrink (μm/min) | 1.784 ± 0.190 | 11 | 0.620* ± 0.148 | 6 | ||||
| Catastrophe (s−1) | 0.007 | 10 | 0.006 | 4 | ||||
| Rescue (s−1) | − | − | 0.002 | 1 | ||||
| Budded | ||||||||
| Grow (μm/min) | 0.446 ± 0.050 | 7 | 0.300* ± 0.033 | 20 | ||||
| Shrink (μm/min) | 0.820 ± 0.118 | 9 | 0.313* ± 0.040 | 19 | ||||
| Catastrophe (s−1) | 0.005 | 5 | 0.003 | 11 | ||||
| Rescue (s−1) | 0.002 | 2 | 0.002 | 6 | ||||
A total of 27 and 29 microtubules were examined for wild-type and dynein-deficient cells, respectively. Rates of growing and shrinking are shown ± SEM.
Rates that are significantly different within a 95% confidence level ot those of wild type.
Frequencies that are significantly different within a 90% confidence level to those of wild type.
Transitions from growing to shrinking, termed catastrophes, occurred at a frequency of 0.006 s−1 and were more frequent than the opposite transitions, known as rescues, which were observed at a frequency of 0.002 s−1 (Table III). Rescue events were rare in budded cells and were not seen in time-lapse sequences examined from unbudded cells. The rarity of rescues might be due to the fact that very short microtubules were hard to follow in time-lapse images; it was difficult to determine whether a microtubule shrank down to a short microtubule, which then resumed growth, or whether a microtubule shrank completely away and a new microtubule subsequently grew in its place.
To determine if interactions between microtubule ends and the cell cortex altered their dynamic properties, we compared dynamic rates between microtubules at the cortex and those free in the cytoplasm. In both unbudded and budded cells, no significant difference was seen in growth or shrinkage rates when microtubules that were in contact with the cell cortex were compared with those that were free in the cytoplasm (data not shown). Although dynamic rates were similar, the majority of microtubules were at the cortical region immediately before a shrinking event (16/20 events). Consistent with this observation, 14/15 catastrophe events occurred only after microtubules grew out to the cortex. In contrast, both observed rescue events occurred when microtubules were short and within the interior of the cell.
Microtubule Properties in Dynein-deficient Yeast Cells
As described above, modulation of both microtubule dynamics and cortical interactions was observed in wild-type cells during different stages of the cell cycle. In conjunction with cytoplasmic microtubules, the microtubule-motor protein, dynein, is also required for normal spindle orientation. To determine how dynein and cytoplasmic microtubules act to position the spindle, we examined microtubule properties in a dynein mutant. A dynein disruption (dyn1:: HIS3) was constructed and the GFP–TUB3 plasmid was introduced into a diploid strain homozygous for the disruption allele (Materials and Methods). As with wild-type cells, fluorescent cytoplasmic microtubules and mitotic spindles were readily visible after induction of GFP–α-tubulin in the dynein disruption strain. Expression of neither α- nor β-tubulin was affected in dynein mutant cells, and GFP–TUB3p expression after induction was similar to that in wild-type cells (data not shown). The fluorescence intensity of both cytoplasmic and spindle microtubules was also similar to that in wild-type cells, suggesting that microtubule number and bundling was not altered in the dynein mutant (data not shown). Both the behavior and dynamics of microtubules were analyzed from time-lapse images as described for microtubules in wild-type cells. As previously described (Yeh et al., 1995), spindle elongation often occurred within the mother cell in the dynein mutants, and the characteristic oscillations across the neck region were not observed (data not shown).
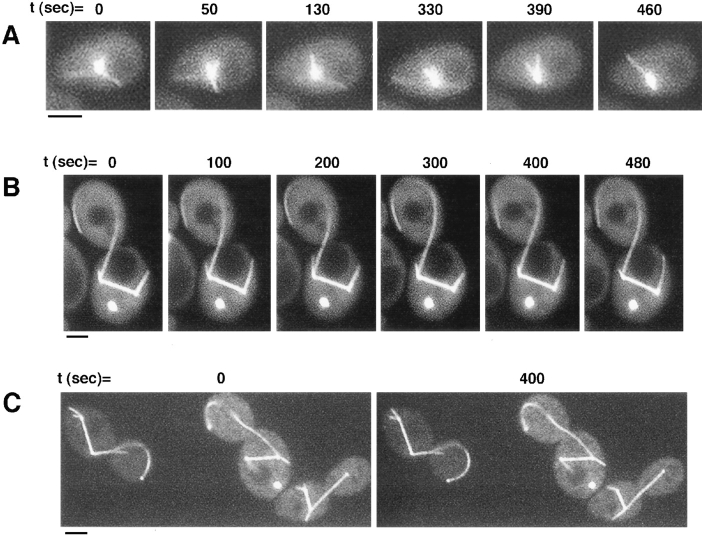
The number of cytoplasmic microtubules was similar in dynein-mutant and wild-type cells. One striking difference was that microtubules in dynein-deficient budded cells were often much longer than those in wild type (Fig. 6, B and C). Most often, these microtubules grew from the bud-proximal spindle pole body, through the neck into the bud, continuing to grow by curving around the bud cortex. On average, the maximum length of cytoplasmic microtubules in budded dynein mutant cells was at least twice as long as in wild-type cells (Table II). In contrast, microtubules in unbudded cells were not significantly longer than in wild-type cells and were never seen to curve around the cell cortex (Fig. 6 A). In both unbudded and budded dynein-deficient cells, microtubules spent twice as much time at the cell cortex than did microtubules in wild type (Table II). There was also a qualitative difference in the types of cortical interactions that occurred with microtubules in the dynein mutant. Approximately the same percentage of dynein-deficient and wild-type cells showed some type of interaction between microtubules and the cortex (Table II, Total cortex interactions), and the same percentage of cells had microtubules that grew to and hit the cell cortex (class 1). However, a lower percentage of dynein-mutant cells had microtubules that continued to grow at the cortex (class 2), mainly due to a decrease in unbudded cells exhibiting this behavior.
Figure 6.
Microtubules show aberrant interactions at the cell cortex in dynein-mutant cells. (A) Cytoplasmic microtubules and the spindle pole body are visualized in an unbudded cell. The length of microtubules in unbudded cells remains comparable to that in wild-type unbudded cells. (B) Budded cells contain longer cytoplasmic microtubules that curve around the cell cortex. In this cell, the spindle remains entirely within the mother cell, and cytoplasmic microtubules grow out from both spindle pole bodies. One long microtubule extends into the bud cell and does not show characteristic dynamic interactions with the cell cortex. (C) Three budded cells are shown containing long spindles, all still within the mother cells. While short cytoplasmic microtubules are present, each cell has at least one unusually long microtubule extending into the bud. A knob can be seen on all three long microtubules. The cytoplasmic microtubules have slower dynamics and show aberrant interactions at the cortex. Bars, 2 μm.
The two classes of cortical interactions that occurred predominantly in budded wild-type cells and resulted in spindle movement were significantly affected in dynein-mutant cells (Table II; class 3 and 4). In mutant cells, long curled microtubules often grew along the cortex, failing to exhibit the end-on cortical interaction characteristic of wild-type cells. While the overall percentage of cells showing sweeping behavior (class 3) was not altered, the percentage of budded cells showing sweeping was reduced by almost twofold compared to wild-type. Thus, in the dynein-mutant cells sweeping occurred equally in unbudded and budded cells and was not cell cycle regulated. A striking difference was seen in the sweeping that did occur in mutant budded cells; in contrast with wild-type cells, most sweeping events were not associated with spindle movement. Only 9% of sweeping showed associated spindle movement, compared with 71% in wild-type cells. Class 4 interactions were also diminished in dynein-mutant cells; no examples of shrinking while remaining associated with the cortex were observed.
Along with changes in microtubule behaviors, microtubule dynamic properties were also affected in dynein mutants. Both growing and shrinking rates were significantly slower in the mutant, with shrinking rates being more dramatically affected (Fig. 5 B and Table III). Both unbudded and budded cells showed significant differences in shrinkage rates, whereas growth rates were significantly slower only in budded cells. Catastrophe frequencies were also reduced by half in mutant cells. As in wild-type cells, no significant difference was seen in dynamic rates between microtubules that were at the cortex vs those in the interior of the cell.
One striking observation in dynein-mutant cells was that microtubules paused almost twice as frequently than did microtubules in wild-type cells. Completely nondynamic microtubules, which were presumably pausing throughout the time-lapse sequence, were sometimes present in cells also containing dynamic microtubules. However, there was also a significant percentage of cells where none of the microtubules were dynamic; these are referred to as “nondynamic cells” in Table II. In contrast with wild-type cells, in which only one nondynamic cell was seen, 14% of mutant cells were nondynamic (Table II). Finally, in budded cells a “knob”, visualized with GFP–TUB3, was sometimes associated with the end of microtubules, occurring more often with longer and less dynamic microtubules (Fig. 6 C).
Discussion
The ability to observe microtubules in living yeast cells has allowed us to make the following conclusions. (a) The images of the yeast microtubule cytoskeleton obtained from fixed cells are representative of what is seen in living cells, with the exception that more cytoplasmic microtubules are observed in living cells. (b) Cytoplasmic microtubules in yeast undergo dynamic instability as observed in animal cells, and microtubule dynamics are regulated during the cell cycle, with more dynamic microtubules being present during interphase. (c) Cytoplasmic microtubules in budded cells spend a significant amount of time in interaction with the cell cortex, and directed movement of the mitotic spindle occurs through these dynein-dependent microtubule–cortex interactions. (d) Deletion of the dynein microtubule motor results in changes in microtubule dynamics. We consider these points below.
The Yeast Microtubule Cytoskeleton In Vivo
The GFP–α-tubulin fusion protein used in these experiments is incorporated into microtubules without an apparent negative effect on microtubule function. Remarkably, fusion of the 27-kD GFP protein to either end of α-tubulin results in an assembly-competent fluorescent tubulin. Because GFP–TUB3 constitutes half of the α-tubulin in expressing cells, we assume that microtubules are able to function normally with half of the tubulin subunits bearing a GFP moiety.
Static images of microtubules visualized with GFP– TUB3 look much like those obtained by immunofluorescence with anti-tubulin antibody (Kilmartin and Adams, 1984). One significant difference is that more cytoplasmic microtubules are observed in vivo than in fixed cells. This is likely due to the relative instability of yeast cytoplasmic microtubules; when yeast cells are chilled or treated with microtubule-depolymerizing drugs, the cytoplasmic microtubules depolymerize well before the spindle microtubules (Stearns, T., unpublished results). This instability might make cytoplasmic microtubules more difficult to fix for immunofluorescence.
Regulated Microtubule Dynamics
Cytoplasmic microtubules in S. cerevisiae exhibit dynamic instability and have rates of polymerization and depolymerization that are 10–40-fold lower than typical animal cell rates (Table IV). Catastrophe and rescue frequencies are 5–15-fold below the lowest frequencies observed in mammalian cells (Table IV). Purified yeast tubulin has slower rates of growth and shrinkage compared with purified animal cell tubulin (Davis et al., 1993); therefore, the difference between yeast and animal rates in vivo might be intrinsic to yeast tubulin proteins. There is, however, abundant evidence in our observations for modulation of microtubule dynamics in yeast cells. First, the rate of depolymerization in vivo is faster than in vitro (Table IV), and pausing of microtubules is observed in vivo, but not in vitro (data not shown) (Davis et al., 1993). Second, microtubule dynamics in yeast change during the cell cycle, with faster rates of depolymerization occurring in unbudded cells. These differences in dynamic properties result in unbudded cells having higher microtubule turnover and more dynamic microtubules.
Table IV.
Comparison of Dynamic Rates in Different Systems
| Organism | Growth rate | Shrinkage rate | Catastrophe frequency | Rescue frequency | ||||
|---|---|---|---|---|---|---|---|---|
| μm/min | s−1 | |||||||
| S. cerevisiae (in vivo)* | 0.491 ± 0.163 | 1.350 ± 0.710 | 0.006 | 0.002 | ||||
| S. cerevisiae (in vivo)‡ | 0.510 ± 0.050 | 0.430 ± 0.043 | 0.001 | − | ||||
| Mammalian§ | 7.6 − 19.7 | 15.0 − 32.2 | 0.035 − 0.046 | 0.033 − 0.133 | ||||
Rates are shown ± SD.
This study.
Rates were converted from dimers/s to μm/min (Davis et al., 1993).
The range of rates from three different cell types is shown (Dhamodharan and Wadsworth, 1995).
Unbudded cells can be considered to represent the interphase state of the yeast cell cycle, as the initiation of DNA synthesis coincides with bud emergence and is followed soon after by the formation of a mitotic spindle (Pringle and Hartwell, 1981). Thus, interphase microtubules are more dynamic than mitotic microtubules in yeast. This is the opposite of the situation in animal cells in which interphase microtubules are less dynamic than mitotic microtubules (Salmon et al., 1984; Saxton et al., 1984; Belmont et al., 1990; Gliksman et al., 1992; Verde et al., 1992). In animal cells, microtubule dynamics are modulated during mitosis via catastrophe and rescue frequencies, whereas changes in yeast microtubule dynamics occur mainly in depolymerization rates. A proposed rationale for the increased microtubule dynamics in animal cell mitosis is that mitotic microtubules must find the kinetochores for attachment to the spindle, and thus need to sample a large cytoplasmic volume (Kirschner and Mitchison, 1986; Belmont et al., 1990). We propose that the same sort of considerations result in exactly the opposite requirements for a yeast cell. During interphase, the cytoplasmic microtubules in yeast must locate the bud site so that the nucleus, and subsequently the spindle, can be oriented. Thus, the cytoplasmic microtubules must search more cytoplasmic space early in the cell cycle.
It is important to note that we have only observed the dynamics of cytoplasmic microtubules in these experiments. The bundled nature of the microtubules in the yeast spindle prevents the resolution of single microtubules by fluorescence microscopy. It may be possible in the future to examine the dynamics of yeast spindle microtubules by photobleaching the GFP fluorophore in labeled microtubules (Cole et al., 1996).
Spindle Orientation and the Cell Cortex
We have shown that cytoplasmic microtubules frequently interact end-on with the cell cortex during mitosis. Although this interaction was inferred previously from images of fixed cells (Snyder et al., 1991), the complexity of the interaction was not appreciated. Microtubule interaction with the cortex is not with a single site, but rather with a broad region of the cortex in both bud and mother cell, and cortical interactions are also cell cycle regulated. In unbudded cells, the microtubules grow out from a single focus, not interacting preferentially with any part of the cell, whereas microtubules in budded cells grow out from either spindle pole body to the cortical regions of both mother and bud. The sweeping and shrinking behaviors of cytoplasmic microtubules at the cortex become more frequent in budded cells. These cortical interactions occur equally at the mother and bud cortex, and they are most often associated with a movement of the spindle toward the site of microtubule attachment. We believe that it is these interactions, and the spindle movements associated with them, that result in proper orientation of the spindle during mitosis.
As discussed above, both microtubule dynamics and cortical interactions are modulated during the cell cycle, and therefore a regulatory transition must occur between the unbudded and budded stage. Although it is possible that microtubules might also be regulated within the budding stage, we have not observed any significant differences in either dynamics or cortical interactions during the course of mitosis. We have observed that changes in microtubule behavior occur as early as bud emergence, with microtubules spending an appreciable time in association with the bud cortex. Although we have not specifically observed the earliest events of bud site recognition by cytoplasmic microtubules, we propose that they involve the same sorts of cortical interactions described here for spindle orientation. This is consistent with the report of a cytoplasmic microtubule being associated with the tip of the bud early in the cell cycle (Snyder et al., 1991).
It is useful to compare our observations of force-generating interactions of microtubules with the cortex with those from work on C. elegans. The rotation of the centrosomes in C. elegans early embryos is prevented by inhibitors of the actin and microtubule cytoskeletons, and by irradiation of the region between the centrosomes and the anterior cortex, which presumably disrupts microtubules in that region (Hyman and White, 1987; Hyman, 1989). These results suggested that microtubules from one of the centrosomes interact with a cortical attachment site on the anterior cortex, resulting in rotation of that centrosome toward that site. Interestingly, actin capping protein localizes to the anterior cortex before centrosome rotation (Waddle et al., 1994), providing evidence for a specialized structure there. As of yet no connection has been made between actin capping protein and the microtubule cytoskeleton, and it seems likely that other proteins are localized to the site and are responsible for the interaction with microtubules. Also, it has not been possible to observe microtubules directly in living C. elegans embryos, and thus the nature of the microtubule–cortex interaction is not known.
If, by analogy with C. elegans, the cortical regions at the ends of the yeast cell are specialized for microtubule interaction, then it could be predicted that cellular factors newly localized to the bud cortex might be responsible for the initial interaction with cytoplasmic microtubules in the bud. These factors might then relocalize to both mother and bud cortical regions, as microtubules interact with both the mother and bud cortex after bud growth. Many proteins are localized to the tip of the bud (Chant, 1994), but many of these are likely to be involved with the polarized growth of yeast cells in which all growth occurs in the bud, rather than with spindle orientation. However, one interesting candidate protein is Num1p, which is localized to the mother cell cortex in budded cells (Farkasovsky and Kuntzel, 1995). Mutations in NUM1 result in spindle orientation defects similar to those found in dynein mutants (Kormanec et al., 1991; Farkasovsky and Kuntzel, 1995). It is possible that Num1p is responsible for microtubule interactions in the mother cell, and that another protein carries out the function in the bud.
The Role of Dynein in Spindle Orientation
We found that both dynamic and behavioral properties of cytoplasmic microtubules are altered in the yeast dynein mutant. In the absence of dynein, rates of polymerization and depolymerization are significantly slower, and catastrophe frequencies are reduced by half. Microtubule dynamics are more severely affected in budded cells, resulting in longer microtubules on average. Interestingly, longer cytoplasmic microtubules have been noted in mutants deleted for either JNM1 or NUM1, both of which are likely to be involved in dynein/dynactin function (McMillan and Tatchell, 1994; Farkasovsky and Kuntzel, 1995). Whether dynein is directly affecting rates of microtubule growth and shrinkage is not clear. It is possible that a dynein- induced change in conformation of the microtubule end results in faster dynamic rates. Alternatively, the presence of dynein on a microtubule might exclude the binding of a microtubule-associated protein that would normally stabilize the microtubule. Regulation of microtubule dynamics by microtubule motors might be a general phenomenon; the kinesin-related protein Kar3p causes destabilization of the minus ends of microtubules (Endow et al., 1994), and the kinesin-related protein XKCM1 causes an increase in plus end catastrophe frequency (Walczak et al., 1996). Dynein may directly affect transition frequencies, in that interaction with dynein at the cortex may trigger catastrophe events resulting in the depolymerization of attached microtubules (see below), and in dynein mutant cells a decrease in catastrophe frequency is seen mainly in budded cells.
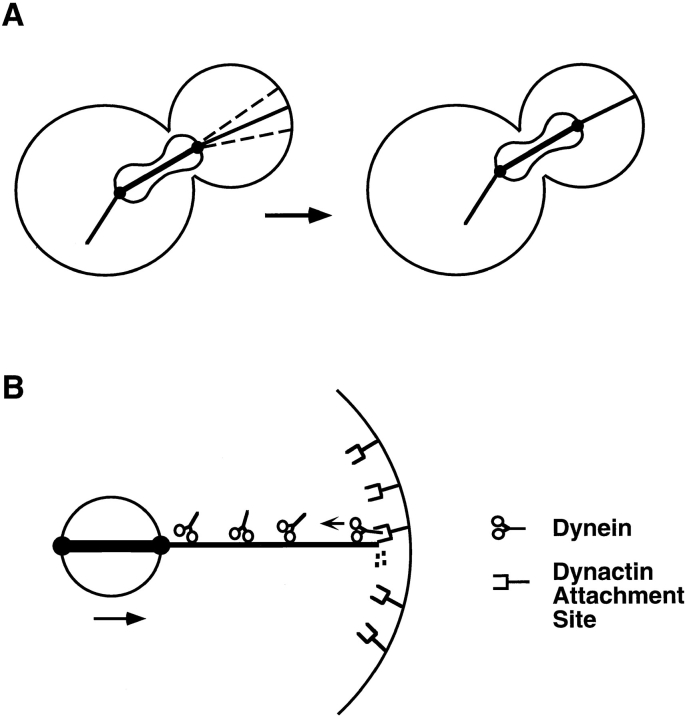
The most dramatic difference in dynein-deficient cells is that cytoplasmic microtubules fail to interact properly with the cortex during mitosis. In particular, the characteristic sweeping and shrinking interactions occur at reduced frequencies and fail to bring about a concerted movement of the spindle toward the cortex (Fig. 7 A). How does the activity of the dynein motor couple cytoplasmic microtubules to the cortex to effect spindle orientation? We propose a model in which microtubule ends are captured by a protein, or complex of proteins, that is localized to the polar regions of the cortex (Fig. 7 B). In this model, dynactin would be a part of the attachment complex, binding to microtubules through the p150Glued dynactin subunit (Waterman-Storer et al., 1995). After the initial microtubule capture, dynactin would interact with cytoplasmic dynein molecules on the microtubule, which are constantly moving toward the minus end. The force to move the spindle toward the cortex could be generated either by the dynein motor itself, coupled to the cortex through dynactin, or by depolymerization of the microtubule while maintaining cortical attachment. The latter method of force generation was originally proposed as a means of moving chromosomes (Koshland et al., 1988) and is particularly attractive in this case because we have observed several cases of spindle movement coordinated precisely with microtubule depolymerization at the cortex. Also, microtubule motors have been shown to be capable of coupling depolymerization energy to movement of objects (Lombillo et al., 1995).
Figure 7.
Model of spindle positioning via dynein-dependent interactions at the cell cortex. (A) Sweeping and shrinking interactions between cytoplasmic microtubules and the cortex lead to movement of the elongated spindle across the neck region. A single microtubule extends to the bud cortex, where it sweeps the cortex, as indicated by the dotted lines (left cell). Microtubule shrinking at the cortex results in an associated movement of the spindle toward the attachment site (right cell). (B) Model for how dynein interacts with cytoplasmic microtubules at the cortex. A dynactin attachment complex is localized at the cortex, binds to microtubules, and subsequently binds to a dynein molecule. Dynein remains bound to this complex and either pulls on the microtubule from the cortex or remains attached to the depolymerizing microtubule, resulting in movement of the spindle pole body and spindle. (Small arrow) Direction of dynein's minus end motor activity; (large arrow) direction of subsequent spindle movement. The small boxes at the end of the microtubule denote the depolymerization of the microtubule.
Dynein has also been proposed to have a role in spindle elongation in yeast because it is required for elongation in cells lacking the major mitotic kinesin motor proteins (Saunders et al., 1995). Given that dynein appears to act strictly through the cytoplasmic microtubules, we might have expected that some of the microtubule/cortex interactions that resulted in movement of the spindle toward the cortex would have resulted in spindle elongation rather than simple spindle movement, but this was never observed. Possibly, it is the absence of the kinesin proteins that allows dynein acting on the cytoplasmic microtubules to elongate the spindle through the application of an outward force on the spindle pole bodies.
Acknowledgments
We thank James Sabry and James Spudich for use of the CCD imaging facility, M. Andrew Hoyt for providing the dyn1::HIS3 deletion construct, and Keith Gull for providing TAT-1 antibody. We also thank James Sabry, Julie Brill, Becket Feierbach, Laura Marschall, and Steve Murphy for helpful discussions and comments on the manuscript, as well as Dave Ehrhardt for help with video imaging. J.L. Carminati was supported by a postdoctoral fellowship from the National Institute of General Medical Science, National Institutes of Health. This work was supported by a grant from the Searle Scholars Program to T. Stearns.
Footnotes
1. Abbreviations used in this paper: CCD, charge-coupled device; GFP, green fluorescent protein.
Please address all correspondence to Tim Stearns, Department of Biological Sciences, Stanford University, Stanford, CA 94305-5021. Tel.: (415) 725-6934. Fax: (415) 725-8309. e-mail: stearns@stanford.edu
References
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Burke D, Gasdaska P, Hartwell L. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. . Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 59–96.
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. . J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Pryer NK, Salmon ED. Real-time observations of microtubule dynamic instability in living cells. J Cell Biol. 1988;107:2223–2231. doi: 10.1083/jcb.107.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science (Wash DC) 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Trends Genet. 1994;10:328–333. doi: 10.1016/0168-9525(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiaeis required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science (Wash DC) 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- Davis A, Sage CR, Wilson L, Farrell KW. Purification and biochemical characterization of tubulin from the budding yeast Saccharomyces cerevisiae. . Biochemistry. 1993;32:8823–8835. doi: 10.1021/bi00085a013. [DOI] [PubMed] [Google Scholar]
- Dhamodharan R, Wadsworth P. Modulation of microtubule dynamic instability in vivo by brain microtubule associated proteins. J Cell Sci. 1995;108:1679–1689. doi: 10.1242/jcs.108.4.1679. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur Mol Biol Organ) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, Van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. S. cerevisiaegenes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliksman NR, Parsons SF, Salmon ED. Okadaic acid induces interphase to mitotic-like microtubule dynamic instability by inactivating rescue. J Cell Biol. 1992;119:1271–1276. doi: 10.1083/jcb.119.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Stearns T. Spindle positioning and cell polarity. Curr Biol. 1992;2:469–471. doi: 10.1016/0960-9822(92)90660-3. [DOI] [PubMed] [Google Scholar]
- Hyman AA, White TJ. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science (Wash DC) 1987;235:312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J, Wright B, Milstein C. Rat monoclonal anti-tubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Kormanec J, Schaaff GI, Zimmermann FK, Perecko D, Kuntzel H. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1protein. Mol Gen Genet. 1991;230:277–287. doi: 10.1007/BF00290678. [DOI] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature (Lond) 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature (Lond) 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiaeis required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner MW. Dynamic instability of microtubule growth. Nature (Lond) 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Neff NF, Thomas JH, Grisafi P, Botstein D. Isolation of the β-tubulin gene from yeast and demonstration of its essential function in vivo. . Cell. 1983;33:211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Koval M, Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. . J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoriagreen-fluorescent protein. Gene (Amst) 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., and L.H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 97–142.
- Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J Cell Biol. 1984;99:2165–2174. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammak PJ, Borisy GG. Direct observation of microtubule dynamics in living cells. Nature (Lond) 1988;332:724–726. doi: 10.1038/332724a0. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiaekinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ, Solomon F, Botstein D. Genetically essential and non-essential α-tubulin genes specify functionally interchangeable proteins. Mol Cell Biol. 1986;6:3722–3733. doi: 10.1128/mcb.6.11.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA, Steuer ER, Scheetz MP. Cytoplasmic dynein is a minus-end directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- Schulze E, Kirschner MW. Dynamic and stable populations of microtubules in cells. J Cell Biol. 1987;104:277–288. doi: 10.1083/jcb.104.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, Kirschner M. New features of microtubule behavior observed in vivo. . Nature (Lond) 1988;334:356–359. doi: 10.1038/334356a0. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. 186 pp.
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. . J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. The green revolution. Curr Biol. 1995;5:262–264. doi: 10.1016/s0960-9822(95)00056-x. [DOI] [PubMed] [Google Scholar]
- Stearns T, Ma H, Botstein D. Manipulating the yeast genome using plasmid vectors. Methods Enzymol. 1990;185:280–296. doi: 10.1016/0076-6879(90)85025-j. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley JH, Minke PF, Bruno KS, Plamann M. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurosporabut required for nuclear distribution. Mol Biol Cell. 1996;7:731–742. doi: 10.1091/mbc.7.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopusegg extracts. J Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegansblastomeres with oriented asymmetric divisions. Development (Camb) 1994;120:2317–2328. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: A Xenopuskinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc Natl Acad Sci USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiaemitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Roghi C, Morris NR. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1995;92:9890–9894. doi: 10.1073/pnas.92.21.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]