Abstract
Membrane trafficking intermediates involved in the transport of proteins between the TGN and the lysosome-like vacuole in the yeast Saccharomyces cerevisiae can be accumulated in various vps mutants. Loss of function of Vps45p, an Sec1p-like protein required for the fusion of Golgi-derived transport vesicles with the prevacuolar/endosomal compartment (PVC), results in an accumulation of post-Golgi transport vesicles. Similarly, loss of VPS27 function results in an accumulation of the PVC since this gene is required for traffic out of this compartment.
The vacuolar ATPase subunit Vph1p transits to the vacuole in the Golgi-derived transport vesicles, as defined by mutations in VPS45, and through the PVC, as defined by mutations in VPS27. In this study we demonstrate that, whereas VPS45 and VPS27 are required for the vacuolar delivery of several membrane proteins, the vacuolar membrane protein alkaline phosphatase (ALP) reaches its final destination without the function of these two genes. Using a series of ALP derivatives, we find that the information to specify the entry of ALP into this alternative pathway to the vacuole is contained within its cytosolic tail, in the 13 residues adjacent to the transmembrane domain, and loss of this sorting determinant results in a protein that follows the VPS-dependent pathway to the vacuole.
Using a combination of immunofluorescence localization and pulse/chase immunoprecipitation analysis, we demonstrate that, in addition to ALP, the vacuolar syntaxin Vam3p also follows this VPS45/27-independent pathway to the vacuole. In addition, the function of Vam3p is required for membrane traffic along the VPS-independent pathway.
The sorting and delivery of vacuolar hydrolases in the yeast Saccharomyces cerevisiae has become an important model system of membrane trafficking that parallels lysosomal biogenesis in animal cells (Kornfeld and Mellman, 1989; Rothman et al., 1989; Mellman, 1990; Conibear and Stevens, 1995). Current data support a model whereby a receptor (Vps10p) recognizes soluble vacuolar hydrolases such as carboxypeptidase (CPY)1 in the late Golgi apparatus. This receptor/ligand complex departs the Golgi apparatus and is delivered to a prevacuolar/endosomal compartment (PVC) where the receptor dissociates from CPY and recycles back to the Golgi apparatus. Soluble hydrolases as well as endocytosed proteins that are delivered to the PVC then move on to the vacuole (for review see Horazdovsky et al., 1995).
Many nonessential genes that play a role in the delivery of proteins to the yeast vacuole have been identified. These VPS genes (Vacuolar Protein Sorting) were identified by isolating yeast mutants that secrete CPY instead of efficiently sorting and delivering it to the vacuole (Bankaitis et al., 1986; Rothman et al., 1989; Stack et al., 1995). Mutations in many of these genes have provided tools to map out the vacuolar biogenesis pathway described above. Loss of function mutations in many of these genes result in the accumulation of the membrane transport intermediates that appear to play a central role in the transport of proteins to the vacuole.
One of these intermediates is a class of transport vesicles controlled by a group of VPS genes termed class D (Raymond et al., 1992), which includes PEP12 (a syntaxin homologue) (Becherer et al., 1996), VPS21/YPT51 (an Rab5 homologue) (Horazdovsky et al., 1994; Singer-Kruger et al., 1994), and VPS45 (an SEC1 homologue) (Cowles et al., 1994; Piper et al., 1994). Since cells with null alleles in any of the above class D genes accumulate many small vesicles within the cytoplasm, these genes are thought to act together to effect a vesicle fusion event along the vacuolar biogenesis pathway. Furthermore, cells carrying a temperature-sensitive allele of VPS45 rapidly accumulate these ∼60–70-nm transport vesicles concomitant with a loss of the ability to sort CPY upon shifting cells to the nonpermissive temperature (Piper et al., 1994). Epistasis studies together with the ability of these accumulated vesicles to trap anterograde traffic en route to the vacuole (Piper, R.C., and T.H. Stevens, unpublished observations) support the idea that these class D Vps proteins allow the fusion of Golgi-derived transport vesicles with the prevacuolar compartment (Conibear and Stevens, 1995; Horazdovsky et al., 1995).
Transit through the PVC appears to be controlled by a different set of VPS genes, the class E VPS genes (Raymond et al., 1992). Cells carrying a temperature-sensitive allele of one of these class E genes, VPS27, rapidly and reversibly accumulate cycling Golgi proteins (such as Vps10p), endocytosed proteins (such as Ste3p), and vacuolar proteins in a large, exaggerated form of the PVC (termed the “class E” compartment) (Piper et al., 1995). The accumulation of the prevacuolar compartments into the characteristic class E compartment results from the decreased ability of proteins to depart this compartment and either to move on to the vacuole or to return to the Golgi (Raymond et al., 1992; Piper et al., 1995). Together, the vps45 and vps27 mutations can be used to define intermediates that lie along the vacuolar biogenesis pathway from the late Golgi to the vacuole and that are responsible for the delivery of soluble hydrolases such as CPY as well as membrane proteins such as Vph1p (a subunit of the vacuolar H+-ATPase).
Yet another route from Golgi to the vacuole is taken by proteins such as Ste2p, Ste3p, and Ste6p (Davis et al., 1993; Berkower et al., 1994). All of these membrane proteins transit to the plasma membrane, whereupon they are endocytosed and delivered to the vacuole (Singer and Riezman, 1990; Davis et al., 1993; Berkower et al., 1994). Whereas the delivery of these endocytosed proteins is not blocked by vps45Δ mutations that accumulate Golgi-derived transport vesicles headed for the prevacuolar compartment, their delivery to the vacuole is blocked by vps27 mutations, which cause proteins that follow this route to be trapped in the class E PVC (Piper et al., 1995).
One curious feature of cells carrying either vps45Δ, vps27Δ, or both mutations is that there remains a discernible vacuole. Previous studies have shown that at least one membrane protein, alkaline phosphatase (ALP), can be immunolocalized to the vacuole in either vps45Δ or vps27Δ cells, whereas other vacuolar membrane proteins are found in the characteristic post-Golgi intermediates (Raymond et al., 1992; Piper et al., 1994). This led us to hypothesize that ALP travels from the late Golgi to the vacuole by an alternative route independent of the vacuolar biogenesis pathway defined to date. In this study we report that ALP is rapidly and faithfully delivered to the vacuole independent of Vps45p or Vps27p function. Unlike the other vacuolar membrane proteins we examined, ALP escaped entrapment in the Golgi-derived transport vesicles in vps45 mutant cells or the prevacuolar/endosomal class E compartment in vps27 mutant cells. This pathway appears to be completely intracellular as we did not detect ALP passing via the plasma membrane in wild-type cells or cells without Vps45p or Vps27p function. Furthermore, we find that the sorting determinant within the ALP protein that allows it to bypass the VPS45/VPS27-dependent pathway is contained within its NH2-terminal cytosolic tail.
Materials and Methods
Materials
Enzymes used in DNA manipulations were from New England Biolabs (Beverly, MA), Boehringer Mannheim Biochemicals (Indianapolis, IN), GIBCO BRL (Gaithersburg, MD), or United States Biochemical Corp. (Cleveland, OH). Glutathione agarose beads were from Sigma Chemical Co. (St. Louis, MO). FITC-conjugated streptavidin, Texas red–conjugated goat anti–rabbit antibodies, and biotin-conjugated goat anti–rabbit antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). The anti-CPY and anti-ALP polyclonal antibodies have been previously described (Raymond et al., 1990; Cooper and Stevens, 1996). The anti-Vma2p mAb, 13D11B2, has been reported (Raymond et al., 1992). The anti-ALP mAb 1D3 (Molecular Probes, Eugene, OR) and the anti–dipeptidyl aminopeptidase (DPAP) B antibody were used as previously described (Cooper and Stevens, 1996; Roberts et al., 1989). The rabbit polyclonal anti-Vam3p antibody was raised against the cytosolic NH2-terminal domain of Vam3p in the context of a glutathione-S-transferase fusion protein expressed from the plasmid pRCP160. Fixed Staphylococcus aureus cells (IgG Sorb) were obtained from The Enzyme Center (Malden, MA). 35S-Express label was from New England Nuclear (Boston, MA). Oxalyticase was from Enzogenetics (Corvallis, OR). All other chemicals were of high purity commercial grade.
Plasmid Construction
Plasmids used in this study are listed in Table I. The GAL1-VPH1 integrating construct, pLG39, was made by first subcloning the DraI fragment of VPH1, (encompassing the VPH1 open reading frame [ORF]) behind the GAL1 promoter in the BamHI site of pCJR52 to yield pLG1. pCJR52 was made by subcloning an EcoRI/BamHI fragment encoding the GAL1 promoter into the CEN-based URA3 plasmid, pSEYC68 (Herman and Emr, 1990). The EcoRI fragment from pLG1 (containing the GAL1 promoter and codons 1–707 of VPH1) was subcloned into pRS306 (Sikorski and Hieter, 1989) to create pLG38. pLG39 was made by religating blunt ends that were created after removing the internal PflM1/HindIII fragment (codons 369–841) from pLG38, thus creating the vph1-Δ369 allele.
Table I.
Strains Used in This Study
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| RPY10 | MATα leu 2-3,112 ura3-52 his4-519 ade6 gal2 | Piper et al., 1995 | ||
| SF838-9D | pep4-3 | Rothman et al., 1989 | ||
| HYY1 | νps27Δ::LEU2 pep4-3 | Piper et al., 1995 | ||
| AACY5 | νps27Δ::LEU2 | Piper et al., 1995 | ||
| RPY107 | νps27(νpl23-5) dap2Δ::LEU2 | This study | ||
| RPY11 | νps45Δ | Piper et al., 1994 | ||
| RPY12 | νps45Δ pep4-3 | Piper et al., 1994 | ||
| NBY68 | νps45Δ pho8-ΔX | This study | ||
| RPY90 | pep4-3 νps27-ts pho8-Δ329::LEU2::GAL1-PHO8 νph1-Δ369::URA3::GAL1-VPH1 | This study | ||
| RPY94 | pep4-3 νps45-ts pho8-Δ329::LEU2::GAL1-PHO8 νph1-Δ369::URA3::GAL1-VPH1 | This study | ||
| RPY111* | pep4-3 pho8-Δ329::LEU2::GAL1-PHO8 νph1-Δ369::URA3::GAL1-VPH1 | This study | ||
| RPY96 | VPS10::LEU2::νps10-10* | This study | ||
| RPY102 | νps45-ts VPS10::LEU2::νps10-10* | This study | ||
| RPY99 | νps45Δ VPS10::LEU2::νps10-10* | Piper et al., 1994 | ||
| RPY110 | pep4-3 VPS10::LEU2::νps10-10* | This study | ||
| RPY103 | pep4-3 vps27-ts VPS10::LEU2::νps10-10* | This study | ||
| CKRY2-8A | pep4-3 νps27Δ::LEU2 pho8Δ::LEU2 | Raymond et al., 1992 | ||
| SEY5016 | MATα sec1-ts leu2-3,112 ura3-52 | Emr et al., 1984 | ||
| RPY108 | MATα νps-45Δ::URA3 sec1-ts leu2-3,112 ura3-52 | This study | ||
| JHRY23-10D | MATα sec1-1 (ts) νps1-1 (vpl1-1) | Rothman and Stevens, 1986 | ||
| RPY95 | MATα vam3Δ | This study |
All strains except SEY5016, RPY108, and JHR23-10D are congenic to RPY10.
The GAL1-PHO8 construct, pRCP132, was derived from pRCP131. pRCP131 was made by inserting a BamHI/XbaI PCR fragment encoding codons 1–567 of PHO8 (bp 1–1,701) into pTS308 behind the GAL1 promoter. pTS306 (generously provided by Dr. T. Stearns, Stanford University, Stanford, CA) contains the GAL1 promoter in a CEN-based URA3 plasmid. pRCP132 was made, creating the pho8-Δ329 allele, by destroying the BglII site of pRCP131 at codon 329 by blunt-end ligation, thus creating a frame shift mutation and a stop codon at codon 338.
The GAL1-PEP4 construct, pRCP39, was made by subcloning a PCR fragment derived from PEP4 encompassing codons 1–405 (bp 1–1,413) into the BamHI/SalI sites of pCJR52, thus placing the PEP4 gene under the control of the GAL1 promoter.
pAIN1, encoding the ALP/DPAP B fusion construct, was made by generating a PCR fragment containing codons 1–32 of PHO8 (encoding ALP) and 29–380 of DAP2 using an oligonucleotide corresponding to codons 1–40 of the resulting fusion protein and containing a BamHI site and an oligonucleotide corresponding to bp 1,140–1,110 of the DAP2 ORF (encoding DPAP B) and containing an SspI site. This PCR product was digested with BamHI/SspI and ligated to the SspI/HindIII fragment (bp 599–2,678) from DAP2 in the BamHI/HindIII of pCJR52 in a three-part ligation. The pAIN1 construct places the ALP/DPAP B ORF (codons 1–32 of ALP and 29–842 of DPAP B) downstream of the GAL1 promoter. pAIN2, encoding the Δ11 ALP/DPAP B fusion construct, was made by generating a PCR fragment amplified from pAIN1 that placed a BamHI site followed by an initiating methionine just before codon 12 of the ALP/DPAP B ORF using the oligonucleotide: CCGGATCCATGACACGTCTTGTTCC in conjunction with the oligonucleotide: GGAC-GTATGCAATATTATTAG. This PCR product was digested with BamHI/ SspI and ligated to the SspI/HindIII fragment (bp 599–2,678) from DAP2 in the BamHI/HindIII of pCJR52 in a three-part ligation.
pNB3 was derived by deleting codons 2–21 of the PHO8 ORF from pSN92 according to the method of Kunkel (Kunkel et al., 1987). pNB4 was similarly derived by deleting codons 2–31 of the PHO8 ORF from pSN92. The vps45Δ::URA3 construct, pRCP133, was made by inserting a HindIII fragment containing the URA3 gene in place of the NheI/Bsu36I fragment of VPS45 in pTS34 to create the same deletion as is carried in the RPY11, RPY12, RPY99, and NBY68 yeast strains (Piper et al., 1994). pTS34 contains an SmaI/SphI fragment encoding VPS45 in pRS316 (Piper et al., 1994). The integrating vps10-Δ10* construct, pRCP95, was made by inserting the SalI/SacI fragment of pTS63 (Cooper and Stevens, 1996) into pRS305. The SalI/SacI fragment of pTS63 encodes a protein that contains a STOP codon at codon 1,425 of VPS10, resulting in a protein that lacks all but the first 10 amino acids of the COOH-terminal cytosolic tail of Vps10p.
The VAM3 gene (bp −204–1,156) was amplified from genomic DNA using the PCR and subcloned into the BamHI/EcoRI sites of pRS316 to generate pCC1. pRCP160 was made by subcloning a PCR fragment encoding amino acids 1–264 of Vam3p into the BamHI/EcoRI sites of pGEX-3X. pRCP161 constitutes the dominant-interfering VAM3 allele, VAM3-TMΔ, comprised of the cytosolic domain of Vam3p under the inducible control of the GAL1 promoter (VAM3-TMΔ). pRCP161 was made by subcloning a fragment encoding amino acids 1–264 of Vam3p into the BamHI/XbaI sites of pCJR52.
Strains
Strains were constructed using standard genetic techniques and grown in rich media (1% yeast extract, 1% peptone, 2% dextrose [YPD]) or standard minimal medium with appropriate supplements. All yeast strains were derived from the SF838–9D strain (Table II), except for SNY31 which was derived from the sister spore SF838–1D (Nothwehr et al., 1995). A congenic PEP4 strain, RPY10, has been described previously (Piper et al., 1995).
Table II.
Plasmids Used in This Study
| Plasmid | Description | Source | ||
|---|---|---|---|---|
| pLG1 | GAL1-VPH1 in CEN:URA3 plasmid, pCJR52 | This study | ||
| pLG39 | Integrating GAL1-VPH1 construct: GAL1 promoter fused to νph1-Δ369 allele in pRS306 | This study | ||
| pRCP131 | GAL1 promoter fused to PHO8 ORF in CEN-URA3 plasmid | This study | ||
| pRCP132 | GAL1-PHO8 integrating plasmid: GAL1 promoter fused to pho8-Δ329 allele in pRS305 | This study | ||
| pRCP39 | GAL1-PEP4 in CEN-URA3 plasmid | This study | ||
| pRCP95 | νps10-10* LEU2 integrating plasmid (VPS10 without cytosolic tail; STOP codon at amino acid 1,425) | This study | ||
| pLC4-8 | sec4-ts integrating plasmid | Nothwehr et al., 1995 | ||
| pSN92 | PHO8 in CEN-URA3 plasmid | Nothwehr et al., 1993 | ||
| pNB3 | PHO8 lacking codons 2–21 in CEN-URA3 plasmid | This study | ||
| pNB4 | PHO8 lacking codons 2–31 in CEN-URA3 plasmid | This study | ||
| pCC1 | VAM3 gene in pRS316 | This study | ||
| pRCP160 | GST-VAM3 fusion construct | This study | ||
| pRCP161 | VAM3-TMΔ allele. GAL1 promoter fused to fragment encoding cytosolic domain of Vam3p | |||
| pAIN1 | ALP/DPAP B fusion construct: codons 1–31 of PHO8 fused to codons 29–842 of DAP2 behind the GAL1 promoter in CEN-URA3 plasmid | This study | ||
| pAIN2 | Δ11 ALP/DPAP B fusion construct: codons 1, 12–31 of PHO8 fused to codons 29–842 of DAP2 behind the GAL1 promoter in CEN-URA3 plasmid | This study | ||
| pSN100 | (F85,F87→ A,A) A-ALP in CEN-URA3 plasmid | Nothwehr et al., 1993 | ||
| pCJR6 | GAL1-DAP2 construct in CEN:URA3 plasmid | Roberts et al., 1989 | ||
| pRCP56 | νps45-ts (νps45-13) integrating plasmid | Piper et al., 1994 | ||
| pKJH2 | νps27Δ::LEU2 disruption plasmid | Raymond et al., 1992 | ||
| pRCP41 | νps45Δ disruption plasmid (loop in/loop out) | Piper et al., 1994 | ||
| pRCP134 | νps45Δ::URA3 disruption plasmid | This study | ||
| pSN111 | pho8-Δ X disruption plasmid (loop in/loop out) | Nothwehr et al., 1995 | ||
| pCJR18 | dap2Δ::LEU2 disruption plasmid | Roberts et al., 1989 |
RPY90, RPY94, and RPY111 were derived from RPY15 (vps27-ts), RPY3 (vps45-ts), and SF838-9D, respectively (Piper et al., 1994, 1995), by creating the pho8-Δ329::LEU2::GAL1-PHO8 and vph1-Δ369::URA3:: GAL1-VPH1 loci. To create the pho8-Δ329::LEU2::GAL1-PHO8 mutation, pRCP132 (pho8-Δ369 allele fused to the GAL1 promoter) was linearized using NcoI and integrated at the PHO8 locus. This creates a tandem integration where one copy of PHO8 is under the control of the GAL1 promoter and the other copy of PHO8 is truncated at codon 329. To create the vph1-Δ369::URA3::GAL1-VPH1 mutation, the plasmid pLG39 (vph1-Δ369 allele fused to the GAL1 promoter) was linearized with BstEII and integrated at the VPH1 locus. This creates a tandem integration where one copy of VPH1 is under the control of the GAL1 promoter and the other copy of VPH1 is truncated at codon 369.
RPY96, RPY110, and RPY103 were derived from RPY10 (wild type), SF838-9D (pep4-3), and RPY3 (vps27-ts) (Piper et al., 1995), respectively, by creating the VPS10::LEU2::vps10-Δ10* locus. To create the VPS10:: LEU2::vps10-Δ10* locus, the plasmid pRCP95 (vps10-Δ10* allele) was linearized with XbaI and integrated at the VPS10 locus. This creates a tandem integration at the VPS10 locus containing the wild-type VPS10 gene and the vps10-Δ10* gene. RPY102 was derived from RPY10 by simultaneously transforming with linearized pRCP95 (vps10-Δ10* construct) and pRCP56 (vps45-ts construct), followed by selection on 5-fluoroorotic acid (5-FOA) to excise the endogenous VPS45 gene (Boeke et al., 1984; Piper et al., 1994). All culture steps for the RPY102 strain were at 22°C.
NBY68 was derived from RPY11 (vps45Δ) by transforming with pSN111 (pho8-ΔX construct) linearized with SalI. Ura+ transformants were plated onto media containing 5-FOA, and pho8-ΔX colonies were identified through Western analysis. RPY107 was derived from MY1885 (Piper et al., 1995) by transforming with the HindIII fragment of pCJR18 (dap2Δ::LEU2 construct). RPY109 was derived from RPY10 by transforming with pLC4-8 followed by selection on 5-FOA at 22°C.
RPY95 (vam3Δ) in which the VAM3 ORF is replaced with the LEU2 gene was made by transforming an ends-out disruption cassette amplified by PCR into the RPY10 strain. The oligos used were: ATTAACAAA-TTGGCCAACTAATATCCACTGCAGAAAGTTGAGATTATTTTG-TACTGAGAGTGCACCAT and GGGCTACCAGAAAGTCTGTG-CTCAATGCGCGTTTAAGGAGATTAGGTATTTCACACCGCATA (where the underlined portion represents the region that primes polymerization from the pRS314 plasmid).
Immunofluorescence
Indirect immunofluorescence microscopy for the localization of ALP, Vph1p, and DPAP B was performed as previously described (Roberts et al., 1991). Cells were grown in YPD at 30°C before fixation unless specified. For cells harboring plasmids, cells were grown to 1 OD/ml in minimal media, and then were diluted 1:1 with YPD and allowed to grow for 2 h before fixation. For experiments involving induction from the GAL1 promoter in temperature-sensitive mutants, cells were grown overnight in standard minimal media containing 2% raffinose at 22°C. Galactose was added upon shifting cells to 37°C; after 45 min at 37°C, cycloheximide was added to a final concentration of 100 μg/ml 10 min before fixation. Cells were prepared for immunofluorescence as described previously (Roberts et al., 1991). Basically, cells were fixed in 3% formaldehyde for 10 min, followed by incubation in 2% paraformaldehyde/50 mM KPO4, pH 7.0, for 18 h. Cells were spheroplasted and permeabilized with 1% SDS for 3 min. Cells were washed in 1.2 M sorbitol and allowed to adhere to poly-l-lysine–coated slides. Incubation of cells with the primary antibody was performed at 4°C overnight followed by 1-h incubations of secondary and tertiary antibodies at 22°C. For experiments requiring labeling of ALP and Vph1p in the same cells, the anti-ALP 1D3 monoclonal was visualized using biotinylated secondary antibody in combination with FITC-labeled streptavidin, and the rabbit anti-Vph1p was visualized using Texas red– labeled goat anti–rabbit antibody. To localize Vam3p in wild-type and vps27Δ cells, cells were transformed with the low copy VAM3 plasmid, pCC1; however, similar results were obtained with nontransformed cells. Images were captured on 35-mm film (TMAX-400; Eastman Kodak Co., Rochester, NY) with a ×100 Zeiss oil immersion lens on a Zeiss Axioplan fluorescence microscope (Zeiss, Oberkochen, Germany). Film negatives were digitized using a Polaroid SprintScan 35 (Polaroid Corp., Cambridge, MA). Images were adjusted with standard settings using Adobe Photoshop™ (Adobe Systems, Inc., Mountain View, CA).
Pulse/Chase Immunoprecipitations
Secretion of newly synthesized CPY was quantified by immunoprecipitations as previously described (Nothwehr et al., 1995; Piper et al., 1995; Cooper and Stevens, 1996). Briefly, yeast cultures were grown overnight in selective synthetic media without methionine to OD600 = 1. Cultures were adjusted to 50 mM KPO4, pH 5.7, containing 2 mg/ml BSA. 0.5 OD600 cells per time point to be analyzed were labeled for 10 min with 100 μCi 35S-Express label and then chased for specified times by the addition of excess unlabeled methionine and cysteine (final concentration of 100 μg/ml). The chase was terminated by chilling the cells to 4°C in the presence of 50 mM sodium azide. Cells were separated from the culture media by centrifugation and then were spheroplasted. CPY was immunoprecipitated from the resulting intracellular (lysed spheroplasts) and extracellular (media supernatant) fractions and analyzed by SDS-PAGE and fluorography.
For immunoprecipitation of ALP, Vps10p, and related proteins, cells were prepared and labeled as above but without the addition of BSA or KPO4 according to previously published procedures (Bryant and Stevens, 1997). After the chase period, cells were spheroplasted and lysed using 1% SDS/8 M urea. Lysates were then adjusted to 0.1% SDS, 0.1% Triton X-100, 0.8 M urea, and 20 mM Tris, pH 8.0, before the addition of 1 μl anti-Vps10p or anti-ALP antisera. After a 1.5-h incubation at 4°C, Staph A cells (IgG Sorb) were added for an additional 1.5 h.
Results
Differential Distribution of ALP and Other Vacuolar Proteins in vps45Δ and vps27Δ Cells
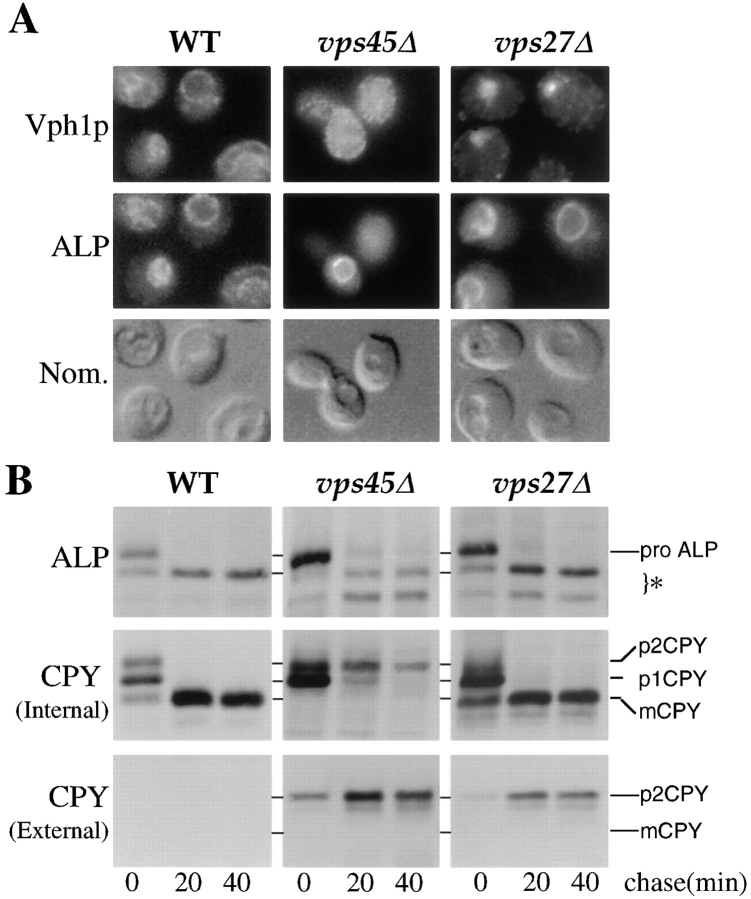
The recent characterization of many VPS genes has enabled the development of genetic tools that provide specific blocks at various stages along the vacuolar biogenesis pathway from the late Golgi to the vacuole. vps45 mutations appear to block the fusion of a class of Golgi-derived transport vesicles that are destined to fuse with a PVC (Cowles et al., 1994; Piper et al., 1994). This block prevents soluble hydrolases such as CPY and membrane proteins such as Vph1p (an integral membrane subunit of the vacuolar H+-ATPase; Manolson et al., 1992) from reaching the vacuole (Piper et al., 1994). Mutations in VPS27 cause the accumulation of an exaggerated form of a PVC (termed the class E compartment) that traps both Vph1p and CPY (Raymond et al., 1992; Piper et al., 1995). The effect of these blocks can be visualized through the localization of Vph1p in vps45 and vps27 null mutants (Fig. 1 A). Wild-type (SF838-9D), vps45Δ (RPY12), and vps27Δ (HYY1) cells were grown at 30°C, fixed, and decorated with anti-Vph1p and anti-ALP antibodies. In contrast with its localization in wild-type cells, Vph1p was not found in the vacuole in vps45Δ cells (defined by Nomarski optics as a large depression). Instead, Vph1p was localized to many small punctate structures consistent with its predicted distribution in the transport vesicles that accumulate in vps45Δ mutants (Cowles et al., 1994; Piper et al., 1995). Similarly, Vph1p was not localized to the vacuole in vps27Δ cells, but instead was found within large perivacuolar class E structures that accumulate when traffic out of the PVC is blocked (Raymond et al., 1992; Piper et al., 1995).
Figure 1.
Differential localization of ALP and other vacuolar proteins. (A) Double label immunofluorescence of Vph1p (upper panels) and ALP (middle panels) is shown for wild-type cells (SF838-9D), vps45Δ cells (RPY12), and vps27Δ cells (HYY1). The depressions visible in the Nomarski image (lower panels) identify the yeast vacuole. (B) Immunoprecipitation of newly synthesized ALP and the soluble vacuolar hydrolase CPY in wild-type cells (RPY10), vps45Δ cells (RPY11), and vps27Δ cells (AACY5). Newly synthesized proteins were labeled for 10 min at 30°C with the addition of 35S-Express. Excess unlabeled methio-nine and cysteine were then added for the indicated times. After centrifugation, internal (pellet) and external (supernatant) fractions were then prepared. CPY was immunoprecipitated from intracellular and extracellular fractions and ALP was immunoprecipitated from intracellular fractions. The PEP4-dependent cleavage products of ALP are indicated (*).
Blocks in the vacuolar biogenesis pathway were also evident in pulse/chase labeling experiments (Fig. 1 B). Newly synthesized proteins were labeled with [35S]methionine/ cysteine for 10 min, after which CPY was immunoprecipitated from intracellular and extracellular fractions after various chase times in the presence of unlabeled methio-nine. Most of the CPY in vps45Δ cells was secreted and recovered in the extracellular fraction. However, the fraction that remained intracellular failed to be converted from the Golgi-modified p2 form to the mature form, indicating that this intracellular CPY had not been exposed to proteolytic processing enzymes in the vacuole. These results agree with previous findings (Cowles et al., 1994; Piper et al., 1994) and, together with the localization of Vph1p, suggest that proteins that exit the Golgi in vps45Δ cells are trapped within transport vesicles. In vps27Δ cells, the block at the prevacuolar compartment was not evident by following the processing of newly synthesized CPY since the intracellular CPY was processed to the mature form in these cells. However, previous studies have shown that this is not indicative of CPY reaching the vacuole since the class E prevacuolar compartment is proteolytically active (Raymond et al., 1992; Piper et al., 1994; Bryant and Stevens, 1997) and CPY can be immunolocalized to the class E compartment (Raymond et al., 1992).
In contrast with the nonvacuolar localization of Vph1p in vps45Δ and vps27Δ cells, another membrane protein, ALP, was found exclusively in the vacuole (Fig. 1 A). These results are consistent with previous studies (Raymond et al., 1992; Piper et al., 1994). Not only was the differential localization of ALP and the vacuolar protein Vph1p observed by immunofluorescence localization studies, but ALP also appeared to be targeted differently when analyzed by pulse/chase analysis. ALP, the product of the PHO8 gene, is a type II membrane protein that undergoes a PEP4-dependent cleavage upon reaching the vacuole (Kaneko et al., 1987; Klionsky and Emr, 1989). This processing event serves as a convenient way to monitor the trafficking of ALP to vacuolar membranes (Klionsky and Emr, 1989). Fig. 1 B shows that, despite the block of intracellular CPY processing in vps45Δ cells, ALP was fully processed with nearly wild-type kinetics into its mature forms (Bryant and Stevens, 1997). In vps27Δ cells, ALP was also processed, although in this case the localization of ALP to the vacuole can only be inferred from the immunolocalization data since the class E prevacuolar compartment is proteolytically active.
Trafficking of ALP and Other Membrane Proteins to the Vacuole Is Independent of Vps45p-dependent Transport Vesicles and the Vps27p-dependent Prevacuolar Compartment
The data presented thus far indicate that, despite the profound blocks in the vacuolar biogenesis pathway that vps45 and vps27 mutations impose, ALP is still localized to the vacuole. One model is that ALP departs the yeast Golgi via a different route than that taken by proteins such as Vph1p and the CPY/CPY receptor (Vps10p) complex. Thus, membrane proteins such as Vph1p as well as vacuolar proteases would reach the vacuole via a VPS-dependent pathway, whereas ALP would reach the vacuole via an alternative bypass pathway. While the data in Fig. 1 are consistent with this interpretation, the immunofluorescence experiments examined only the steady state distribution of ALP and Vph1p in vps27 and vps45 mutant cells, which may not accurately reflect any kinetic effects on newly synthesized proteins. The pulse/chase analyses also may be difficult to interpret strictly since ALP is compared to a soluble vacuolar hydrolase, CPY, which may well have a different set of requirements for reaching the vacuole and becoming proteolytically processed since it is a soluble protein that is carried through part of the vacuolar biogenesis pathway by a recycling membrane protein receptor (Marcusson et al., 1994). Finally, this analysis was performed in cells carrying null mutations that not only result in the depletion of the processing enzymes resulting in a Pep− phenotype (Jones, 1990), but also may lead to indirect effects and to induce cryptic trafficking pathways as has been observed with chc1Δ cells and other mutations (Seeger and Payne, 1992b ; Nothwehr et al., 1995).
To address these concerns, we used cells carrying temperature-sensitive alleles of either VPS45 or VPS27 that rapidly lose their function in cells shifted from 22°C to 37°C. This strategy ensured that the vacuolar biogenesis pathway(s) suffered no long-term defects and that vacuolar morphology and function were normal. We then followed a wave of newly synthesized ALP and Vph1p using immunofluorescence localization in cells that had undergone rapid inactivation of Vps45p or Vps27p function (Fig. 2). For these experiments, the VPH1 and PHO8 (ALP) open reading frames were placed under the control of the inducible GAL1 promoter. This was accomplished by creating a tandem integration at both the VPH1 and PHO8 loci, which truncated the endogenous copy of the gene while inserting a new copy of the gene containing the GAL1 promoter. Within 40 min of adding galactose, the level of induced protein was sufficient to follow newly synthesized protein by immunofluorescence localization as shown in Fig. 2 (bottom) where ALP and Vph1p labeling were only discernible in cells induced by galactose.
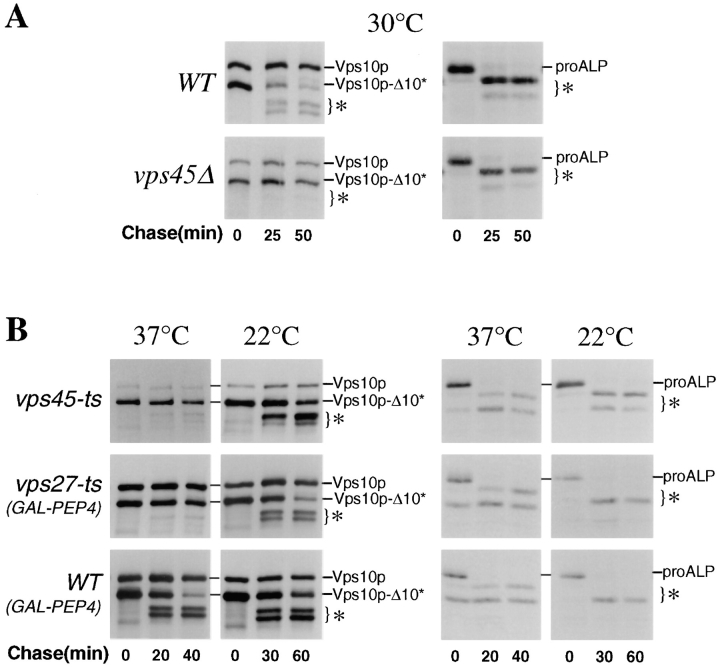
Figure 2.
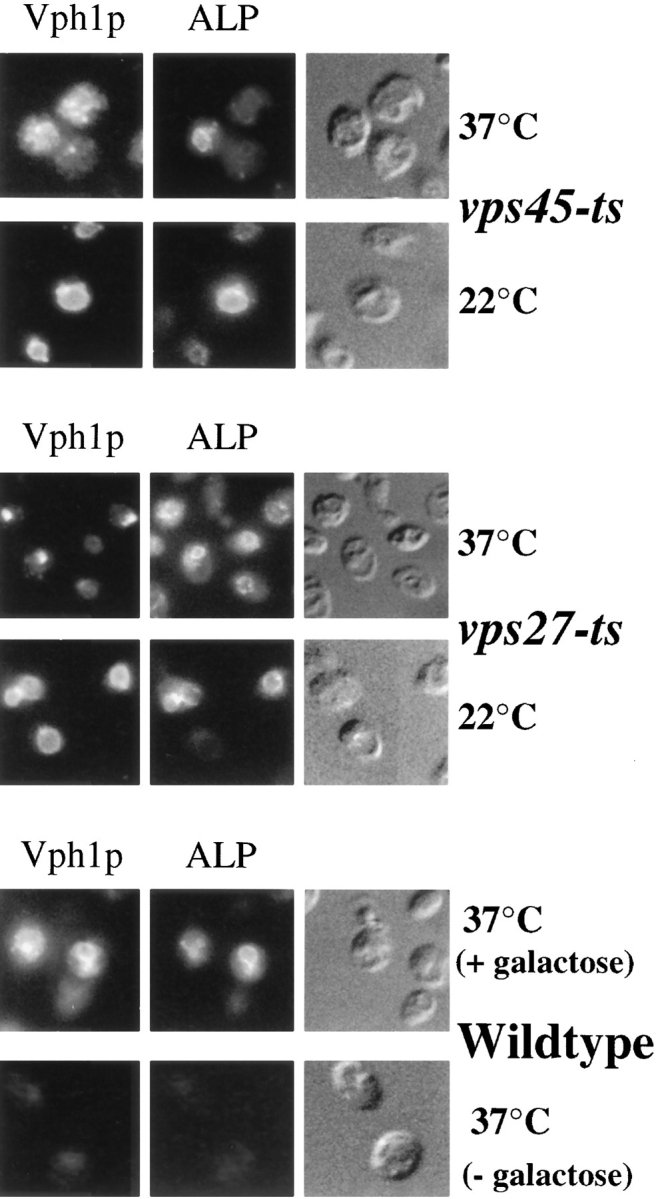
Differential targeting of ALP and Vph1p in temperature-sensitive vps mutants. The targeting of newly synthesized Vph1p and ALP was monitored in vps45-ts cells (RPY94; top panels), vps27-ts cells (RPY90; middle panels), and wild-type cells (RPY111; bottom panels). The production of both ALP and Vph1p in RPY94, RPY90, and RPY111 cells was under the control of the galactose-inducible GAL1 promoter. Cells were grown overnight at 22°C in the absence of galactose. Galactose was then added, and cells were incubated for 45 min at 22°C or immediately shifted to 37°C as indicated. Cycloheximide was then added (100 μg/ml) for an additional 10 min before fixation and double labeled for Vph1p and ALP using indirect immunofluorescence. Vph1p was labeled with rabbit anti-Vph1p antibodies and Texas red–conjugated secondary antibody (left panels); ALP was labeled with anti-ALP 1D3 mAb with biotinylated secondary and FITC-conjugated streptavidin. Shown as a control are wild-type cells (RPY111) incubated at 37°C in the presence or absence of galactose (lower panels).
A 45-min wave of newly synthesized protein was induced from the GAL1 promoter in vps27-ts or vps45-ts cells that were maintained either at 22°C or raised to 37°C concomitant with galactose addition. Fig. 2 shows that at 37°C Vph1p did not reach the vacuole in vps45-ts cells (RPY94), but instead was found in many punctate structures similar to its distribution in vps45Δ cells and consistent with its distribution in transport vesicles. In these same cells, however, newly synthesized ALP was only found in the vacuole. In three experiments, examination of >300 double-labeled cells that showed the vesicular distribution of Vph1p showed that ALP was vacuolar, and no ALP could be detected in vesicular-like structures.
Similar experiments were performed on vps27-ts cells (RPY90). Previous studies have shown that the characteristic morphology of the class E prevacuolar compartment forms well within 15–20 min after raising vps27-ts cells to the nonpermissive temperature (Piper et al., 1995; Bryant and Stevens, 1997). At 37°C, newly synthesized Vph1p was found in class E prevacuolar compartments and did not reach the vacuole. While Vph1p had been trapped in the rapidly formed class E compartment, newly synthesized ALP was found in the vacuole. In four experiments, >400 double-labeled cells that showed Vph1p within the class E prevacuolar compartment were examined; all showed ALP within the vacuole as outlined by Nomarski optics. No ALP was observed colocalized with Vph1p in class E prevacuolar structures.
To show that the vacuolar biogenesis pathway was fully functional before the temperature shift, the synthesis of ALP and Vph1p was induced at 22°C in both vps45-ts and vps27-ts cells. Fig. 2 shows that both ALP and Vph1p reach the vacuole as in wild-type cells (data not shown). To show that the differential temperature-sensitive effects on the sorting of ALP and Vph1p were solely due to the vps27-ts or vps45-ts alleles, wild-type cells carrying the GAL1-VPH1 and GAL1-ALP alleles (RPY111) were examined and found to deliver efficiently both proteins to the vacuole at 37°C (Fig. 2).
PEP4-dependent Processing of ALP But Not Another Vacuolar Membrane Protein, Vps10p-Δ10*, Occurs Despite Blocks in the VPS-dependent Pathway
We also used a pulse/chase analysis to assess the transport route of newly synthesized membrane proteins to the vacuole. In order to compare ALP with another membrane protein that undergoes a PEP4-dependent processing, we chose to follow the fate of the vacuolar protein Vps10p-Δ10*. The Vps10-Δ10* protein is a mutant form of the CPY receptor, Vps10p, which lacks all but 10 amino acids of its COOH-terminal cytosolic tail (Cooper and Stevens, 1996). Several features make this a useful membrane protein marker to monitor the function of the vacuolar biogenesis pathway: firstly, all of the known information in the cytosolic tail that localizes Vps10p to the Golgi has been eliminated, resulting in a protein that is localized to the vacuole with rapid kinetics (Cooper and Stevens, 1996); secondly, it undergoes a PEP4-dependent cleavage upon reaching the vacuole (Cereghino et al., 1995; Cooper and Stevens, 1996); finally, the Vps10-Δ10* protein localizes to the class E prevacuolar compartment in vps27Δ cells, indicating that it reaches the vacuole via the VPS-dependent pathway (Cooper, A.A., and T.H. Stevens, unpublished observations).
In wild-type cells (RPY96) Vps10p-Δ10* is rapidly processed with a half-time of ∼20 min, consistent with previous measurements (Cooper and Stevens, 1996; Bryant and Stevens, 1997). The full-length Vps10 protein was also followed in these cells to serve as a convenient internal control. In wild-type cells Vps10p is quite stable, owing to tyrosine-based signals within its cytosolic domain that confer its retrieval to the Golgi apparatus and prevent its delivery to the vacuole (Cereghino et al., 1995; Cooper and Stevens, 1996) (Fig. 3). In vps45Δ cells, Vps10p-Δ10* failed to undergo a PEP4-dependent cleavage (Fig. 3 A), consistent with its being trapped in transport vesicles and thus prevented from reaching the vacuole. These data demonstrate that Vps10p-Δ10* can be used to monitor blocks along the VPS-dependent pathway.
Figure 3.
Trafficking of ALP and Vps10p-Δ10* in temperature-sensitive vps mutants. (A) The rate of PEP4-dependent processing of Vps10p-10* (left panels) and ALP (right panels) was measured in wild-type cells (RPY96) and vps45Δ cells (RPY99) by pulse/chase analysis. Cells were labeled with 35S-Express for 10 min at 30°C and chased for the indicated times. Cell lysates were divided and subjected to immunoprecipitation with anti-ALP and anti-Vps10p antibodies. Both the full-length Vps10p and the truncated Vps10p-Δ10* were present. The PEP4-dependent cleavage products of Vps10p-Δ10* and ALP are indicated (*). (B) The rate of PEP4-dependent cleavage of Vps10p-Δ10* and ALP was measured in vps45-ts cells (RPY102). Cells were grown overnight at 22°C and then either maintained at 22°C or shifted to 37°C 10 min before labeling with 35S-Express for 10 min. The chase times used are indicated. ALP and Vps10p-Δ10* were immunoprecipitated as described above. The rate of PEP4-dependent cleavage of Vps10p-Δ10* and ALP was measured in vps27-ts cells (RPY103) and wild-type cells (RPY110) each carrying the GAL1-PEP4 plasmid, pRCP39. Cells were grown in galactose-containing media for 24 h at 22°C and then shifted to glucose-containing media for 24 h before the pulse/chase immunoprecipitation scheme used for vps45-ts cells (RPY102) above. Cells were either maintained at 22°C or shifted to 37°C 10 min before labeling.
Using the cleavable membrane protein Vps10p-Δ10* as a reporter, we examined the trafficking of ALP to the vacuole in cells that had sustained recent and abrupt blocks in the VPS-dependent pathway. In vps45-ts cells at 22°C, the delivery of ALP and Vps10p-Δ10* to the vacuole showed the same kinetics as that observed in wild-type cells (Fig. 3 B). However, in vps45-ts cells shifted to 37°C for 10 min before labeling, Vps10p-Δ10* did not undergo PEP4-dependent cleavage, indicating that it was blocked in a compartment preceding the vacuole. In contrast, ALP in these cells was rapidly processed, indicating that despite the profound block in membrane traffic imposed by the vps45-ts allele, ALP was faithfully delivered to the vacuole.
To perform a similar analysis in vps27-ts cells, measures were taken to prevent the newly formed class E prevacuolar compartment from becoming proteolytically active as is typically observed (Piper et al., 1995). This was to allow proteins caught in the class E compartment to be distinguished from those delivered to the vacuole by following their PEP4-dependent cleavage. To accomplish this, wild-type and vps27-ts cells carried a plasmid in which the PEP4 open reading frame was placed under the control of the inducible GAL1 promoter (pRCP39). Cells grown in media containing galactose would be Pep+; however, when cells are shifted to glucose to shut off production of Pep4p, active vacuolar proteases would be flushed from PVCs, thus allowing the newly formed class E prevacuolar compartment to remain incapable of PEP4-dependent processing. Under this strategy, the vacuole itself would remain proteolytically active as a result of the phenomenon of phenotypic lag, in which the autocatalytic activation cycle of protease B results in this and other proteases remaining active for many cell divisions after elimination of Pep4p (Zubenko et al., 1982; Jones, 1991). Western analysis of wild-type cells transformed with pRCP39 grown on glucose demonstrated no PEP4-dependent processing of CPY or ALP. However, in cells grown in galactose, or in cells shifted from galactose to glucose for 24 h, all of the ALP and CPY was found in the processed form (data not shown), indicating that the GAL1-PEP4 plasmid could be used to exert tight control over PEP4 expression.
To monitor the delivery of ALP and Vps10p-Δ10* to the vacuole in cells lacking Vps27p function, vps27-ts GAL1-PEP4 cells were grown at 22°C in media containing 2% raffinose and 2% galactose for 24 h, after which cells were diluted in media containing 2% glucose and grown at 22°C for an additional 24 h. Cells were then shifted to 22°C or 37°C for 10 min and subjected to pulse/chase analysis to follow the fate of both newly synthesized ALP and newly synthesized Vps10p-Δ10* as described above. Fig. 3 B shows that at 22°C both ALP and Vps10p-Δ10* were cleaved with normal kinetics, indicating that both had reached the vacuole. These data show that, as expected, the vacuole in these cells is fully capable of processing both ALP and Vps10p-Δ10* despite the fact that there had been no Pep4p production for several generations. At 37°C, the VPS-dependent pathway was blocked as demonstrated by the severe inhibition of Vps10p-Δ10* processing. These data indicate that Vps10p-Δ10* was trapped within a proteolytically inactive form of the class E prevacuolar compartment unable to reach the vacuole. In contrast, ALP within the same cells was processed with the same kinetics as that observed in wild-type cells. To further demonstrate the efficacy of the GAL1-PEP4 technique, wild-type cells carrying the GAL1-PEP4 plasmid were also analyzed. The processing of ALP and Vps10p-Δ10* was rapid at both 37°C and 22°C (Fig. 3); this rate was indistinguishable from wild-type cells carrying the wild-type PEP4 gene (data not shown).
ALP Is Transported to the Vacuole by an Intracellular Route and Does Not Transit through the Plasma Membrane
These results strongly support a model in which ALP is delivered to the vacuole from the Golgi by a route that bypasses VPS45-dependent Golgi-derived transport vesicles and the VPS27-controlled PVC. One other well-described route to the vacuole is for proteins to transit through the plasma membrane before being endocytosed to the vacuole. This route via the cell surface is taken by the Ste3p, Ste2p, and Ste6p membrane proteins, and their delivery to the vacuole requires function of the late secretory pathway (i.e., Sec4p) and the endocytic pathway (i.e., End4p) (Berkower et al., 1994; Nothwehr et al., 1995). ALP does not follow this route in wild-type cells since the delivery of ALP to the vacuole is independent of Sec4p function and End4p function, which are required for secretory vesicle fusion and endocytosis from the plasma membrane, respectively (Novick et al., 1981; Raths et al., 1993). It is also unlikely that such a route is induced by mutations in VPS27 since if ALP was transported to the vacuole via endocytosis it would likely become trapped in the Vps27p-controlled class E PVC (Piper et al., 1995). Thus, ALP likely takes an alternative intracellular route to the vacuole, perhaps entering a new class of Golgi-derived vesicles.
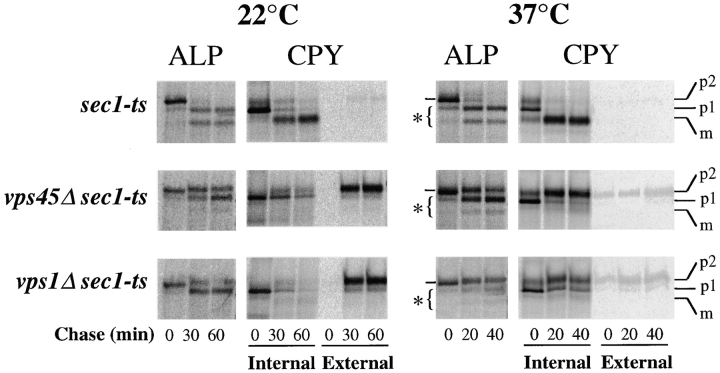
The above studies do not exclude the formal possibility that loss of Vps45p function induces a novel pathway for ALP to the vacuole via the cell surface. This activation of a cell surface cryptic pathway has been observed with vps1 mutant cells, where Golgi proteins and vacuolar proteins including ALP are delivered to the vacuole only after being diverted to the cell surface (Nothwehr et al., 1995). This effect is understandable since Vps1p is predicted to act in vesicle formation in the late Golgi apparatus and would be required to segregate and sort proteins away from the secretory system (Wilsbach and Payne, 1993; Conibear and Stevens, 1995). In contrast, the proposed role of Vps45p is to effect vesicle fusion based on its mutant phenotype and the fact that it is a Sec1p-like homologue (Aalto et al., 1992). Thus, it is unlikely that loss of Vps45p function would modify traffic out of the Golgi, especially given the data in Figs. 2 and 3 B, where a rapid onset of temperature-sensitive alleles of VPS45 was used. To eliminate this possibility, we confirmed that the trafficking of ALP to the vacuole in vps45Δ cells was also independent of the late secretory pathway. Fig. 4 shows that ALP was processed with normal kinetics in sec1-ts cells at 37°C in the presence or absence of VPS45. Immunoprecipitation of CPY from intracellular and extracellular fractions of the same cells showed that secretion was blocked by the sec1-ts allele at 37°C. In contrast with the ability of ALP to route intracellularly to the vacuole in the absence of Vps45p function, we found that cells without Vps1p function cannot deliver ALP to the vacuole without Sec1p function as observed in previous studies (Nothwehr et al., 1995). Together these data support the view that ALP is not rerouted to the cell surface by vps45Δ mutations. Rather, ALP travels from the Golgi apparatus to the vacuole via an alternative intracellular route that bypasses both the Vps45p-controlled transport vesicle step and the Vps27p-controlled PVC.
Figure 4.
ALP does not travel to the cell surface in vps45Δ cells. The rate of PEP4-dependent cleavage of ALP was measured in wild-type cells (RPY96), sec1-ts cells (RPY109), sec1-ts vps45Δ cells (YM), and sec1-ts vps1Δ cells (SNY31). Cells were grown overnight at 22°C and either maintained at 22°C or shifted to 37°C for 15 min before labeling for 10 min with 35S- Express. Label was chased for the indicated times. After centrifugation, internal (pellet) and external (supernatant) fractions were prepared, divided, and subjected to immunoprecipitation with anti-CPY and anti-ALP antibodies.
ALP Transport to the Vacuole Is Dependent on the Vacuolar Syntaxin Vam3p
The above studies show that the VPS-dependent pathway defined by the functions of VPS45 and VPS27 is responsible for delivery of a subset of membrane proteins excluding ALP to the vacuole. One model is that ALP is sorted into a separate class of vesicles at the TGN that later fuse with the vacuole and bypass the prevacuolar compartment. While the specific blocks imposed by the vps45-ts and vps27-ts alleles demonstrate that ALP transport bypasses the prevacuolar compartment, we sought to characterize further the alternative pathway by identifying machinery that controls the delivery of ALP to the vacuole.
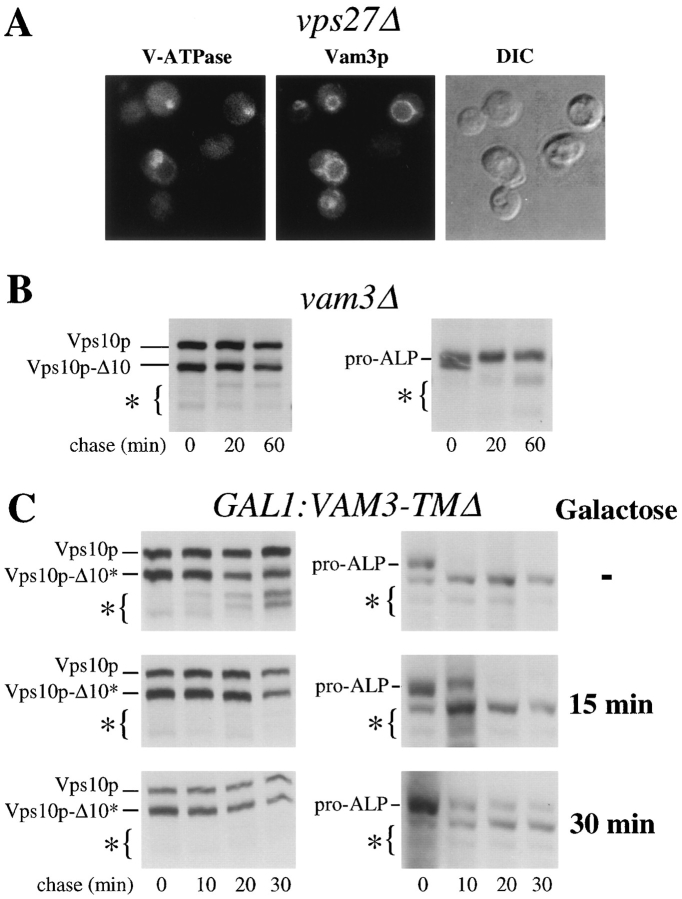
One such candidate was the syntaxin family member, Vam3p. Vam3p has been localized to the yeast vacuole and plays a role in the homotypic fusion of vacuoles (Nichols et al., 1997), and vam3 mutants were originally isolated in a screen for yeast with abnormal vacuolar morphology (Wada et al., 1992). As a proposed vacuolar syntaxin, we reasoned that Vam3p might also catalyze other fusion events with the vacuole, allowing proteins from the VPS-dependent pathway and/or the alternative pathway to be delivered to the vacuole. We also reasoned that any proteins that catalyzed the fusion of ALP-containing vesicles with the vacuole would themselves have to be delivered to the vacuole in a manner independent of the VPS-dependent pathway. Fig. 5 shows that Vam3p fits these criteria. Consistent with the results of Nichols et al. (1997), we found Vam3p was localized to the vacuole in wild-type cells (data not shown), and, interestingly, we also found that Vam3p was localized to the vacuole in vps27Δ cells, suggesting that Vam3p itself follows the alternative pathway to the vacuole.
Figure 5.
The vacuolar syntaxin Vam3p is required for the alternative bypass pathway. (A) Localization of Vam3p in vps27Δ cells. Vam3p was labeled with rabbit anti-Vam3p antibodies, biotinylated secondary and FITC-conjugated streptavidin (middle panels). As a marker of the class E prevacuolar compartment, the 60-kD subunit of the vacuolar ATPase, Vma2p, was localized with the mAb 13D11B2 and Texas red–conjugated secondary antibody (left panels). (B) The rate of PEP4-dependent processing of Vps10p-10* (left panels) and ALP (right panels) was measured in vam3Δ cells (RPY95). Cells were labeled with 35S-Express for 10 min at 30°C and chased for the indicated times. Cell lysates were divided and subjected to immunoprecipitation with anti-ALP and anti-Vps10p antibodies. (C) The rate of PEP4-dependent processing of Vps10p-10* (left panels) and ALP (right panels) was measured in wild-type cells (RPY96) carrying the dominant-interfering allele VAM3-TMΔ (pRCP161) under the inducible control of the GAL1 promoter. pRCP161 expresses the cytosolic domain of Vam3p in response to the addition of galactose. Cells were grown overnight at 30°C in raffinose. Galactose was then added for the indicated times (no addition, 15 min, or 30 min) before labeling cells with 35S-Express for 10 min at 30°C. Label was chased for the indicated times and cell lysates were divided and subjected to immunoprecipitation with anti-ALP and anti-Vps10p antibodies.
Null mutants of VAM3 display fragmented vacuoles typical of class B vps mutants (Wada et al., 1992). Pulse/chase analysis shows that in vam3Δ mutants both ALP and the VPS-dependent pathway marker protein, Vps10p-Δ10*, were not processed (Fig. 5). These data indicated that Vam3p function is required for delivery of proteins from both the VPS-dependent pathway and the alternative pathway. Such a dual function for Vam3p would be consistent with the recent finding that Vam3p is also required for vacuolar homotypic fusion (Nichols et al., 1997). The block in both ALP processing and the processing of Vps10p-Δ10* was also evident in other class B vps mutants including the ypt7 (YPT7 encodes a rab7 homologue), vam7 (VAM7 encodes a protein bearing homology with SNAP-25), and the class C vps mutant, vps33 (VPS33 encodes a Sec1p-like protein). The common phenotypes displayed by these mutants suggest that the molecules they represent all constitute part of the fusion machinery that allows the delivery of proteins by both the VPS-dependent and the alternative pathways.
To test this possibility we constructed a conditional allele of VAM3. As a syntaxin homologue, the cytosolic domain of Vam3p would be predicted to bind a host of other proteins (e.g., v-SNARE, Sec17p, and a Sec1p-like protein) to fulfill its function. Overexpression of the cytosolic tail of Vam3p might compete with the wild-type Vam3p and thus exert a dominant-interfering effect. Thus, we placed a fragment encoding the entire Vam3p cytosolic domain (VAM3-TMΔ) behind the inducible GAL1 promoter and monitored the effects of expression at various times after induction with galactose. Production of the cytosolic domain of Vam3p resulted in a block in the processing of both ALP and Vps10p-Δ10*. This processing delay was apparent after 15 min of induction with galactose and was more pronounced after 30 min of induction with galactose. Production of the Vam3p cytosolic domain was also monitored in the pulse/chase analysis and showed that high levels of synthesis were achieved by 30 min of galactose treatment (data not shown).
ALP Contains Information in its Cytosolic Tail That Targets It to the Alternative Bypass Pathway
With the exception of ALP and Vam3p, all of the vacuolar membrane proteins thus far examined travel to the vacuole by the VPS-dependent pathway. This includes not only proteins such as Vph1p and DPAP B, which function as vacuolar proteins, but also proteins that lack specific targeting information such as the Vps10p-Δ10* and other late Golgi proteins with mutations in their cytosolic tail, Golgi localization determinants (Nothwehr et al., 1993; Cooper and Stevens, 1996; Bryant and Stevens, 1997). Thus, at the outset, it seemed plausible that there would be a positive acting signal within the ALP protein responsible for its sorting into the alternative bypass pathway.
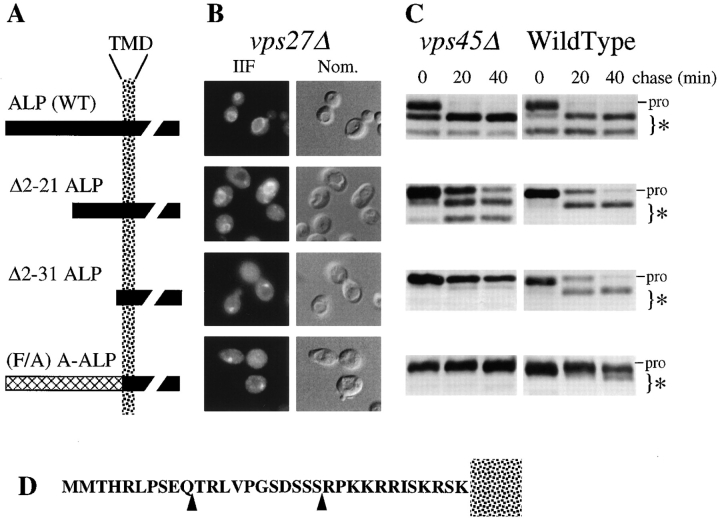
Previous analyses in vps27Δ cells showed that replacement of the cytosolic NH2-terminal region of ALP with that of a Golgi-localized membrane protein, Ste13p, as well as mutant cytosolic tails of Ste13p, resulted in a protein that is localized to the class E compartment (Raymond et al., 1992; Bryant and Stevens, 1997). Therefore, we reasoned that at least part of the sorting information was within the cytosolic tail of ALP. To test this theory, we analyzed the targeting of mutant ALP proteins bearing deletions of the first 21 and 31 amino acids (ALP Δ2–21, ALP Δ2–31). We assessed whether these proteins would localize to the vacuole or instead would be found within the class E prevacuolar compartment in vps27Δ cells, which would indicate whether these proteins had entered the VPS-dependent pathway. To complement this approach, we used pulse/chase immunoprecipitation analysis to determine whether these proteins were susceptible to the block in membrane traffic in vps45Δ cells or, like the full-length ALP, whether they still contained the information necessary to bypass this block. Fig. 6 shows that both the ALP and the ALP Δ2–21 protein are delivered to the vacuole in vps27Δ cells and vps45Δ cells. Both proteins were found exclusively in the vacuole by immunofluorescence in vps27Δ cells, and both proteins were processed in vps45Δ cells with the same kinetics as that observed in wild-type cells. However, the ALP Δ2–31 protein, which lacks the first 31 amino acids of the ALP cytosolic domain (a truncation resulting in a cytosolic tail of about three amino acids), was localized to the class E prevacuolar compartment as shown by immunofluorescence localization. As a control, we also analyzed the targeting of an Ste13/ALP fusion protein derivative designated (F/A) A-ALP, which follows the VPS-dependent pathway to the vacuole. This protein consists of the transmembrane and luminal domains of ALP fused to the cytosolic tail of the Golgi protein Ste13p, in which the Golgi retrieval targeting motif FXFXD has been ablated (Nothwehr et al., 1993). In wild-type cells the (F/A) A-ALP protein is localized to the vacuole, and in vps27Δ cells it is localized to the class E prevacuolar compartment (Bryant and Stevens, 1997).
Figure 6.
The cytosolic domain of ALP is necessary for entry into the alternative bypass pathway. (A) Schematic description of the ALP mutant proteins analyzed. Truncations in the NH2-terminal cytosolic tail of ALP were made by deleting the first 21 or 31 codons of the ALP open reading frame to generate pNB3 (Δ2–21 ALP) and pNB4 (Δ2–31 ALP). The (F/A) A-ALP protein consists of the luminal and transmembrane domains of ALP fused to the cytosolic tail of Ste13p in which the Golgi localization motif FXFXD has been ablated by the mutation AXAXD. (B) The distribution of the mutant ALP proteins (wild-type ALP, Δ2–21, Δ2–31, and [F/A] A-ALP) was assessed in vps27Δ pho8 cells (CKRY2-8A) transformed with CEN-URA3 plasmids, pSN92 (ALP), pNB3 (Δ2–21-ALP), pNB4 (Δ2–31-ALP), and pSN100 ([F/A] A-ALP) using immunofluorescence. Transformants were fixed and labeled with anti-ALP antibodies together with biotinylated secondary antibodies and FITC-conjugated streptavidin. (C) The PEP4-dependent cleavage of the mutant ALP proteins (wild-type ALP, Δ2–21, Δ2–31, and [F/A] A-ALP) was assessed in vps45Δ pho8Δ cells (NBY68) using pulse/chase analysis. Transformants were labeled for 10 min with 35S-Express and chased for the indicated times, after which time lysates were subjected to immunoprecipitation with anti-ALP antibodies. (D) Schematic representation of the cytosolic tail of ALP. (Arrowheads) Second residue (after methionine) of the deletion constructs.
Consistent with these results, the PEP4-dependent processing of ALP Δ2–31 was blocked in vps45Δ cells, indicating that this protein had lost information necessary for entry into the alternative bypass pathway (Fig. 6). As predicted, processing of the (F/A) A-ALP protein was also blocked in vps45Δ cells, while both the ALP and the ALP Δ2–21 protein were processed with kinetics similar to that observed in wild-type cells. In wild-type cells, all ALP-based proteins were correctly targeted to the vacuole as shown by immunofluorescence (data not shown) and pulse/chase immunoprecipitation analysis (Fig. 6). These data show that the amino acids near the transmembrane domain of the cytosolic tail are necessary for entry of ALP into the alternative pathway.
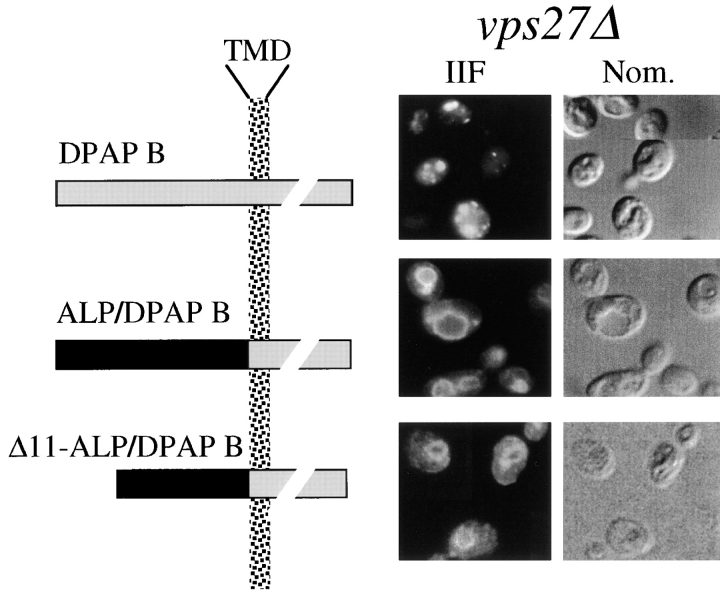
To determine whether the cytosolic domain of ALP contained sufficient sorting information for the alternative pathway, we analyzed a chimeric protein composed of the cytosolic domain of ALP fused to the transmembrane and luminal domains of another type II membrane protein, DPAP B (Roberts et al., 1989). To assay the targeting of these proteins, the ALP/DPAP B chimera and DPAP B were expressed using the GAL1 promoter and immunolocalized in vps27Δ cells. DPAP B was found in class E–like prevacuolar structures, indicating that DPAP B traveled through the VPS-dependent pathway since it was unable to bypass the vps27Δ-imposed block (Fig. 7). Approximately 94% of labeled cells examined showed labeling of DPAP B exclusively in class E prevacuolar structures. In contrast, the ALP/DPAP B chimera was found exclusively in the vacuole in vps27Δ cells. The ALP/DPAP B chimera was not detected in class E prevacuolar structures in >400 cells examined. Deletion of the NH2-terminal amino acids 2–11 of the ALP/DPAP B chimera did not affect its targeting to the vacuole, consistent with the deletion analysis in Fig. 6. Together these data show that the cytosolic tail of ALP contains information in the sequence RPKKRRISKRSK adjacent to the transmembrane domain that constitutes the sorting domain for targeting into the alternative bypass pathway.
Figure 7.
The cytosolic domain of ALP can confer sorting into the alternative bypass pathway. The schematic (right) shows the wild-type DPAP B protein (DPAP B), a chimeric protein comprised of the cytosolic domain of ALP fused to the transmembrane and luminal domains of DPAP B (ALP/DPAP B), and an ALP/DPAP B chimeric protein with a deletion of amino acids 2–11 of the ALP cytosolic domain. All constructs (pCJR6, pAIN1, and pAIN2, respectively) were under the control of the GAL1 promoter. The targeting of these proteins was analyzed by immunofluorescence (left) in vps27 dap2Δ cells (RPY107) transformed with pCJR6 or pAIN1. Transformants were grown overnight in raffinose, shifted to 2% galactose for 1 h, fixed, and then processed for immunofluorescence labeling using anti–DPAP B antibodies, biotinylated secondary antibody, and FITC-conjugated streptavidin.
Discussion
Two Intracellular Pathways for Yeast Vacuolar Membrane Proteins
Proteins that enter the secretory pathway transit through the ER and successive Golgi compartments. Although this process has now been recognized as selective at each vesicle transport stage, all anterograde traffic appears to travel through the same pathway (Pelham and Munro, 1993; Rothman and Wieland, 1996; Schekman and Orci, 1996). Once proteins enter the TGN they face a very complex decision. In mammalian cells, proteins can leave the TGN in one of many vesicles including basolaterally or apically targeted secretory vesicles bound for the plasma membrane, regulated secretory granules, or clathrin-coated vesicles that transport them to endosomal/lysosomal compartments (Bauerfeind et al., 1994; Jahn and Sudhof, 1994; Robinson, 1994; Ikonen et al., 1995; Hunziker and Geuze, 1996). This level of complexity is also seen in other eukaryotic cells such as Saccharomyces cerevisiae. The “yeast TGN” can be defined as the last functional compartment of the Golgi (Graham and Emr, 1991; Redding et al., 1991). Proteins that enter the yeast TGN can depart either by being retrieved to earlier compartments (Harris and Waters, 1996), by entering vesicles bound for a PVC subsequently allowing proteins to reach the vacuole (Conibear and Stevens, 1995; Horazdovsky et al., 1995), or by entering one of two vesicle populations that will ultimately deliver cargo to the plasma membrane (Govidan et al., 1995; Harsay and Bretscher, 1995). The experiments reported here underscore the complexity of protein sorting at the yeast TGN by establishing an additional pathway from the Golgi to the vacuole that is distinct from the established vacuolar biogenesis pathway defined by various VPS gene functions.
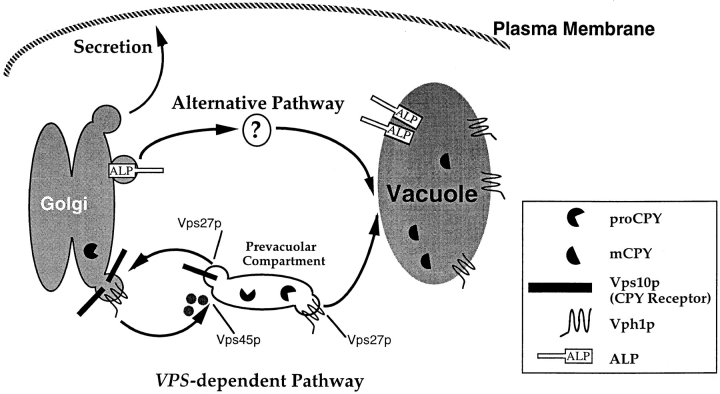
A current model for how CPY is delivered to the vacuole along the vacuolar biogenesis pathway has been proposed previously (Conibear and Stevens, 1995; Horazdovsky et al., 1995) and is presented in Fig. 8. Membrane proteins depart the Golgi by entering transport vesicles that fuse with a PVC. From this compartment, proteins can either recycle back to the Golgi, as is the case for the TGN proteins Ste13p and Vps10p, or travel on to the vacuole, as is the route taken by the vacuolar H+-ATPase subunit, Vph1p. Although this model is largely based on work in mammalian cells that examines the trafficking of the mannose-6-phosphate receptor, direct evidence for this model comes from fractionation experiments that purified PVCs (Vida et al., 1993) as well as from work that has used specific mutant alleles in certain VPS genes to accumulate membrane trafficking intermediates (Raymond et al., 1992; Piper et al., 1995).
Figure 8.
Model for the VPS-dependent pathway and the alternative bypass pathway to the vacuole. Proteins such as the CPY receptor and Vph1p enter vesicles in the late Golgi (TGN) of yeast that fuse with a PVC. This fusion event is controlled by Vps45p. The CPY receptor then recycles back to the Golgi while proteins such as Vph1p and CPY are delivered to the vacuole. These latter steps are controlled by Vps27p. ALP bypasses these intermediates and neither enters the Vps45p-controlled Golgi-derived transport vesicles nor transits through the PVC. ALP may enter a specialized transport vesicle that fuses directly with the vacuole or may be transported via a yet unidentified intermediate compartment (?).
In this study we have used both vps45-ts and vps27-ts mutations to impose specific blocks along the vacuolar biogenesis pathway. Loss of Vps45p function prevented newly synthesized Vph1p and Vps10p-Δ10* from reaching the vacuole, and Vph1p was found within small vesicular structures, consistent with its being trapped within transport vesicles that are observed by EM (Cowles et al., 1994; Piper et al., 1994). Loss of Vps27p function also resulted in blocking Vph1p and Vps10p-Δ10* from reaching the vacuole. In this case, newly synthesized Vph1p was found in a characteristic class E compartment that previous studies have demonstrated to represent an exaggerated form of the PVC (Raymond et al., 1992; Piper et al., 1995; Rieder et al., 1996). Despite the rapid and profound blocks imposed by loss of Vps45p and Vps27p function, the vacuolar protein ALP was able to reach the vacuole. These data not only suggest that ALP leaves the Golgi via a different route than the Golgi-derived transport vesicles that carry Vph1p and Vps10p, but also that ALP is not delivered to the PVC. Our model proposes that ALP is carried directly to the vacuole; however, a series of yet unknown trafficking intermediates may well be involved. Although exactly which membrane intermediate(s) is involved in ALP's transit to the vacuole remains unclear, the simplest hypothesis is that ALP enters a different set of transport vesicles that are distinct from those of the VPS-dependent pathway or those destined for the plasma membrane. Further studies focused on isolating and characterizing the intermediates along the alternative pathway will resolve these issues.
Previous localization studies have indicated that ALP could follow a different route to the vacuole (Raymond et al., 1992; Piper et al., 1994). Similarly, indications that ALP follows a different pathway out of the Golgi have been extrapolated from differential processing defects of ALP and soluble vacuolar hydrolases in a number of vps mutants (Fig. 1) (Herman et al., 1991; Horazdovsky et al., 1994, 1996; Becherer et al., 1996). However, interpretation of these processing differences alone may be misleading. Such differences observed in null mutants may not accurately reflect delivery to the vacuole, as exemplified by the processing of CPY in vps27Δ mutants where CPY is processed without necessarily reaching the vacuole (Raymond et al., 1992; Piper et al., 1994) (Fig. 1). Also, in vps9 mutant cells, differential kinetic delays in CPY processing and ALP processing are only seen after rapid inactivation of temperature-sensitive alleles but not in null mutants (Burd et al., 1996). Other complications are also evident. For example, Vps45p is proposed to work in conjunction with two other class D VPS genes, PEP12/VPS6 (syntaxin) and VPS21/YP51 (Rab5) (Singer-Kruger et al., 1994; Horazdovsky et al., 1994, 1995; Becherer et al., 1996). Like vps45Δ cells, pep12Δ cells and vps21Δ cells exhibit a punctate vesicular-like labeling pattern for Vph1p, whereas ALP is clearly localized to the vacuole (Piper, R.C., N.J. Bryant, and T.H. Stevens, unpublished results). However, pep12Δ mutants have been reported to have severe processing defects in both ALP and CPY (Becherer et al., 1996), whereas vps21Δ/ypt51 mutants have a slight or no defect in processing of either ALP or CPY (Horazdovsky et al., 1994; Singer-Kruger et al., 1994). Thus, because of possible secondary effects due to null mutations, it has not been possible to conclude confidently that ALP follows a different pathway to the vacuole. The studies reported here circumvent these difficulties by using both immuno-localization and pulse/chase immunoprecipitation techniques to follow the trafficking of newly synthesized ALP and two other membrane proteins as they travel to the vacuole in cells subjected to acute blocks in membrane traffic.
How the Alternative Pathway Might be Controlled
We have characterized a sorting determinant in the cytosolic tail of ALP that allows ALP to enter the alternative pathway. This signal acts in a positive fashion to divert proteins into the alternative pathway that would otherwise transit through the VPS-dependent pathway. The necessary determinants of the signal itself were found near the transmembrane domain within amino acids 20–31 of the NH2-terminal cytosolic tail. It is not yet known which features contained in the sequence SRPKKRRISK are necessary for the ALP sorting domain. Although this region is highly basic, similar stretches of amino acids are found near the transmembrane domain of the Kex2p, DPAP B, and Vps10p tails.
We have also found that the syntaxin protein, Vam3p, also appears to follow a route distinct from the VPS-dependent pathway since it is localized to the vacuole in vps27Δ cells. Further characterization of other proteins that are transported via the alternative pathway will be helpful in determining the function of this new post-Golgi transport step. It is difficult to speculate at this time what unique function this protein transport pathway provides that could not otherwise be fulfilled by the VPS-dependent pathway. One possibility is that the alternative pathway may intersect other transport pathways to the vacuole such as the autophagy pathway or the process of importing cytosolic proteins into the vacuole as defined by the CVT and VID genes (for review see Scott and Klionsky, 1997).
Further studies on the cis-sorting sequence of ALP and Vam3p may help identify the protein machinery that acts at the level of the yeast TGN to allow exit via the alternative pathway. Of particular interest will be characterization of the coat proteins responsible for sorting ALP out of the Golgi. Although the identity of such coat proteins and associated factors is far from clear, one protein, Vps1p, does seem directly involved in this process. In cells lacking Vps1p function, vacuolar proteins that normally route through the VPS-dependent pathway as well as ALP are shunted to the cell surface before being endocytosed to the vacuole (Nothwehr et al., 1995). This is thought to be due to the inhibition of vesicle formation at the TGN, which in turn forces proteins into secretory vesicles (Conibear and Stevens, 1995; Nothwehr et al., 1995). The profound effect of vps1 mutations on both pathways suggests a dual role for Vps1p. Importantly, mutations that block the fusion of TGN-derived transport vesicles in the VPS-dependent pathway (Fig. 4) or mutations such as vps34 and vps15, which are believed to block TGN vesicle formation, do not appear to divert ALP to the cell surface (Piper, R.C., and T.H. Stevens, unpublished observations). As a dynamin homologue, Vps1p is believed to work in the scission of clathrin-coated pits into coated vesicles (Robinson, 1994; De Camilli et al., 1995), and there is evidence that loss of clathrin function causes a small amount of ALP to appear at the cell surface (Seeger and Payne, 1992a ). Thus, there may be two types of clathrin-coated vesicles, each targeted differently as is the case for clathrin-coated AP-1 and AP-2 vesicles in animal cells (Robinson, 1994). Alternatively, given that the effects of clathrin mutations on ALP trafficking are slight, there may well be an additional role for Vps1p other than its proposed role with clathrin.
At the other end of the alternative pathway lies the machinery responsible for the final delivery of ALP to the vacuole. We found that the vacuolar syntaxin Vam3p may constitute part of the fusion machinery that is responsible for the delivery of ALP to the vacuole. Expression of the cytosolic domain of Vam3p imposed a dominant-interfering effect and resulted in a significant delay in ALP processing. This is consistent with our finding that in vam3 null mutants ALP processing is blocked. We also find that the VPS-dependent pathway, marked by the processing of the Vps10p-Δ10* protein, is also blocked either in vam3Δ mutants or in cells carrying the conditional dominant-interfering VAM3-TMΔ allele. These data indicate that Vam3p has a variety of roles by allowing membrane traffic from a number of different routes to fuse with the vacuole. These functions would allow the alternative pathway and the VPS-dependent pathway to converge and to use the same fusion machinery as that responsible for vacuolar homotypic fusion (Nichols et al., 1997). We have also observed similar phenotypes in other mutants. For instance, ypt7 mutations block the maturation of ALP (Wichmann et al., 1992) and our preliminary results with ypt7 and other mutants displaying a class B vps phenotype indicate that the PEP4-dependent processing of both ALP and Vps10-Δ10* is blocked (Bryant, N.J., R.C. Piper, and T.H. Stevens, unpublished observations). Like Vam3p, Ypt7p has an additional role in vacuolar homotypic fusion (Haas et al., 1995) and appears to have a role in transport from the late endosome to vacuole (Schimmoller and Riezman, 1993). Yet another protein that may participate in the fusion of ALP-containing vesicles with the vacuole is the class C Vps protein, Vps33p/Slp1p, which belongs to the Sec1p family of proteins and is hypothesized to bind to a t-SNARE–related protein. vps33 mutants also fail to process ALP (Banta et al., 1990) and Vps10-Δ10* (Bryant, N.J., R.C. Piper, and T.H. Stevens, unpublished observations). One curious feature of yeast mutants that have blocks in the late stages of the vacuolar biogenesis pathway (e.g., ypt7 and vam3) is that intracellular CPY is proteolytically cleaved, whereas the Vps10p-Δ10* protein is not (Wichmann et al., 1992; Nichols et al., 1997; Bryant, N.J., R.C. Piper, and T.H. Stevens, unpublished observations). This underscores the difficulty in interpreting the processing of CPY as discussed above. Exactly where along the vacuolar biogenesis pathway CPY is matured is not strictly known. One explanation is that CPY could be susceptible to proteases present in late endosomal compartments, which are likely to accumulate in some mutants. This is supported by the observations of Schimmoller and Riezman, who found that late endosomal compartments isolated from ypt7 mutants contained active vacuolar proteases (Schimmoller and Riezman, 1993).
The present study has used mutant cells to show that the pathway taken by ALP to the vacuole is distinct from that taken by previously well-characterized vacuolar proteins. Perhaps of greater interest will be determining how the alternative pathway functions in wild-type cells. One focus for future studies will be to isolate biochemically and characterize the vesicles that carry ALP to the vacuole and to determine how many sorting steps lie between the TGN and vacuole within this pathway. Such characterization will likely elucidate the overall function of this pathway in eukaryotic cells.
Acknowledgments
We thank Antony Cooper and Greg Payne for helpful discussions and for sharing unpublished data; Laurie Graham for supplying anti-Vph1p antibody; and Kathryn Hill for supplying affinity-purified anti-ALP antibody. We also thank Charles Cary for making pCC1 and RPY95.
This work was supported by grants from the National Institutes of Health (GM32448 [to T.H. Stevens] and GM16601-01 [to R.C. Piper]).
Abbreviations used in this paper
- ALP
alkaline phosphatase
- CPY
carboxypeptidase Y
- DPAP
dipeptidyl aminopeptidase
- 5-FOA
5-fluoroorotic acid
- ORF
open reading frame
- PVC
prevacuolar/endosomal compartment
- YPD
1% yeast extract, 1% peptone, 2% dextrose
Note Added in Proof
The identification of an alternative pathway from the TGN to the vacuole in yeast has recently been reported by Cowles et al. (Cowles, C.R., W.B. Snyder, C.G. Burd, and S.D. Emr. 1997. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO (Eur. Mol. Biol. Organ.) J. 16: 2769–2782). The vast majority of our findings are in agreement with this study, except those concerning the ALP targeting signal. Taken together, the two studies suggest that the 33–amino acid ALP cytosolic domain may contain two redundant elements, each sufficient for targeting into the alternative pathway.
Footnotes
Please address all correspondence to Tom H. Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403-1229. Tel.: (541) 346-5884. Fax: (541) 346-4854. e-mail: stevens@molbio.uoregon.edu
R.C. Piper and N.J. Bryant contributed equally to this work.
References
- Aalto MK, Keranen S, Ronne H. A family of proteins involved in intracellular transport. Cell. 1992;68:181–182. doi: 10.1016/0092-8674(92)90462-l. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Vida TA, Herman PK, Emr SD. Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol Cell Biol. 1990;10:4638–4649. doi: 10.1128/mcb.10.9.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerfeind R, Ohashi M, Huttner WB. Biogenesis of secretory granules and synaptic vesicles. Facts and hypotheses. Ann NY Acad Sci. 1994;733:233–244. doi: 10.1111/j.1749-6632.1994.tb17273.x. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homolog, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. . Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluororotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol. 1996;16:2369–2377. doi: 10.1128/mcb.16.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell. 1995;83:513–516. doi: 10.1016/0092-8674(95)90088-8. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Emr SD, Horazdovsky BF. Mutations in the VPS45 gene, a SEC1homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- Davis NG, Horecka JL, Sprague GF., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Takei K, McPherson P. The function of dynamin in endocytosis. Curr Opin Neurobiol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- Emr SD, Schauer I, Hanson W, Esmon P, Schekman R. Invertase beta-galactosidase hybrid proteins fail to be transported from the endoplasmic reticulum in Saccharomyces cerevisiae. . Mol Cell Biol. 1984;4:2347–2355. doi: 10.1128/mcb.4.11.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govidan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18(NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiaeis required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO (Eur Mol Biol Organ) J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Waters M. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–988. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman P, Emr S. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuolar segregation. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Busch GR, Emr SD. VPS21encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO (Eur Mol Biol Organ) J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, DeWald DB, Emr SD. Protein transport to the yeast vacuole. Curr Biol. 1995;7:544–551. doi: 10.1016/0955-0674(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Horazdovsky B, Cowles C, Mustol P, Holmes M, Emr S. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem. 1996;271:33607–33615. doi: 10.1074/jbc.271.52.33607. [DOI] [PubMed] [Google Scholar]
- Hunziker W, Geuze H. Intracellular trafficking of lysosomal membrane proteins. Bioessays. 1996;18:379–389. doi: 10.1002/bies.950180508. [DOI] [PubMed] [Google Scholar]
- Ikonen E, Tagaya M, Ullrich O, Montecucco C, Simons K. Different requirements for NSF, SNAP, and Rab proteins in apical and basolateral transport in MDCK cells. Cell. 1995;81:571–580. doi: 10.1016/0092-8674(95)90078-0. [DOI] [PubMed] [Google Scholar]
- Jahn R, Sudhof T. Synaptic vesicles and exocytosis. Annu Rev Neurosci. 1994;17:219–246. doi: 10.1146/annurev.ne.17.030194.001251. [DOI] [PubMed] [Google Scholar]
- Jones EW. Vacuolar proteases in yeast Saccharomyces cerevisiae. . Methods Enzymol. 1990;185:372–386. doi: 10.1016/0076-6879(90)85033-k. [DOI] [PubMed] [Google Scholar]
- Jones EW. Three proteolytic systems in the yeast Saccharomyces cerevisiae. . J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- Kaneko Y, Hayashi N, Toh-e A, Banno I, Oshima Y. Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. . Gene (Amst) 1987;58:137–148. doi: 10.1016/0378-1119(87)90036-9. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO (Eur Mol Biol Organ) J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivoassembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and antigen processing. Semin Immunol. 1990;2:229–237. [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature (Lond) 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. . J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, O'Hara PJ, Eichinger G, Rothman JH, Stevens TH. Molecular analysis of the yeast VPS3gene and the role of its product in vacuolar protein sorting and vacuolar segregation during the cell cycle. J Cell Biol. 1990;111:877–892. doi: 10.1083/jcb.111.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vpsmutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2p protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. . J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson MS. The role of clathrin, adaptors and dynamin in endocytosis. Curr Opin Cell Biol. 1994;6:538–544. doi: 10.1016/0955-0674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Stevens TH. Protein sorting in yeast: mutants defective in vacuolar biogenesis mislocalize proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science (Wash DC) 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science (Wash DC) 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Riezman H. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J Cell Sci. 1993;106:823–830. doi: 10.1242/jcs.106.3.823. [DOI] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. Nonclassical protein sorting. Trends Cell Biol. 1997;7:225–229. doi: 10.1016/S0962-8924(97)01050-7. [DOI] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO (Eur Mol Biol Organ) J. 1992a;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO (Eur Mol Biol Organ) J. 1992b;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Riezman H. Detection of an intermediate compartment involved in transport of α-factor from the plasma membrane to the vacuole in yeast. J Cell Biol. 1990;110:1911–1922. doi: 10.1083/jcb.110.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B, Stenmark H, Dusterhoft A, Philippsen P, Yoo JS, Gallwitz D, Zerial M. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vammutants. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- Wichmann H, Hengst L, Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP-binding protein (Ypt7p) Cell. 1992;71:1131–1142. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko G, Park F, Jones E. Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. . Genetics. 1982;102:679–690. doi: 10.1093/genetics/102.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]