Abstract
Molecules that couple the actin-based cytoskeleton to intracellular signaling pathways are central to the processes of cellular morphogenesis and differentiation. We have characterized a novel protein, the actin-binding LIM (abLIM) protein, which could mediate such interactions between actin filaments and cytoplasmic targets. abLIM protein consists of a COOH-terminal cytoskeletal domain that is fused to an NH2-terminal domain consisting of four double zinc finger motifs. The cytoskeletal domain is ∼50% identical to erythrocyte dematin, an actin-bundling protein of the red cell membrane skeleton, while the zinc finger domains conform to the LIM motif consensus sequence. In vitro expression studies demonstrate that abLIM protein can bind to F-actin through the dematin-like domain. Transcripts corresponding to three distinct isoforms have a widespread tissue distribution. However, a polypeptide corresponding to the full-length isoform is found exclusively in the retina and is enriched in biochemical extracts of retinal rod inner segments. abLIM protein also undergoes extensive phosphorylation in light-adapted retinas in vivo, and its developmental expression in the retina coincides with the elaboration of photoreceptor inner and outer segments. Based on the composite primary structure of abLIM protein, actin-binding capacity, potential regulation via phosphorylation, and isoform expression pattern, we speculate that abLIM may play a general role in bridging the actin-based cytoskeleton with an array of potential LIM protein-binding partners. The developmental time course of abLIM expression in the retina suggests that the retina-specific isoform may have a specialized role in the development or elaboration of photoreceptor inner and outer segments.
Terminal differentiation and morphogenesis of many vertebrate cell types involve a coordinated program of intercellular interactions, transcriptional regulation, and intracellular movements (Watanabe and Raff, 1990; Altshuler et al., 1991; Saha et al., 1992; Williams and Goldwitz, 1992; Adler, 1993; Beebe, 1994). Photoreceptor cells, in particular, undergo a complex process of cellular morphogenesis that segregates the phototransduction apparatus from other cellular components and results in a highly polarized adult morphology. This morphogenetic process is largely reiterated in the daily renewal cycle of outer segment membrane structure throughout adult life (Young, 1967). The central importance of dynamic cytoskeletal interactions in both the initial assembly of the photoreceptor outer segment and the daily membrane renewal cycle of photoreceptor cells (Williams et al., 1988; Vaughan and Fisher, 1989) suggests that structural elements such as microfilaments and/or microtubules might act in combination with photoreceptor-specific cytoskeletal proteins to control and coordinate membrane movements. An initial search for photoreceptor-specific cytoskeletal elements in the retina led to the discovery of a cytoskeletal protein that is highly concentrated near the sites of membrane assembly (Roof et al., 1991) and is related to erythrocyte dematin (Rana et al., 1993).
Dematin and dematin-like proteins have previously been proposed to participate in dynamic regulation of cellular processes through reversible modulation of the actin-associated membrane skeleton (Koury et al., 1989; Friederich et al., 1992; Mahajan-Miklos and Cooley, 1994). Both the 48- and 52-kD subunits of erythrocyte dematin contain a headpiece domain, similar to the headpiece domain of villin, that binds actin filaments (Rana et al., 1993; Azim et al., 1995). In addition, the dematin headpiece domain is essential for actin-bundling activity, which can be reversibly modulated by phosphorylation (Husain-Chishti et al., 1988). Such activity has been proposed to play a key role in erythroid development. These potential roles for dematin-like proteins in cytoskeletal regulation and cellular morphogenesis prompted us to search for dematin homologues in the vertebrate retina using a molecular cloning strategy.
We report here the characterization of a novel retinal protein, which we term the actin-binding LIM (abLIM)1 protein. abLIM protein has a complex primary structure, with a cytoskeletal dematin-like domain that is fused to a second domain composed of four double zinc finger motifs. The double zinc fingers are cysteine-rich domains that conform to the consensus sequence previously described for LIM motifs: CX2CX16–23HX2CX2CX2CX16–21CX2C/H/ D (Schmeichel and Beckerle, 1994). LIM motifs, which were named for the three proteins in which they were originally identified: Lin-11 (Freyd et al., 1990), Isl-1 (Karlsson et al., 1990), and Mec-3 (Way and Chalfie, 1988), have been identified in > 35 proteins to date (see Sadler et al., 1992; Crawford et al., 1994; Taira et al., 1995; Dawid et al., 1995). The potential importance of the LIM motifs is suggested by the properties of other LIM-containing proteins. Like the three original members of the LIM family, other members of the LIM/homeodomain family such as Xlim-1,-2,-3 (Taira et al., 1993, 1994), apterous (Cohen et al., 1992), LH-2 (Xu et al., 1993), P-Lim/Lhx3 (Bach et al., 1995; Sheng et al., 1996), lmx-1 (German et al., 1992), Isl-2 (Tsuchida et al., 1994), LIM1 (Tsuchida et al., 1994; Shawlot and Behringer, 1995), and LIM3 (Tsuchida et al., 1994) are all involved in the implementation of a diverse array of developmental programs. However, it is now known that LIM motifs are also found in non-homeodomain proteins. Among these non-homeodomain LIM proteins are the LIM-only proteins rhombotin2 (Ttg) (Warren et al., 1994) and MLP (Arber et al., 1994), two proteins that are explicitly involved in cellular differentiation in erythroid and muscle cells, respectively. Another category of non-homeodomain LIM proteins includes paxillin (Salgia et al., 1995) and zyxin (Sadler et al., 1992; Crawford et al., 1994), two proteins that associate with the cytoskeleton through specific interactions with vinculin/ talin or α-actinin and are thought to modulate cell adhesion–stimulated changes in gene expression. Furthermore, the expression of a wide range of other LIM proteins has been implicated in cellular senescence and/or malignant transformation (Royer-Pokora et al., 1991; Wang et al., 1992; Shibanuma et al., 1994; Kiess et al., 1995; Weiskirchen et al., 1995). In view of the wide phylogenetic distribution of LIM proteins, from yeast (Mueller et al., 1994) to humans, and the involvement of many LIM proteins in development, differentiation, and/or growth control, it has been suggested that LIM motifs may play a fundamental role as cellular regulatory sequences, through modulation of a variety of protein–protein interactions (Schmeichel and Beckerle, 1994). The discovery of a novel protein with a dematin-like domain in combination with multiple LIM motifs opens the possibility that abLIM protein may play multiple roles in establishing and maintaining cellular structure through a bifunctional cross-linking with multiple LIM-binding partners.
Materials and Methods
cDNA Cloning
An adult human retinal cDNA library (Clontech, Palo Alto, CA) was initially screened at low stringency by standard hybridization methods (Sambrook et al., 1989) using a 32P-labeled full-length dematin cDNA probe (Rana et al., 1993). Additional rounds of library screening were performed using abLIM gene sequences. Selected cDNA inserts were either subcloned into pBluescript (Stratagene, La Jolla, CA) or amplified by PCR. Both sense and antisense strand sequences were obtained for the entire abLIM coding region. 5′ rapid amplification of cDNA ends (RACE; Frohman et al., 1988) was used to obtain the extreme 38 bp at the 5′ end of the transcript. The RACE procedure was carried out on retina- and brain-derived polyA+ RNA using the 5′-ampliFINDER RACE and Marathon cDNA amplification kits (Clontech) as per the manufacturer's protocols. RACE products were either sequenced directly after purification or, in some cases, subcloned into a plasmid vector followed by sequencing of multiple clones.
Nucleotide and amino acid sequences were analyzed using DNAsis/HIBIO software (Hitachi Software Engineering America, San Bruno, CA). BLAST sequence homology searches were performed against the SwissProt, PIR, DDBJ, and GenBank databases (Altschul et al., 1990). With one exception, all numbering of nucleotide and amino acid sequences is referenced to the longest abLIM isoform sequence (abLIM-l). A different numbering is assigned to the extreme 5′ untranslated region (UTR) of abLIM-s, which is unique to this isoform and is identical to a previously identified transcript. (These sequence data are available from DDBJ under accession number D31883.) The unique 5′ UTR consists of the following sequence inserted 5′ to nucleotide (nt) 1,012 in the abLIM-l sequence: (−164) GAGAGAAGCCTGATATGTAACCCAGGCGGGTGGGAGCCTCAGTCTGTCGGGCTGAGGTCTGGCATCTACAAAGCCTCTTGGCCGTGTTCTGAACTTGAAGCCTGGAGGAGTTCTCTGCTCAGC-ACAGCCAAGGAACAGAATTAGAAGAAAAGGAACCCTGGCCT (−1). The numbering of the inserted unique 5′ UTR sequence is indicated in parentheses.
Northern Blots
RNA preparations from human liver, cardiac muscle, skeletal muscle, brain, and retina (Clontech) were initially analyzed on Northern blots using standard procedures (Sambrook et al., 1989) and a 32P-labeled abLIM probe (nt 1,687–2,587). RNA from selected tissues was further analyzed using two shorter probes. One of the short probes included nt −144 of the unique abLIM-s 5′ UTR sequence (see above) through nt 1,028 of the abLIM sequence. This is identical to a sequence (nt 21–180) from the cDNA previously identified as accession number D31883 (DDBJ). A second short probe included nt 106–501 from the abLIM sequence. These two short probes were designed to detect sequences present in either abLIM-s or abLIM-l RNAs, respectively.
Reverse Transcription–PCR on Tissue RNAs
First strand cDNAs were synthesized from polyA+ RNAs from various tissues or cultured cells with oligo(dT) priming using Superscript reverse transcriptase (GIBCO BRL, Gaithersburg, MD). The 369-bp product amplified specifically from abLIM-s sequences (designated PCR 1) was generated using a nested set of primer pairs. First round amplification primers corresponded to abLIM-s sequences: (sense) nt −114 through nt −95 and (antisense) nt 1,656–1,682. Second round amplification primers corresponded to abLIM sequences (sense) nt 1,407–1,426 and (antisense) nt 1,057–1,084. Sequences of primer pairs used to amplify the 729-bp product derived exclusively from the abLIM-l sequence (designated PCR 2) are: (sense) nt 285–312 and (antisense) nt 983–1,014.
Tissue Preparation, SDS-PAGE and Western Immunoblots
Freshly dissected samples of mouse organs (liver, kidney, lung, brain, cardiac muscle, and retina) were minced and placed into 5 mM Hepes, pH 7.5, 1 mM DTT, and a protease inhibitor cocktail previously described (Roof et al., 1991). The tissue was immediately homogenized and briefly sonicated. An equal volume of 68% sucrose in 10 mM Tris, pH 7.5, 1 mM MgCl2, 60 mM NaCl was added and the sample was centrifuged at 1,600 g, at 4°C for 5 min. The nuclear pellet was discarded, and the supernatant was diluted with an equal volume of 1% Triton X-100 in 10 mM Tris, pH 7.5, 1 mM MgCl2, 60 mM NaCl and centrifuged at 52,000 g for 15 min at 4°C. The cytoskeletal extract was washed in 1% Triton X-100, 10 mM Tris, pH 7.5, 1 mM MgCl2, 60 mM NaCl, resuspended in sample buffer (Laemmli, 1970), and cleared by brief low speed centrifugation.
Retinal rod inner and outer segment fractions were prepared as previously described (Papermaster and Dreyer, 1974; Roof et al., 1991). The respective fractions were collected by high speed centrifugation and either dissolved in SDS-PAGE sample buffer or resuspended in 0.1 M Tris, pH 8.0, 1 mM PMSF for subsequent alkaline phosphatase digestion. Polypeptides were separated using 7.5% SDS-PAGE (Laemmli, 1970) and electrophoretically transferred to polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA) (Towbin et al., 1979). Gel lanes that were not blotted were stained either with silver (Wray et al., 1981) or Coomassie brilliant blue R250. Blots were incubated in primary antisera or preimmune sera at 1:5,000–1:10,000 and developed according to the manufacturer's specifications using a peroxidase-conjugated chemiluminescent secondary antibody system (Boehringer Mannheim Biochemicals, Indianapolis, IN).
Frozen human tissue samples with postmortem intervals up to 11 h were obtained from the National Disease Research Interchange (Bethesda, MD). Extracts from liver, kidney, skeletal muscle, and brain (white matter) were processed as described above, analyzed by SDS-PAGE, and immunoblotted using abLIM-specific antibodies. Specific abLIM polypeptides were identified on immunoblots as a ladder of multiple high mobility bands, a pattern characteristic of extensive proteolysis. Because of this apparent proteolysis, human tissue was not used for further analysis.
Enzymatic Digestion of Retinal Membranes
Inner segment–enriched fractions of retinal membranes were digested with 20 U of alkaline phosphatase (calf intestine; New England Biolabs, Beverly, MA) in 20 μl of 0.1 M Tris, pH 8.0, 1 mM PMSF. Control membranes were digested in an equivalent volume of 0.1 M sodium phosphates, pH 8.0, 1 mM PMSF. Samples were incubated for 15 min, 30 min, or 1 h at 30°C and subsequently analyzed by SDS-PAGE and immunoblotting.
Equivalent amounts of inner segment preparations were digested with either endoglycosidase H at 2 U per sample (Boehringer Mannheim Biochemicals) or endoglycosidase F/N-glycosidase F at 0.2 U per sample (Boehringer Mannheim Biochemicals) in 0.02 M sodium acetate buffer, pH 5.3, 0.1 M NaCl, 0.5 mM MgCl2, and 0.5% Triton X-100. Glycosidase-treated samples and an untreated control sample were incubated for 3 h at 30°C. Samples were analyzed by SDS-PAGE and immunoblotting. To validate the effectiveness of the enzyme treatments on a known glycosylated substrate, rhodopsin, the immunoblots initially stained with abLIM antibodies were washed and restained with anti-rhodopsin mAbs (rho 1D4) (MacKenzie et al., 1984). The apparent shift to higher mobility of the opsin polypeptide indicated complete deglycosylation under the conditions used for abLIM analysis (data not shown).
In Vitro Expression of abLIM Polypeptides
Two glutathione-S-transferase (GST) fusion constructs were generated by subcloning fragments of the abLIM sequence in frame into the pGEX4T-2 plasmid expression vector (Pharmacia Biotech Inc., Piscataway, NJ). One construct (GST–dem) corresponds to amino acids 585–778 of the abLIM sequence and a second construct (GST–LIM) corresponds to abLIM amino acids 58–294. Fusion proteins were expressed in Escherichia coli (Smith and Johnson, 1988) and purified using glutathione agarose according to the manufacturer's instructions (Pharmacia Biotech Inc.). Aliquots of the fusion proteins were checked by digestion with thrombin. Each expressed protein had the correct expected mobility on SDS polyacrylamide gels both before and after thrombin digestion. A GST fusion construct that included the entire abLIM-l coding sequence was also generated and transformants were identified. However, no expressed protein could be recovered from these transformants.
A construct containing the entire coding sequence of abLIM-l with ∼0.2 kb of the 3′ UTR was prepared from two overlapping phage inserts cloned into pBluescript SK(+) (Stratagene). abLIM-m and -s clones were derived from the abLIM-l clone by appropriate modification of the 5′ end of the cloned insert. Clones containing the correct abLIM-l, -m, and -s fragments were identified on the basis of insert size and PCR amplification using sequence specific primers. The identity of the clones was confirmed by sequencing across the vector–insert junction into the NH2-terminal coding region. DNA from each of the clones containing abLIM-s, -m, or -l sequences was purified using CsCl equilibrium gradient centrifugation (Sambrook et al., 1989). Polypeptides derived from the abLIM-l, -m, and -s constructs were synthesized and labeled with [35S]methionine using the TNT-coupled reticulocyte lysate system (Promega, Madison, WI) with or without added microsomal membranes.
Actin-binding Assay
Actin was isolated from rabbit skeletal muscle (Spudich and Watt, 1971) and further purified by gel filtration on a Sephacryl 300 column. The actin-binding assay was performed as previously described (Azim et al., 1995) in a total vol of 60 μl with ∼2.5–8 μg of each fusion protein added to each assay mixture. Actin was added to a final concentration of 240 μg/ml. The actin-binding assay was also performed using 35S-labeled expression products derived from the abLIM-s, -m, and -l clones (see above). Crude TNT reaction mixes containing the expressed polypeptides were precleared by centrifugation and 2.5 μl of the supernatant was added to a total vol of 60 μl, which included a 1:100 dilution of protease inhibitor solution (Pefabloc, leupeptin; Boehringer Mannheim Biochemicals).
Quantitative analysis of actin binding to the carboxy-terminal region of abLIM (amino acids 585–778) was performed using the GST–dem fusion protein with expressed GST as a negative control. Assay mixtures (60 μl) contained a total of fusion protein (either GST–dem or GST) equal to: 0, 1.8, 2.7, 3.5, 10.6, 14.2, or 21.3 μg. The relative amount of protein present in the appropriate bands in the supernatant and pellet fractions was quantitated by scanning the Coomassie-stained SDS gel. The amount of GST– dem in the pellet fraction was used to determine the concentration of fusion protein bound to F-actin. Values for the amount of free GST–dem in each fraction were calculated by subtraction of the bound abLIM values from the known total amount of GST–abLIM added to each reaction tube, assuming 100% recovery in the pellet plus supernatant fractions. The amount of F-actin recovered in each pellet fraction was constant and equivalent to 11.6 μg. Binding parameters, including the maximal binding stoichiometry of GST–dem to F-actin and equilibrium dissociation constant (K d), were calculated using the program Enzfitter (Biosoft, Ferguson, MO).
Antibody Preparation and Immunofluorescence
The purified GST–dem fusion protein was used to prepare antisera in two rabbits (Eastacres Biologicals, Southbridge, MA). Selected blots and tissue sections were stained with serum that was preabsorbed against crude liver membranes to reduce nonspecific background staining, or preabsorbed with an excess of purified expressed GST protein to rule out possible reactivity with the GST portion of the fusion protein. Preabsorption of the serum with the GST–dem fusion protein eliminated antibody binding to tissue sections and to abLIM polypeptides on immunoblots. To preserve antigenicity, samples of adult mouse eyes and cardiac muscle were embedded without fixation directly into Tissue-Tek O.C.T. compound (Miles Inc., Elkhart, IN) and 6-μm cryostat sections were prepared. Sections were processed for immunofluorescence as previously described (Roof et al., 1991). mAbs specific for α-actinin were obtained commercially (clone EA-53; Sigma Chemical Co., St. Louis, MO). Cy3-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). BODIPY FL™ phallacidin and BODIPY™-conjugated anti–mouse IgG were obtained from Molecular Probes, Inc. (Eugene, OR). All samples were viewed with indirect fluorescence and differential interference contrast optics and photographed using TMAX400 film (Eastman Kodak Co., Rochester, NY). Selected samples of retina and cardiac muscle were viewed using a confocal laser scanning microscope (TCS4D; Leica, Malvern, PA).
Results
Molecular Cloning of abLIM
To obtain clones that encode dematin-like polypeptides in the retina, a human retinal cDNA library was screened at reduced stringency using a probe corresponding to the full-length coding sequence of human erythrocyte dematin (Rana et al., 1993). A group of overlapping clones was isolated that represented a novel transcript with homology to dematin. In the first round of screening, two cDNA inserts were sequenced and one of these spanned the junction between the dematin-like and LIM domains. A second round of library screening using sequence-specific probes at high stringency generated four additional clones that spanned the dematin/LIM domain junction. A third round of screening using sequence-specific probes provided three additional clones that confirmed the dematin/LIM domain junction, two clones that contained the putative translation start site, and one clone that was subsequently found to contain all but the ultimate 38 bp at the extreme 5′ end of the cDNA sequence.
Approximately 40 bp at the extreme 5′ end of the putative transcript were not present in any of the cDNA clones and were provided by subsequent 5′ RACE (Frohman et al., 1988) using sequence-specific primers (Fig. 1, arrow). The site of translation initiation is inferred from the presence of an in-frame stop codon 5′ to the putative methionine initiation codon. An open reading frame of 2,334 bp encodes a polypeptide of 778 amino acids with a predicted mass of 87.7 kD (Fig. 1). This polypeptide was named actin-binding LIM, or abLIM, protein based on two features of its primary sequence (see below).
Figure 1.

Nucleotide and derived amino acid sequences of abLIM. Translation initiation sites for the three isoforms are indicated by the circled methionine residues. The abLIM-s isoform contains a unique 5′ UTR that consists of 164 nt inserted 5′ to nt 1,012 and numbered −1 to −164 (see Materials and Methods). The four LIM domains are shaded, the sequence that is deleted in the abLIM-m isoform is underlined, and the 38–amino acid polymorphism (variably present in all isoforms) is indicated by a dashed underline. (Arrow) Sequence provided by 5′ RACE. These sequence data are available from GenBank/EMBL/DDBJ under accession number AF005654.
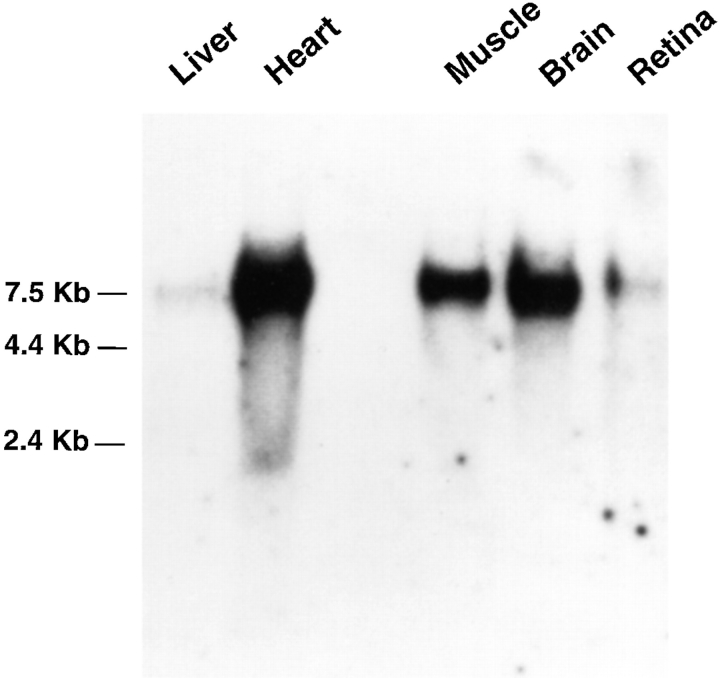
An RNA species of ∼7.7 kb is detected on Northern blots from liver, heart, skeletal muscle, brain, and retina using a probe derived from the COOH-terminal half of the abLIM coding sequence (Fig. 2). This is consistent with a coding sequence for the translated protein of ∼2.4 kb with a large (∼5.1 kb) 3′ UTR.
Figure 2.
Northern blot of abLIM sequences. PolyA+ RNAs from liver, heart, skeletal muscle, brain, and retina were loaded at 0.5 μg per lane and probed with a 32P-labeled abLIM cDNA sequence shared by all abLIM isoforms (nt 1,687–2,587). Size markers are indicated.
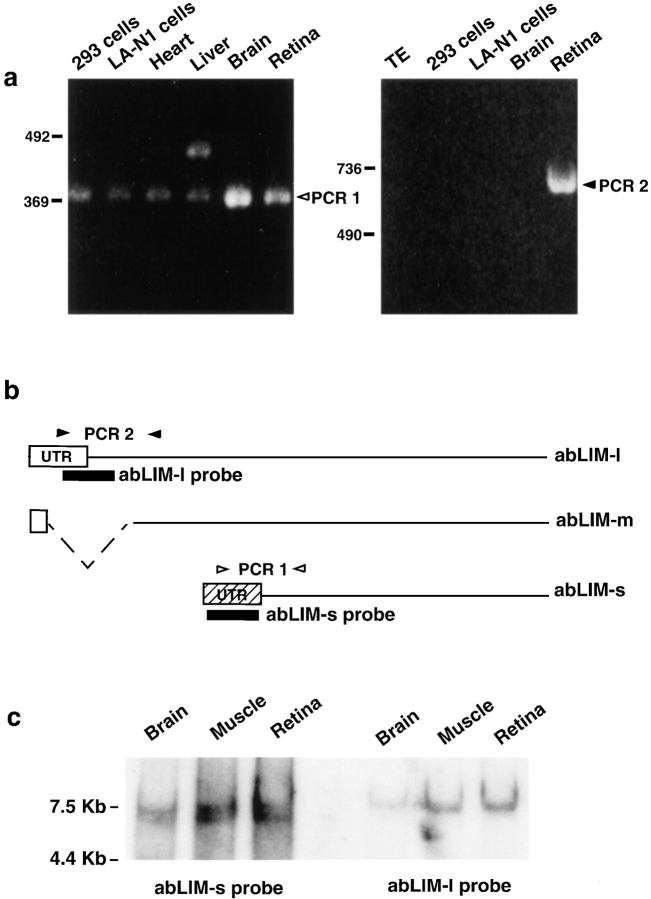
No related transcripts were identified during the retinal library screens. However, subsequent analysis provided evidence that at least two such additional transcripts exist. Homology search of the available nucleotide databases identified a cDNA sequence that is derived from an immature myeloid cell line and is nearly identical to the COOH-terminal part of the retinal abLIM cDNA, from nucleotide position 1012 through the 3′ end of the transcript. The abLIM sequence differs from the previously reported sequence (DDBJ, accession number D31883) at only five, presumably polymorphic, sites within the coding region. The D31883 sequence also includes a large (∼5 kb) 3′ UTR for which the complete sequence is available and a unique region of 5′ UTR (for complete 5′ UTR sequence, see Materials and Methods). The abLIM and D31883 matching sequences encode a polypeptide of 461 amino acids that corresponds to the dematin-like region of abLIM, excluding the LIM motifs. To test the hypothesis that this is a bona fide transcript representing a short isoform of abLIM, we used primers derived from the unique 5′ UTR of the D31883 cDNA (see Materials and Methods) to analyze reverse-transcribed cDNAs in a variety of tissues and cell lines (Fig. 3 a). The reverse transcription (RT)–PCR analysis verified the presence of the small isoform transcript (Fig. 3 a, PCR 1) in every tissue and cell line tested, including retina. The deduced polypeptide was subsequently designated abLIM-s to distinguish it from abLIM-l, the isoform originally cloned from the retinal library. A second PCR product of ∼450 bp was amplified from liver, suggesting that there may be additional, as yet undiscovered, abLIM isoforms or homologues in some tissues.
Figure 3.
Characterization of multiple abLIM transcripts. (a) Isoform-specific primer pairs were used in RT-PCR analysis of mRNA isolated from retina, brain, heart, liver, and two different cell lines, LA-N1 (Seeger et al., 1977) and 293 (Graham et al., 1977) cells. Primers that specifically amplify a sequence from abLIM-s (PCR 1) are indicated by open arrowheads in b. All tissues and cell lines tested gave the expected 369-bp PCR 1 product (a, open arrowhead, left), indicating the widespread presence of the abLIM-s transcript. A product of ∼450 bp was amplified from liver, suggesting that there may be additional abLIM isoforms or homologues in some tissues. Primers that specifically amplify a sequence from abLIM-l (PCR 2) are indicated by filled arrowheads in b. The appropriate 729-bp PCR 2 product is amplified only from retina (a, filled arrowhead, right). Exact primer sequences for PCR 1 and PCR 2 are given in Materials and Methods. (c) Northern blot of polyA+ RNAs (∼1 μg per lane) from human brain, skeletal muscle, and retina using probes unique to the abLIM-s or -l transcripts. Left three lanes were incubated with the probe specific for abLIM-s sequences (described in Materials and Methods). Right three lanes were probed with sequences present only in the abLIM-l isoform. Size markers are indicated to the left of each gel (bp). Primer sequences used to generate the probes are given in Materials and Methods. TE, Tris/EDTA buffer.
abLIM-s expression in selected tissues was further analyzed using Northern blots probed with a sequence that is unique to the 5′ UTR of abLIM-s (Fig. 3 b, abLIM-s probe). Fig. 3 c (left) shows a closely spaced doublet of ∼6.7 kb that corresponds to abLIM-s RNAs. The levels of abLIM-s transcript are approximately equivalent in brain, skeletal muscle, and retina. The two closely spaced bands in the doublet probably represent a polymorphism that was originally detected among the various retinal cDNA clones (see Fig. 1, dashed underline).
The tissue distribution of the abLIM-l transcript was examined using RT-PCR with multiple primer pairs selected to span the portion of the 5′ end of the abLIM-l sequence that is unique to this isoform. Fig. 3 a (right) shows an example of the results using a specific primer pair that amplified the appropriate sequence, designated PCR 2, in cDNA derived from human retina, but not from brain or the two cell lines. Northern blot analysis using a probe derived exclusively from the abLIM-l sequence (Fig. 3 b, abLIM-l probe) identifies an RNA species with slightly lower mobility than the abLIM-s RNA (Fig. 3 c, right). The predicted size difference between the abLIM-s and abLIM-l RNAs is ∼0.9 kb, consistent with their relative mobilities on the Northern blot (Fig. 3 c). Also consistent with the PCR analysis is the finding that abLIM-l is a relatively minor transcript within brain RNAs. However, the abLIM-l transcript is apparently not unique to the retina, but is also present in substantial amounts in other tissues, including skeletal muscle.
Evidence for the existence of yet another abLIM transcript of intermediate length comes from direct sequencing of products derived from 5′ RACE. A RACE product smaller than the predicted size of abLIM-l was amplified from brain-derived cDNAs using a 5′ anchor primer and a gene-specific primer complementary to abLIM nucleotides 1,077–1,096. To determine whether this intermediate-sized product represents a genuine transcript in vivo, the sequence was subsequently amplified from both retina and brain cDNAs using a 5′ anchor primer and an abLIM-specific primer complementary to nt 984–1,014 (Fig. 1). The sequence of the amplified product corresponds to an intermediate-sized abLIM isoform, designated abLIM-m, which is missing nt 21–501 near the abLIM-l NH2 terminus (see Fig. 1 a, underlined sequence). Since abLIM-m and abLIM-l share sequences within the extreme 5′ UTR, they are most likely products derived via alternative splicing.
Homologies of abLIM Protein to Known Functional Domains
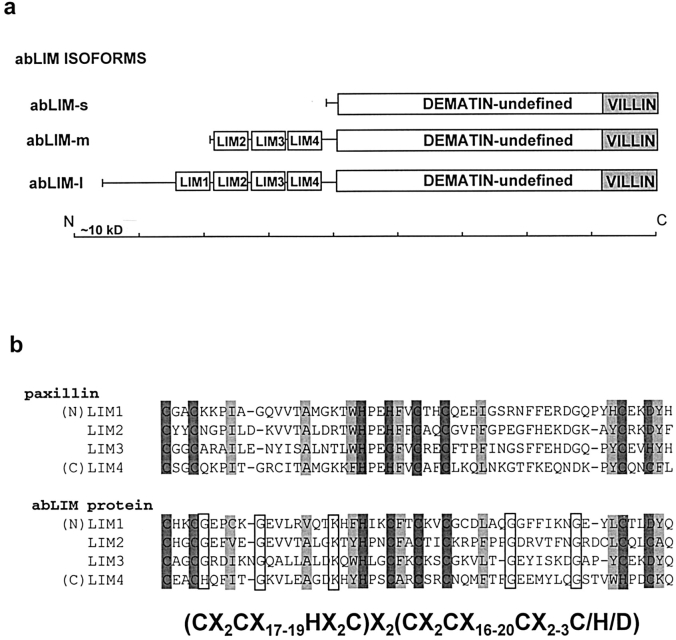
The general structure of the two largest abLIM isoforms deduced from the cDNA sequences is a linear combination of two structurally and functionally distinct domains, each representing about half of the total length of the protein (Fig. 4 a). The COOH-terminal domain of abLIM-m and -l isoforms, as well as the complete sequence of abLIM-s, are ∼50% identical to dematin, an actin-bundling protein from the human erythrocyte membrane skeleton (Rana et al., 1993) (Fig. 4 a). Only a few specific functional aspects of the dematin sequence appear to be conserved in abLIM. A portion of the actin-binding region of dematin at the extreme COOH terminus (dematin amino acid residues 340–383; Rana et al., 1993) is highly conserved in the abLIM polypeptides (abLIM amino acid residues 735–778; Fig. 4 a, VILLIN). This region corresponds to a previously described sequence within the headpiece of villin (Friederich et al., 1992) that shares some homology with the product of a Drosophila villin-like gene, quail (Mahajan-Miklos et al., 1994). These sequences are thought to be directly involved in the binding of both dematin and villin to actin filaments. In contrast, most of the other key features of the dematin primary sequence are not conserved in the abLIM proteins (Rana et al., 1993). The ability of dematin to form trimers is believed to be mediated by a single essential cysteine residue at amino acid position 194 of the dematin sequence. This cysteine is not conserved in the abLIM sequence, which has no cysteine residues within the region of dematin homology. Other features of the dematin sequence that are absent from the abLIM protein primary sequence are a PEST sequence that may be involved in rapid intracellular turnover of dematin and a putative polyglutamate nuclear localization sequence.
Figure 4.
Structure of the abLIM polypeptides. (a) Diagrams of the protein structures of three abLIM isoforms. Features indicated in the protein sequence are: VILLIN, homology to both dematin and the villin headpiece; DEMATIN-undefined, homology to the “undefined” domain of dematin; LIM1–4, LIM motifs. Together, the VILLIN + DEMATIN-undefined regions make up the dematin-like region of abLIM. (b) Sequence comparison of the four LIM domains from paxillin and abLIM protein. The LIM consensus sequence is indicated at the bottom of the figure. Conserved C/H/D residues within the LIM backbone are indicated by dark shading, conserved hydrophobic residues are indicated by light shading, and residues conserved only within the four abLIM protein LIM domains are indicated by unshaded boxes.
The amino-terminal domains of abLIM-m and abLIM-l are highly cysteine rich, moderately glycine rich, and contain three or four copies, respectively, of a cysteine and histidine–containing domain termed the LIM motif. The LIM motif has the backbone consensus sequence indicated at the bottom of Fig. 4 b. The four LIM motifs from abLIM protein are compared with each other and with LIM domains from paxillin (Fig. 4 b), another cytoskeleton-associated LIM protein that also contains four LIM domains. In contrast with paxillin and other cytoskeletal LIM proteins with multiple COOH-terminal LIM domains (Sadler et al., 1992; Salgia et al., 1995; Kuroda et al., 1996), the LIM motifs in abLIM are clustered near its amino terminus. Three of the four LIM motifs from abLIM show perfect agreement with the Cys/His/Asp backbone consensus sequence. The carboxy-terminal-most LIM domain is imperfect at the penultimate Cys residue, which is a His residue in this variant. Comparison with a wide range of LIM motifs from a variety of species and tissues suggests that there is general conservation of hydrophobic residues at the six additional positions indicated in Fig. 4 b. Further conservation of four out of six glycines and a single lysine is observed among the four LIM domains of abLIM protein.
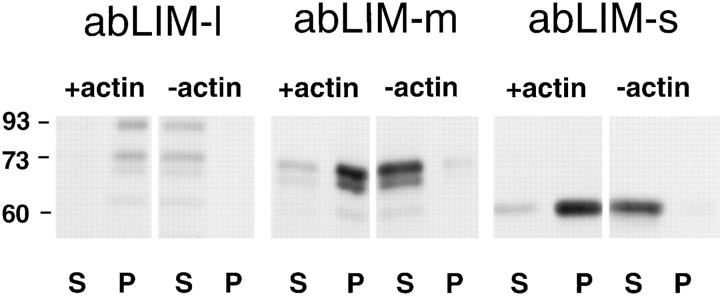
All Three abLIM Isoforms Can Bind to Actin Filaments
The function of abLIM protein, particularly within some restricted subcellular locales, may be related to its ability to bind to actin filaments. This property is strongly predicted from abLIM sequence homology with the known actin-binding proteins, erythrocyte dematin and villin. To verify that abLIM indeed binds actin, all three abLIM isoforms were expressed in vitro using a reticulocyte lysate system, and the actin-binding ability of each expressed isoform was tested independently (Fig. 5). In each case, the presence of actin in the reaction mixture shifted the expressed polypeptides from the supernatant to the pellet fraction. This indicates that all three abLIM isoforms have the capacity to bind to actin filaments in vitro.
Figure 5.
Actin binding to three abLIM isoforms expressed in vitro. Polypeptides expressed in vitro using constructs corresponding to abLIM-l, abLIM-m, and abLIM-s were tested for their ability to bind to actin filaments. All three isoforms cosediment with F-actin (+actin) and are found primarily in the respective pellet fractions (P). Each isoform is found in the supernatant fraction (S) when actin is not present in the assay mixture (−actin). Nonspecific binding to the GST-only polypeptide was negligible at all concentrations (data not shown). Approximate M r (kD) of each isoform is indicated at left.
The Dematin Homology Region of abLIM Binds Actin
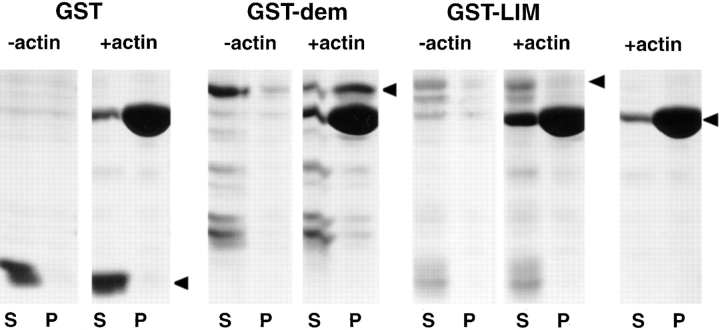
The sequence homology between abLIM and other actin-binding proteins predicts that it is the dematin-like domain of abLIM that is responsible for the observed actin binding. To test this prediction, two different sequences derived from abLIM protein, one that contained the dematin/villin headpiece region and one that included all four LIM domains (see Materials and Methods), were expressed as GST fusion proteins in E. coli. The two purified fusion proteins as well as the GST-only expression protein were tested for binding to actin filaments in vitro. In this assay, GST and the two fusion proteins are all found in the supernatant fraction when actin is not present in the assay mix (Fig. 6). However, only the abLIM construct derived from the dematin/villin homology region (GST–dem) is found in the pellet fraction when the samples are coincubated with actin under polymerizing conditions (Fig. 6). Neither the LIM region fusion protein (GST–LIM) nor GST showed significant F-actin–binding ability. This demonstrates a relatively specific interaction between F-actin and the dematin-like sequences of abLIM protein.
Figure 6.
Actin binding to GST–abLIM fusion proteins. Three different polypeptides expressed in E. coli and purified by affinity chromatography were tested for the ability to associate with F-actin: GST only (GST), GST fused to part of the dematin region of abLIM (GST–dem), and GST fused to part of the LIM domain of abLIM (GST–LIM). Each polypeptide was incubated with (+actin) or without (−actin) actin under conditions that favor actin polymerization. After centrifugation to sediment F-actin, each sample was separated into a supernatant (S) and pellet (P). Samples are indicated at the top of the SDS gel. Control incubations with actin but without added fusion proteins were run in parallel (far right panel). Mobilities of each fusion protein with the appropriate mass are indicated by filled arrowheads (right). Note the presence of each fusion protein in the supernatant fraction in −actin samples (left). Only GST–dem is enriched in the +actin pellet fraction, indicating cosedimentation of GST–dem, but not GST or GST–LIM, with F-actin.
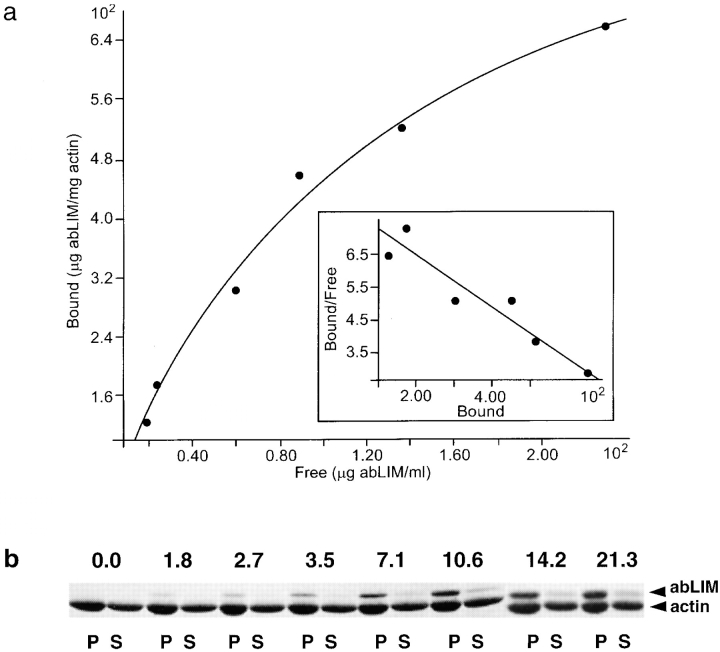
To date, it has not been possible to express the full-length abLIM sequence in vitro as a GST fusion protein. However, the actin-binding affinity and stoichiometry of the shorter expressed GST–dem construct were estimated by quantitative application of the sedimentation assay described above (Fig. 7). The calculated dissociation constant for the abLIM/F–actin interaction is ∼2.6 μM, assuming a single binding site and a molecular mass for abLIM equal to the abLIM (amino acids 585–778) plus the GST polypeptide. The maximal binding stoichiometry of abLIM is approximately one GST–abLIM monomer per 1.1 actin molecules.
Figure 7.
Quantitation of abLIM binding to actin filaments. Assay mixtures containing different total amounts of GST–abLIM polypeptide (putative actin-binding domain of abLIM, amino acids 585–778) were processed to separate bound abLIM in each pellet fraction (P) from free abLIM in the supernatant fractions (S). The derived values are plotted in a. The total amount of abLIM added (μg) in each 60-μl reaction mixture is indicated above each pair of gel lanes in b.
Comparison of abLIM Polypeptides Expressed In Vitro and In Vivo
To further define the expression pattern and size of all abLIM isoforms, the three isoforms expressed in vitro were compared with their counterparts expressed in vivo in various tissues (Fig. 8 a). The tissue variants of abLIM were detected on Western blots using an antibody that is specific for the COOH-terminal dematin homology sequence that is common to all abLIM proteins (amino acids 585–778). The abLIM-specific antibody used in these studies was tested for reactivity with human erythrocyte dematin to exclude any possible cross-reactivity on Western blots with either the 48- or 52-kD dematin polypeptides (data not shown). Although the antibody was prepared using a GST fusion protein derived from the human abLIM sequence, the deduced murine abLIM amino acid sequence was subsequently found to be highly conserved (93% identical to the human sequence) within this region.
Figure 8.
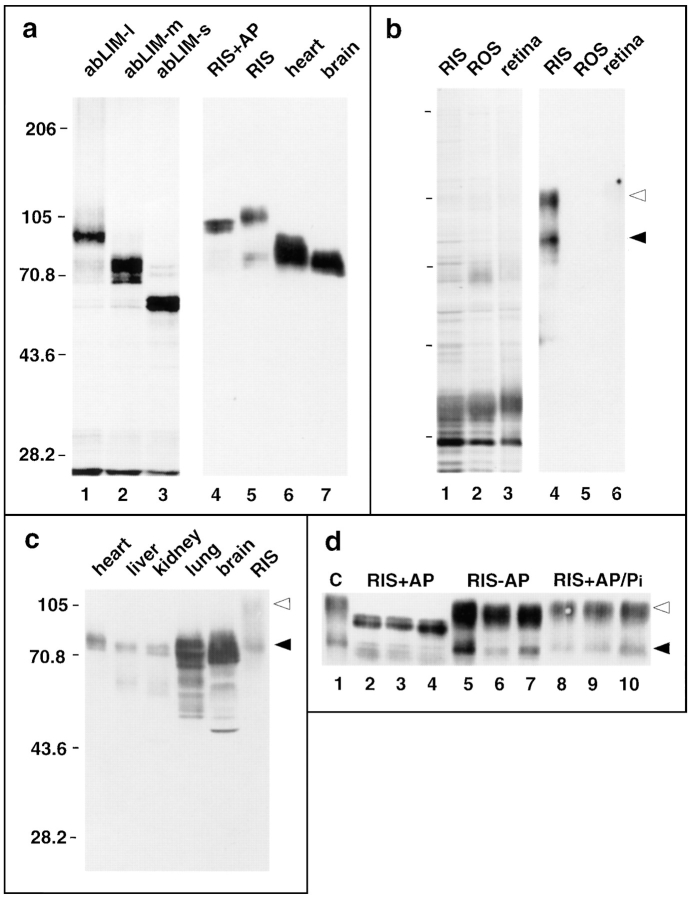
Characterization of expressed abLIM polypeptides. (a) Comparison of abLIM polypeptides expressed in vitro and in vivo. 35S-labeled polypeptides expressed in vitro in reticulocyte lysates (a, lanes 1–3) are compared with abLIM polypeptides in immunoblots of extracts from various tissues (a, lanes 5–7). The additional bands below the major products in lanes 1–3 are probably due to internal initiation products from methionines 213, 324, 446, 582, and a cluster of nine methionines near the COOH terminus (between amino acids 696–771). Both sets of polypeptides were run on the same gel and prestained mobility standards were used to align the Western blot and the 35S-fluorogram. (b) A silver-stained SDS gel profile (lanes 1–3) and immunoblot (lanes 4–6) of retinal fractions. (c) Immunoblot of cytoskeletal extracts from various tissues. The SDS gel was overloaded to show minor polypeptide species. (d) Immunoblot of retinal extracts incubated for 15 min (lanes 2, 5, and 8), 30 min (lanes 3, 6, and 9), or 1 h (lanes 4, 7, and 10) with/without the inclusion of alkaline phosphatase. Tissue extracts in all panels are cytoskeletal fractions from heart, liver, kidney, lung, and brain. Mobility standards (kD) are indicated at left of each panel. (Filled arrowhead) abLIM-m; (open arrowhead) abLIM-l at ∼70 and ∼105 kD, respectively. abLIM-specific antiserum was used at 1:5,000. No staining was observed at 1:5,000 using preserum or serum absorbed with the GST–dem fusion protein (data not shown). RIS, rod inner segments; ROS, rod outer segments; retina, whole retina; C, control (untreated) rod inner segment extract; RIS+AP, RIS incubated with alkaline phosphatase; RIS–AP, RIS incubated without addition of alkaline phosphatase; RIS+AP/Pi, RIS treated with alkaline phosphatase in the presence of excess inorganic phosphate.
The expressed human abLIM polypeptides are compared in Fig. 8 a (lanes 1–3) with the respective murine abLIM polypeptides in retina, heart, and brain extracts (Fig. 8 a, lanes 5–7). On Western blots of all tissue extracts, abLIM-m is detected as a polypeptide doublet of ∼75 kD. It seems likely that the two polypeptides of the doublet may arise from a short 114 base sequence that is apparently variably spliced into the dematin region of abLIM protein (see Fig. 1 a, dashed underline).
In contrast, a polypeptide of ∼105 kD was identified exclusively in membrane extracts from murine retinas (Fig. 8 a, RIS). The 105-kD polypeptide, which corresponds to the largest identified cDNA sequence, is not detected in brain, lung, liver, kidney, or cardiac muscle extracts (Fig. 8, a and c). Further separation of retinal extracts into photoreceptor inner and outer segment fractions suggests that abLIM-l is not present in the light-sensing organelle, the rod outer segment, but is enriched in rod inner segment (RIS) extracts (Fig. 8 b). Thus, despite the apparent lack of tissue specificity at the level of expression of the abLIM-l RNA transcript (Fig. 3 c), the abLIM-l polypeptide is detected only in extracts from retinal cells.
The smallest isoform of abLIM protein, which contains only the dematin homology domain and none of the LIM motifs, is not expressed at comparable levels in extracts from murine heart, brain, retina (Fig. 8 a), liver, kidney, or lung (Fig. 8 c). It is likely that abLIM-s is expressed at low abundance in these tissues, consistent with the presence of ∼60-kD products on overloaded immunoblots (Fig. 8 c). However, the presence of additional minor proteolytic products, particularly in the extracts from brain and lung, obscures the further analysis of a putative abLIM-s polypeptide.
Posttranslational Modifications of abLIM
The three isoforms of abLIM protein have predicted masses of 52.7, 70.6, and 87.7 kD, respectively. It is notable that the apparent sizes of the abLIM-l, -m, and -s polypeptides expressed using the rabbit reticulocyte lysate system are larger than the predicted sizes of the respective isoforms (Fig. 8 a, lanes 1–3). At least part of this difference is expected by comparison with dematin polypeptides: the predicted sizes of the two dematin subunits are 43 and 45 kD as compared with the actual M r on SDS-PAGE of 48 and 52 kD, respectively (Rana et al., 1993; Azim et al., 1995).
There is a further discrepancy between the size of abLIM-m and -l expressed in tissues compared with the respective polypeptides expressed in vitro (Fig. 8 a). To test the possibility that the difference between the predicted mass (87.7 kD) and experimentally determined mass (∼105 kD) of the large isoform of abLIM results from posttranslational modifications to the abLIM-l primary sequence, membrane extracts from murine rod inner segments were treated with enzymes to specifically alter either abLIM protein glycosylation or phosphorylation. Although abLIM glycosylation cannot be completely ruled out, no shift in polypeptide mobility was observed after exhaustive treatment with endoglycosidase F/N-glycosidase and endoglycosidase H (data not shown). The general effectiveness of the glycosidase treatment was monitored by a concomitant shift in the mobility of rhodopsin, which is also present in the RIS extract and is known to bear carbohydrate chains. In addition, the inclusion of microsomal membranes in the in vitro translation mixture (data not shown) had no effect on the size of the expressed abLIM products, suggesting that abLIM is not a good substrate for glycosylation. This is consistent with the null effect of deglycosylating agents on the tissue polypeptides.
In contrast, treatment of membrane extracts from murine rod inner segments with alkaline phosphatase causes a large shift in the apparent mass of abLIM-l from 105 to ∼95 kD (Fig. 8 a, lane 4, and Fig. 8 d, RIS+AP). This suggests that abLIM-l is highly phosphorylated in vivo in adult light-adapted retinas and is consistent with the predicted presence of up to 36 potential phosphorylation sites within the deduced primary amino acid sequence.
Dephosphorylation of the putative abLIM-l polypeptide in rod inner segment preparations (Fig. 8 d, lanes 2–4) brings the mobility of the putative abLIM-l isoform close to that of the expressed variant. However, all of the in vitro expressed polypeptides remain uniformly 2–3 kD smaller than their respective counterparts in tissue extracts. This small residual size discrepancy is probably due to a species difference since the polypeptides were expressed from the human sequence, while the extracts were prepared from mouse tissues. We have subsequently determined the complete cDNA sequence of the murine abLIM-l isoform (data not shown) and find that the deduced murine amino acid sequence is 2.6 kD larger than its human counterpart, largely due to an insertion of 28 amino acids between P348 and T349 of the human sequence. This insertion is within the dematin homology region and would be expected to affect the size of all three isoforms, as observed in Fig. 8 a. Excluding the insert in the murine sequence, the murine and human abLIM sequences are colinear and show a 90% overall amino acid identity.
Subcellular Localization of abLIM
The biochemical fractionation of abLIM-l into the photoreceptor inner segment fractions (Fig. 8 b) corresponds well with the immunofluorescence staining pattern of the abLIM COOH-terminal antigen in retina that shows abLIM antigenicity concentrated in the inner segment and outer plexiform layers of both adult murine and human retinas (Fig. 9, c and f). This staining is not present in control sections treated either with preimmune rabbit serum (Fig. 9 a) or abLIM-specific antiserum that has been preabsorbed with the GST–abLIM fusion protein (Fig. 9 b). Nor does the abLIM-specific antibody staining pattern change when the antiserum is preabsorbed with the GST-only portion of the fusion protein (Fig. 9 e).
Figure 9.
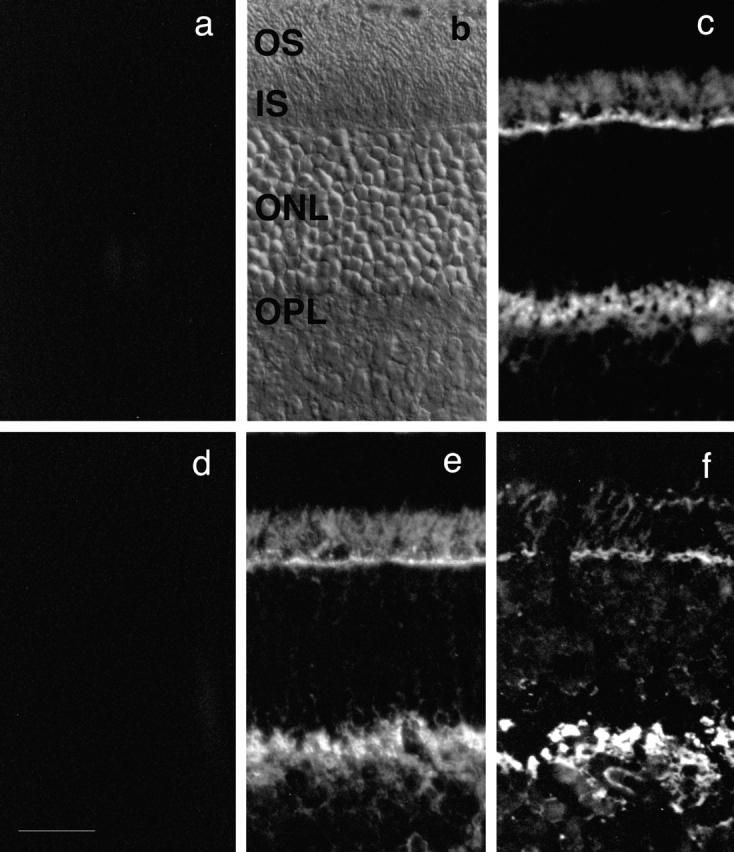
Localization of abLIM within retinal tissue sections. Unfixed cryostat sections prepared from adult murine retinas (a–e) or an adult human retina (f). Sections in c and f were treated with abLIM-specific antiserum at 1:500. Staining is prominent in the inner segment layer (IS) and outer plexiform layer (OPL). Staining is abolished in control sections treated with preimmune rabbit serum (a) or abLIM-specific serum preabsorbed with the GST–abLIM fusion protein (d). Staining is intact in control sections treated with abLIM-specific antiserum preabsorbed with the GST-only portion of the GST–abLIM fusion protein (e). All primary sera were used at 1:500. Secondary antibodies are Cy3-conjugated anti–rabbit antibodies. Nomarski view of unstained adult mouse retina is shown for reference. OS, outer segment layer; ONL, outer nuclear layer. Bar, 34 μm.
Subcellular localization of abLIM protein within striated muscle shows a regular and repetitive expression pattern. abLIM protein within cardiac myocytes is regularly spaced in a striped pattern approximately every 1.8 μm along the myofibril axis in areas that contain relatively relaxed muscle fibers (Fig. 10 d, arrow). This pattern of abLIM stripes coincides with the midpoint of the actin-containing portion of the muscle fibers, a position equivalent to the Z disk, as defined by the faint black stripe that bisects the brightly stained F-actin stripes (Fig. 10 d, arrow). The localization of abLIM immunoreactivity to the Z disk of striated muscle is confirmed by the observed coincidence of abLIM staining with the α-actinin Z repeats (Fig. 10, g–i). For comparison, actin costaining of retinal sections (Fig. 10, a–c) shows that abLIM protein colocalizes with actin in regions of high actin concentration, such as the adherens junctions of the outer limiting membrane (Fig. 10 a, arrow). Thus, in both retina and striated muscle, abLIM colocalizes with areas of high actin concentration, consistent with the presence of a putative actin-binding domain in this protein.
Figure 10.
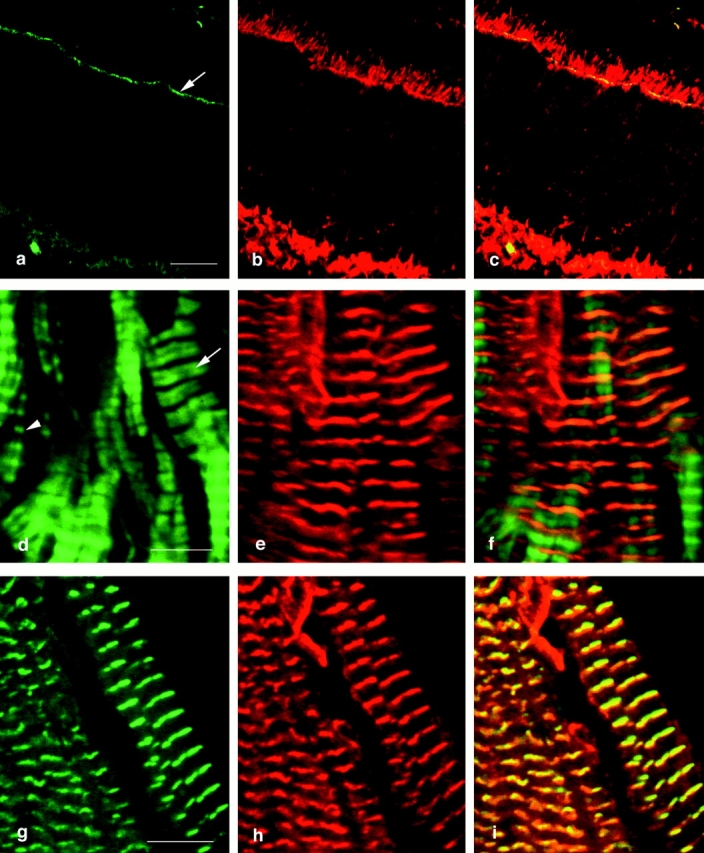
Colocalization of abLIM proteins with cytoskeletal proteins in adult murine retina and cardiac muscle. Murine retinal sections (a–c) were used to colocalize F-actin and abLIM. F-actin was visualized using BODIPY FL-conjugated phallacidin (a, green) and abLIM was visualized using specific primary antibodies followed by Cy3-conjugated secondary antibodies (b, red). Superimposed confocal microscope images identify coincident F-actin and abLIM staining in the actin-rich outer limiting membrane (c). F-actin (green) and abLIM (red) are similarly stained in unfixed sections of murine cardiac muscle (d–f). The periodicity of actin repeats varies across the section with areas of highly contracted muscle at left (arrowhead) and relatively relaxed muscle fibers at right (arrow). In the areas of the more relaxed fibers, abLIM is centered over the broad band of actin staining. Costaining of cardiac muscle with antibodies specific for mouse anti–α-actinin (g, green) and rabbit anti-abLIM (h, red) confirms the coincidence of the two proteins at the Z disks in cardiac muscle (i). Bars: (a–c) 20 μm; (d–f) 5.2 μm; (g–i) 5.6 μm.
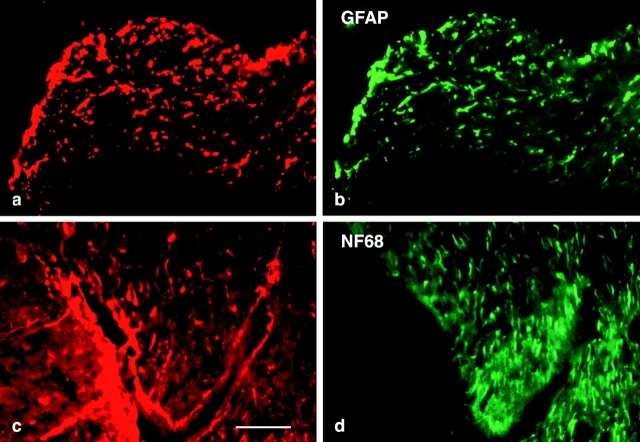
In murine brain, abLIM protein is present in cellular processes located along the subpial and periventricular surfaces of the brain, suggesting a particular concentration within astrocyte processes. This is confirmed by costaining with antibodies specific for glial fibrillary acidic protein (GFAP), which is a relatively specific marker for astrocytes (Fig. 11 b). The nerve fiber layer of the retina, which stains positively for GFAP, also stains strongly with abLIM-specific antibodies (Fig. 12; data not shown), suggesting that abLIM protein is a component of retinal astrocytes as well. However, in contrast with its localization within both glial cells and photoreceptor neurons in the retina, abLIM protein expression within the brain appears to be largely restricted to glial cell processes. abLIM staining is not coincident with neuronal processes as indicated by costaining with antibodies specific for the 68-kD neurofilament polypeptide (Fig. 11, c and d, NF68).
Figure 11.
Costaining of abLIM proteins and intermediate filament polypeptides in murine brain. Cryostat sections of murine brain were costained with the following primary antibodies: (a and c) rabbit polyclonal anti-abLIM, (b) mouse monoclonal anti-GFAP, and (d) mouse monoclonal anti-neurofilament 68-kD subunit (NF68). GFAP and abLIM are both localized to putative astrocyte processes at the periventricular surface (a and b), while abLIM and NF68 are localized within distinct types of processes. Bar, 34 μm.
Figure 12.
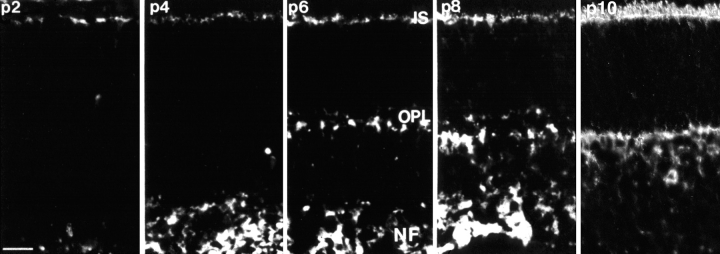
Developmental time course of abLIM expression in murine retinas. Unfixed cryostat sections of developing mouse retinas at postnatal days 2, 4, 6, 8, and 10 were stained with abLIM-specific antibodies. Outer plexiform layer (OPL) staining is present after postnatal day 6 (p6) and staining of dots in the outer limiting membrane becomes apparent within the inner segment layer (IS) at postnatal days 8–10 (p8–p10). NF, nerve fiber layer. Bar, 20 μm.
abLIM Expression during Retinal Development
The developmental expression pattern of abLIM protein within the photoreceptor layers of the retina suggests possible ways in which the actin-binding activity may be coordinated with retinal development. At birth, murine photoreceptor precursors express only small amounts of abLIM protein. Between postnatal days 4–10, abLIM protein expression is detected at increasing levels within the elongating photoreceptor inner segments and within the developing outer plexiform layer (Fig. 12). After postnatal day 8, abLIM is also expressed at high concentrations within the outer limiting membrane. Thus, abLIM expression roughly coincides with some important developmental landmarks in retinal morphogenesis: the elongation of inner segments, the formation of the photoreceptor synaptic layer, and the initiation of outer segment formation subsequent to the formation of the adherens junction comprising the outer limiting membrane.
Discussion
We have identified a novel protein with multiple isoforms expressed in a wide range of cells and tissues. The properties of this protein suggest that it could play a role in establishing or altering cell morphology through actin binding and/or participate in a signaling pathway involved in cellular differentiation.
Several lines of evidence suggest that abLIM protein can function as an actin-binding protein in vivo: all variants of abLIM show strong sequence homology to a previously described actin-binding protein, dematin, with conservation of the specific dematin-like sequences that have been implicated in its actin binding function (Azim et al., 1995). An abLIM fusion protein that contains sequences from the dematin homology region binds with high affinity and high stoichiometry to actin filaments in vitro. Each of the three identified abLIM isoforms expressed in vitro can bind to actin filaments, showing that the amino-terminal LIM domains do not inhibit actin binding through the abLIM carboxy-terminal domain. Finally, the tissue localization patterns of abLIM protein coincide with regions of high actin concentration. For example, in the retina, abLIM colocalizes with actin at adherens junctions in the outer limiting membrane. In cardiac muscle, abLIM protein is highly enriched at or very close to the Z disk, an area where actin filaments are anchored within the sarcomere.
Comparison of the primary sequences of abLIM and dematin suggest that, while the actin-binding site(s) are likely to be similar in the two molecules, if abLIM asserts higher order control over actin cross-linking, this is achieved by a different mechanism than for dematin. Dematin bundles actin filaments by trimer formation putatively involving a critical cysteine residue (Azim et al., 1995). This cysteine is not conserved in the abLIM primary sequence. However, since only the minimal backbone of a single LIM domain is necessary and sufficient to achieve LIM protein interactions both in vitro and in vivo (Feuerstein et al., 1994; Arber and Caroni, 1996), any of the four NH2-terminal LIM domains could dimerize. Thus, although we have shown that the LIM domains from abLIM protein cannot themselves bind to actin in vitro, LIM domains acting in combination with a cytoskeleton-binding domain could permit more complex actin associations in vivo and therefore mediate changes in cell shape.
The mechanism by which abLIM could modulate cell shape may be analogous to the dynamic changes attributed to dematin during erythropoiesis (Azim et al., 1995), which are putatively controlled through reversible phosphorylation by cAMP kinase (Husain-Chishti et al., 1988, 1989). abLIM is a substrate for intracellular kinase(s) and is highly phosphorylated in the retina under some physiological conditions, such as light adaptation. Thus, although the molecular mechanisms by which dematin and abLIM can achieve actin cross-linking are probably distinct, reversible cytoskeletal modulation may be a property of both molecules.
In addition to potential homodimer associations that would cross-link actin filaments, the four abLIM double zinc finger domains could also bridge the actin cytoskeleton to an array of cytoplasmic targets. LIM proteins have previously been shown to bind both to heterologous LIM proteins via LIM–LIM interactions (Sadler et al., 1992) as well as to LIM-binding proteins that do not contain LIM domains (Agulnick et al., 1996; Kuroda et al., 1996). This suggests that abLIM could serve a more complex role in cellular signaling pathways, perhaps similar to other LIM proteins that belong to the class of LIM proteins that are relatively large (>60 kD), that contain two to four LIM domains, and that are found within the cell cytoplasm. The two best characterized cytoplasmic LIM proteins from this group are paxillin (Turner et al., 1991; Turner and Miller, 1994; Salgia et al., 1995) and zyxin (Sadler et al., 1992; Crawford et al., 1994), each of which has a clearly defined role in cellular differentiation and/or malignant transformation. Paxillin, a vinculin- and talin-binding protein (Crawford et al., 1994; Salgia et al., 1995), has been placed within a signaling pathway that couples growth factor receptors via tyrosine kinases and SH2/SH3 binding sequences to the control of cell growth and maintenance of the differentiated state (Fuortes et al., 1994; Turner and Miller, 1994; Salgia et al., 1995). Zyxin, in combination with another LIM protein to which it binds (CRP), is postulated to participate in a regulatory or signaling pathway that is initiated via association with adhesion plaques through binding to α-actinin (Sadler et al., 1992; Crawford et al., 1994). It is probable that abLIM protein is also a bifunctional protein–protein linker and, like zyxin and paxillin, could participate in an undiscovered signaling pathway either through heterologous LIM/LIM or LIM/LIM-binding protein interactions in combination with a cytoskeletal association.
It is also possible that abLIM directly mediates the interaction of other LIM proteins with the actin cytoskeleton. To date, abLIM is the only LIM protein with a demonstrated ability to bind directly to actin filaments. As such, abLIM could act as the central mediator for the interaction of many different LIM proteins with microfilaments, particularly for the class of small LIM-only cytoskeleton-associated proteins that apparently lack a separate cytoskeleton-binding domain (Arber et al., 1994; Arber and Caroni, 1996; Stronach et al., 1996).
The ways in which such LIM-mediated interactions are coordinated to produce specific developmental programs and to determine specific cell fates are only beginning to be understood. It appears that the coordinated expression of several distinct LIM proteins from a multigene family is central in the determination of cell fate for several classes of motor neurons, both in developing chick (Tsuchida et al., 1994) and zebrafish (Appel et al., 1995) embryos. Based on these data, a general model for the determination of cell fate by LIM homeodomain proteins has been proposed (Tsuchida et al., 1994; Lumsden, 1995): closely related, but distinct, genes are expressed in unique cell-specific combinations during differentiation of particular cell lineages. In a similar way, the coordinated expression of two distinct variants from the multigene family of CRP-like proteins has been postulated to control different facets of myogenesis in Drosophila embryos (Stronach et al., 1996).
The three abLIM isoforms could serve as a variation on this basic theme by which LIM protein function is regulated in vivo. It has been demonstrated, for a diverse set of LIM domains, that the addition of a single LIM “cassette” to a LIM-containing polypeptide can dramatically alter the targeting and, presumably, function of the expressed polypeptide in situ (Arber and Caroni, 1996). abLIM is the first LIM protein to be discovered with multiple isoforms that differ only by the insertion/deletion of one or more LIM cassettes. Our data suggest that the abLIM-m polypeptide, which contains three LIM domains, is expressed ubiquitously and at relatively high levels in all adult tissues, including the retina. Superimposed upon this baseline expression of abLIM-m in the retina is the photoreceptor-specific expression of the abLIM-l isoform, which contains one additional LIM cassette. We do not know the functional consequences of this cell-specific expression, but it is likely that within photoreceptors abLIM-l could compete for functional binding sites with the more widely expressed abLIM variants, particularly abLIM-m. This could have profound consequences for the activation or inhibition of any signaling pathway in which either variant participates. A molecule such as abLIM protein, which associates with the actin-rich adherens junctions as they are being formed between developing photoreceptor cells and Mueller glia, could coordinate the formation of these junctions with subsequent steps in the morphogenesis of photoreceptors, including the formation of the light-sensing organelle, the outer segment. The coordinated expression of three or more abLIM isoforms at different times during development and/or in different tissues could provide a mechanism by which abLIM proteins function in more general aspects of cellular differentiation.
As probes specific for each abLIM isoform become available and additional abLIM-binding partners are identified, it will be possible to define both the potential role of abLIM in general cellular regulatory pathways as well as a specific role in photoreceptor development and/or morphogenesis. Ultimately, the cellular consequences of genetic ablation of each abLIM isoform will reveal the importance of such abLIM-associated pathways in vivo.
Acknowledgments
We thank Guy Jirawuthiworavong for assistance with the confocal microscopy, the Schepens Eye Research Institute for generously allowing us to use the confocal microscope facility, and the National Disease Research Interchange for supplying valuable human tissue specimens. We also thank Dr. Jeff Johnston and Dr. Meredithe Applebury for many helpful discussions.
This work was supported by research grants from the National Institutes of Health (EY11692 and HL51445), Foundation Fighting Blindness, and Massachusetts Lions Research Fund. A.H. Chisti is an established investigator of the American Heart Association.
Abbreviations used in this paper
- abLIM
actin-binding LIM
- GFAP
glial fibrillary acidic protein
- nt
nucleotide
- RACE
rapid amplification of cDNA ends
- RIS
rod inner segment
- ROS
rod outer segment
- RT
reverse transcription
- UTR
untranslated region
Footnotes
Please address all correspondence to Dorothy J. Roof, Howe Laboratory, Department of Ophthalmology, Harvard Medical School, 243 Charles Street, Boston, MA 02114. Tel.: (617) 573-3416. Fax: (617) 573-3615.
References
- Adler R. Determination of cellular types in the retina. Invest Ophthalmol Vis Sci. 1993;34:1677–1682. [PubMed] [Google Scholar]
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature (Lond) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altshuler, D.M., D.L. Turner, and C.L. Cepko. 1991. Specification of cell type in the vertebrate retina. In Development of the Visual System: Proceedings of the Retina Research Foundation Symposia, Vol III. D.M-K. Lam, and C.J. Schatz, editors. The MIT Press, Cambridge, MA. 37–58.
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned homeobox gene expression in embryonic zebrafish. Development (Camb) 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes & Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Azim AC, Knoll JHM, Beggs AH, Chishti AH. Isoform cloning, actin binding, and chromosomal localization of human erythroid dematin, a member of the villin superfamily. J Biol Chem. 1995;270:17407–17413. doi: 10.1074/jbc.270.29.17407. [DOI] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse RV, II, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DC. Homeobox genes and vertebrate eye development. Invest Ophthalmol Vis Sci. 1994;35:2897–2900. [PubMed] [Google Scholar]
- Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes & Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci. 1995;318:295–306. [PubMed] [Google Scholar]
- Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. . Nature (Lond) 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell. 1992;70:81–92. doi: 10.1016/0092-8674(92)90535-k. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene–specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuortes M, Jin WW, Nathan C. β2 integrin-dependent tyrosine phosphorylation of paxillin in human neutrophils treated with tumor necrosis factor. J Cell Biol. 1994;127:1477–1483. doi: 10.1083/jcb.127.5.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MS, Wang J, Chadwick RB, Rutter WJ. Synergistic activation of the insulin gene by a LIM-homeo domain protein and a helix-loop-helix protein: building a functional insulin minienhancer complex. Genes & Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Husain-Chishti A, Levin A, Branton D. Abolition of actin-bundling by phosphorylation of human erythrocyte protein 4.9. Nature (Lond) 1988;334:718–721. doi: 10.1038/334718a0. [DOI] [PubMed] [Google Scholar]
- Husain-Chishti A, Faquin W, Wu CC, Branton D. Purification of erythrocyte dematin (protein 4.9) reveals an endogenous protein kinase that modulates actin-bundling activity. J Biol Chem. 1989;264:8985–8991. [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature (Lond) 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kiess M, Scharm B, Aguzzi A, Hajnal A, Klemenz R, Schwarte-Waldhoff I, Schaefer R. Expression of ril, a novel LIM domain gene, is down-regulated in HRAS-transformed cells and restored in phenotypic revertants. Oncogene. 1995;10:61–68. [PubMed] [Google Scholar]
- Koury ST, Koury MJ, Bondurant MC. Cytoskeletal distribution and function during the maturation and enucleation of mammalian erythroblasts. J Cell Biol. 1989;109:3005–3013. doi: 10.1083/jcb.109.6.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A. A ‘LIM code' for motor neurons? . Curr Biol. 1995;5:491–495. doi: 10.1016/s0960-9822(95)00100-x. [DOI] [PubMed] [Google Scholar]
- MacKenzie D, Arendt A, Hargrave P, McDowell JH, Molday RS. Localization of binding sites for carboxyl terminal specific anti-rhodopsin monoclonal antibodies using synthetic peptides. Biochemistry. 1984;23:6544–6549. doi: 10.1021/bi00321a041. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. The villin-like protein encoded by the Drosophila quailgene is required for actin bundle assembly during oogenesis. Cell. 1994;78:291–301. doi: 10.1016/0092-8674(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Mueller L, Zu G, Wells R, Hollenberg CP, Piepersberg W. LRG1 is expressed during sporulation in Saccharomyces cerevisiaeand contains motifs similar to LIM and rho/rac GAP domains. Nucleic Acids Res. 1994;22:3151–3154. doi: 10.1093/nar/22.15.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papermaster DS, Dreyer WJ. Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- Rana AP, Ruff P, Maalouf GJ, Speicher DW, Chishti AH. Cloning of human erythroid dematin reveals another member of the villin family. Proc Natl Acad Sci USA. 1993;90:6651–6655. doi: 10.1073/pnas.90.14.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D, Hayes A, Hardenbergh G, Adamian M. A 52 kD cytoskeletal protein from retinal rod photoreceptors is related to erythrocyte dematin. Invest Ophthalmol Vis Sci. 1991;32:582–593. [PubMed] [Google Scholar]
- Royer-Pokora B, Loos U, Ludwig W-D. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11) Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha MS, Servetnick M, Grainger RM. Vertebrate eye development. Curr Opin Genet Dev. 1992;2:582–588. doi: 10.1016/s0959-437x(05)80176-5. [DOI] [PubMed] [Google Scholar]
- Salgia R, Li J, Lo SH, Brunkhorst B, Kansas GS, Sobhany ES, Sun Y, Pisick E, Hallek M, Ernst T, et al. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL . J Biol Chem. 1995;270:5039–5047. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Seeger RC, Rayner SA, Banerjee A, Chung H, Laug WE, Neustein HB, Benedict WF. Morphology, growth, chromosomal pattern, and fibrinolytic activity of two new human neuroblastoma cell lines. Cancer Res. 1977;37:1364–1371. [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1in head-organizer function. Nature (Lond) 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Zhadanov AB, Mosinger B, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Sing-Ping H, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science (Wash DC) 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- Shibanuma M, Mashimo J-I, Kuroki T, Nose K. Characterization of the TGFβ1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene (Amst) 1988;61:31–49. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Stronach BE, Siegrist SE, Beckerle MC. Two muscle-specific LIM proteins in Drosophila. . J Cell Biol. 1996;134:1179–1195. doi: 10.1083/jcb.134.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira M, Hayes WP, Otani H, Dawid IB. Expression of LIM class homeobox gene Xlim-3 in Xenopus development is limited to neural and neuroendocrine tissues. Dev Biol. 1993;159:245–256. doi: 10.1006/dbio.1993.1237. [DOI] [PubMed] [Google Scholar]
- Taira M, Otani H, Saint-Jeannet J-P, Dawid IB. Role of the LIM class of homeodomain protein Xlim-1 in neural and muscle induction by the Spemann organizer in Xenopus. . Nature (Lond) 1994;372:677–679. doi: 10.1038/372677a0. [DOI] [PubMed] [Google Scholar]
- Taira M, Evrard JL, Steinmetz A, Dawid IB. Classification of LIM proteins. Trends Genet. 1995;11:431–432. doi: 10.1016/s0168-9525(00)89139-8. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldasarre M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Turner CE, Miller JT. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak binding region. J Cell Sci. 1994;107:1583–1591. doi: 10.1242/jcs.107.6.1583. [DOI] [PubMed] [Google Scholar]
- Turner CE, Kramarcy N, Sealock R, Burridge K. Localization of paxillin, a focal adhesion protein, to smooth muscle dense plaques, and the myotendinous and neuromuscular junctions of skeletal muscle. Exp Cell Res. 1991;192:651–655. doi: 10.1016/0014-4827(91)90090-h. [DOI] [PubMed] [Google Scholar]
- Vaughan DK, Fisher SK. Cytochalasin D disrupts outer segment disc morphogenesis in situ in rabbit retina. Invest Ophthalmol Vis Sci. 1989;30:339–342. [PubMed] [Google Scholar]
- Wang X, Lee G, Liebhaber SA, Cooke NE. Human cysteine-rich protein: a member of the LIM/double-finger family displaying coordinate serum induction with c-myc. J Biol Chem. 1992;267:9176–9184. [PubMed] [Google Scholar]
- Warren AJ, Colledge WH, Carlton MBL, Evans MJ, Smith AJH, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein Rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Raff MC. Rod photoreceptor development in vitro: intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron. 1990;4:461–467. doi: 10.1016/0896-6273(90)90058-n. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Pino JD, Macalma T, Bister K, Beckerle MC. The cysteine-rich protein family of highly related LIM domain proteins. J Biol Chem. 1995;270:28946–28954. doi: 10.1074/jbc.270.48.28946. [DOI] [PubMed] [Google Scholar]
- Williams DS, Linberg KA, Vaughan DK, Fariss RN, Fisher SK. Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J Comp Neurol. 1988;272:161–176. doi: 10.1002/cne.902720202. [DOI] [PubMed] [Google Scholar]
- Williams RW, Goldwitz D. Lineage versus environment in embryonic retina: a revisionist perspective. Trends Neurosci. 1992;15:368–373. doi: 10.1016/0166-2236(92)90181-7. [DOI] [PubMed] [Google Scholar]
- Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz EM, Yancopoulos GD, Jessell TM, Alt FW. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc Natl Acad Sci USA. 1993;90:227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]