Abstract
Elevational variation in species richness is ubiquitous and important for conservation, but remains poorly explained. Numerous studies have documented higher species richness at mid-elevations, but none have addressed the underlying evolutionary and biogeographic processes that ultimately explain this pattern (i.e. speciation, extinction and dispersal). Here, we address the evolutionary causes of the mid-elevational diversity hump in the most species-rich clade of salamanders, the tropical bolitoglossine plethodontids. We present a new phylogeny for the group based on DNA sequences from all 13 genera and 137 species. Using this phylogeny, we find no relationship between rates of diversification of clades and their elevational distribution, and no evidence for a rapid ‘species pump’ in tropical montane regions. Instead, we find a strong relationship between the number of species in each elevational zone and the estimated time when each elevational band was first colonized. Mid-elevation habitats were colonized early in the phylogenetic history of bolitoglossines, and given similar rates of diversification across elevations, more species have accumulated in the elevational zones that were inhabited the longest. This pattern may be widespread and suggests that mid-elevation habitats may not only harbour more species, but may also contain more phylogenetic diversity than other habitats within a region.
Keywords: amphibians, biodiversity, elevation, phylogeny, speciation, species richness
1. Introduction
Species richness shows important variation in two dimensions, both horizontally (e.g. the latitudinal diversity gradient) and vertically (e.g. elevational diversity patterns). Although much attention has focused on latitudinal patterns in species richness (e.g. Gaston & Blackburn 2000; Willig et al. 2003; Lomolino et al. 2005), elevational variation in diversity is widespread and may be more relevant for conservation planning at the scale of single countries or regions.
Numerous studies have documented a hump-shaped elevational pattern in species richness in many groups of organisms and in many different regions (e.g. Rahbek 1995, 1997; McCain 2005; Oomen & Shanker 2005). Thus, for a given group of organisms within a given region, species richness is relatively low at low elevations, highest at intermediate elevations and lowest at the highest elevations.
The ultimate causes of this widespread pattern remain unclear. Although some studies have addressed correlations between elevational species richness patterns and climatic variables (e.g. McCain 2005; Oomen & Shanker 2005), patterns of species richness must ultimately be explained in terms of the processes that directly change the number of species in a region, namely speciation, extinction and biogeographic dispersal (e.g. Ricklefs 2004; Wiens & Donoghue 2004). Climate and other factors may still play a critically important role (and may be tightly correlated with species numbers), but they must act on these three processes to directly change the species numbers within a region. Even local-scale patterns of elevational diversity ultimately depend on these three processes.
Several previous authors have suggested that montane regions may have high species richness because their topographic heterogeneity (or other factors) drives high rates of speciation relative to rates of extinction (species-pump model; Moritz et al. 2000; Rahbek & Graves 2001). Such elevational differences in diversification rate (=speciation rate−extinction rate) could have many underlying causes, including increased speciation rates at mid-elevations or increased extinction rates at the lowest and highest elevations. An alternative hypothesis is that rates of diversification are similar at different elevations, and that intermediate elevations might have higher species richness because (for a given group) they have been colonized for longer periods of time than lowland or extreme high elevations, and there has been more time for new species to arise and accumulate at these elevations (time-for-speciation effect; Stephens & Wiens 2003). These two general hypotheses may potentially be distinguished by combining data on phylogeny, elevational ranges of species and divergence time estimates, but no previous studies have done so.
In this paper, we address the underlying evolutionary causes of elevational species richness patterns in tropical bolitoglossine salamanders. Tropical bolitoglossines contain 13 currently recognized genera and 228 species (AmphibiaWeb 2006), and include nearly half of all salamander species. Most species and genera occur in MesoAmerica (Mexico to Panama) rather than South America (South America has only two genera and 28 species, with no endemic genera and only 26 endemic species; IUCN et al. 2004; AmphibiaWeb 2006). Studies of elevational species richness patterns of salamanders in MesoAmerica suggest that they show a hump-shaped pattern (Wake & Lynch 1976; Campbell 1999). Although there have been many recent phylogenetic analyses within genera of tropical bolitoglossines (García-París & Wake 2000; García-París et al. 2000; Parra-Olea & Wake 2001; Parra-Olea 2002, 2003; Parra-Olea et al. 2002, 2004, 2005), no previous studies have attempted to address the generic-level relationships with extensive taxon sampling.
We present a phylogenetic analysis of tropical bolitoglossines and use this phylogeny to test these alternate hypotheses for the evolutionary causes of the hump-shaped pattern of elevational species richness. First, we reconstruct relative divergence times of clades, estimate their rates of diversification (speciation rate−extinction rate), and test for a relationship between elevational ranges of clades and their diversification rates, given the hypothesis that diversification rates should be higher in clades that inhabit mid-elevation regions. Then, we estimate the relative timing of colonization of each elevational zone, and test for a relationship between the number of species in each zone and how long tropical bolitoglossines appear to have been present there. Our results suggest that the hump-shaped pattern of species richness is the result of early colonization of middle elevations and later colonization of higher and lower elevations (time-for-speciation effect; Stephens & Wiens 2003), and is not caused by differences in diversification rates at different elevations.
2. Material and methods
(a) Elevational patterns of species richness
The taxonomy used follows the AmphibiaWeb (2006) database. Data on species elevational ranges were obtained largely from the Global Amphibian Assessment (GAA; IUCN et al. 2004), which incorporates information from the previous literature. Ambiguous elevational ranges were resolved with reference to Campbell (1999) and museum localities. Elevational data for taxa not included in the GAA were obtained from the original species descriptions. Elevational data were checked for errors, and five species (out of 228) had seemingly erroneous values in the GAA database, and were corrected with reference to the original literature and personal observations. A table of elevational data and associated references is available as electronic supplementary material 1.
Elevational patterns of species richness were summarized using elevational bands of 500 m width (i.e. 0–500 m.a.s.l., 501–1000 m) and tallying the number of species occurring in each band (based on their estimated elevational range). Given that most genera and species of bolitoglossines are endemic to MesoAmerica, we focused our analyses of species richness patterns on this region, and excluded South American species and the primarily North American genus Batrachoseps.
We also tested the observed patterns of species richness relative to a mid-domain model (Colwell & Hurtt 1994), in which ranges of taxa are randomly shuffled among elevations. We used the program Mid-Domain Null (McCain 2004) to simulate the random placement of geographical midpoints within the two extreme elevations observed (0–5000 m), and given the observed elevational ranges of species. Simulations (50 000 replicates) were used to generate 95% upper and lower confidence intervals for the number of species expected at each 500 m elevational band, for comparison to the observed pattern. Sampling with and without replacement gave similar results, and only those from sampling without replacement are shown. We tested the fit between the observed values and the expected values under the mid-domain model (midpoint of upper and lower confidence intervals) for each elevational band using linear regression.
(b) Phylogeny
Previous phylogenetic and taxonomic studies within tropical bolitoglossine genera have generated extensive DNA sequence data for portions of the mitochondrial cytochrome b and large subunit ribosomal RNA (16S) genes (e.g. García-París & Wake 2000; García-París et al. 2000; Parra-Olea & Wake 2001; Parra-Olea 2002, 2003; Parra-Olea et al. 2002, 2004, 2005). We obtained data for all available species for these genes from GenBank. Furthermore, new sequences for an additional segment of 16S were generated for at least one representative of each bolitoglossine genus by G.P.O. using standard methods (Parra-Olea 2002), along with new sequences for some taxa for the other two gene regions. GenBank numbers from new and previously published sequences are available in the electronic supplementary material 2. We did not use every available sequence for every species, but instead tried to represent each species with a single individual for which data from both genes were available; in general, data available for multiple individuals of a species were used to help delimit species prior to the present study. For most species, data from cytochrome b (609 bp, 319 parsimony-informative characters (pic)) and one segment of 16S were available (570 bp, 281 pic). Data from another segment of 16S (644 bp (157 excluded), 261 pic) were available for only 46 species, but simulations (Wiens 1998) and analyses of empirical datasets (Wiens et al. 2005b) suggest that adding sets of characters which are incompletely sampled among taxa can still improve phylogenetic results relative to excluding these characters entirely. We also included several outgroup taxa, including representatives of Batrachoseps (the other major clade within Bolitoglossinae), and the other subfamilies of Plethodontidae (Hemidactylinae, Plethodontinae, Spelerpinae, sensu Chippindale et al. 2004).
Alignment of cytochrome b was performed using default options in Clustal X (Thompson et al. 1994) and was unambiguous. For 16S, we used default parameters and tested two additional gap-opening penalties (12.5 and 17.5; default=15). Characters showing different alignment under different gap-opening penalties were excluded. However, given that sequences were short to begin with (and to avoid excluding most potentially informative variation), we examined only a limited set of parameters and did not exclude some positions that differed only slightly between gap-opening penalties.
The primary estimate of phylogeny was based on a Bayesian analysis of all characters combined, but results also were confirmed by uniformly weighted parsimony analysis. Combined analysis is straightforward given that mitochondrial genes should share the same phylogenetic history. Prior to combining datasets, we identified the best-fitting model for each gene using likelihood-ratio tests and the Akaike information criterion (using MrModeltest, v. 2.0; Nylander 2004). We also tested whether there was support for recognizing additional partitions within each gene, using comparison of Bayes factors (Nylander et al. 2004; Brandley et al. 2005). Analyses of model fit were based on a subset of taxa. These analyses selected the GTR+I+Γ model (general time reversible with a proportion of sites invariable and rates at other sites varying according to a gamma distribution) for all three gene regions, and supported the use of separate partitions within each dataset (stems and loops for 16S and codon positions for cytochrome b). Stem and loop regions within 16S were identified using the European ribosomal database (http://www.psb.ugent.be/rRNA/) with Plethodon yonahlossee as the model.
We conducted a replicated pair of Bayesian searches using MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001), each using 8.0×106 generations. Trees generated prior to achieving stationarity were discarded as burn-in, and stationarity was identified based on (i) plots of log likelihoods over time, (ii) similarity in topologies, branch support (posterior probabilities, Pp) and likelihoods between trees from each replicate, and (iii) average standard deviation of split frequencies between runs.
Phylogenetic analyses were based entirely on mitochondrial data. Thus, some phylogenetic results could be an artefact of discordance between mitochondrial and species phylogenies, through introgression or incomplete lineage sorting (Maddison 1997). However, our results show strong concordance with previous generic-level taxonomy based on morphology, suggesting that such incongruence (if present) has not obscured overall phylogenetic patterns within this group.
(c) Divergence date estimates
Estimates of divergence dates were used for calculating rates of diversification and for determining the relative timing of colonization of different elevational bands. These analyses depend on the relative ages of clades, but not the absolute ages. We used the penalized likelihood (PL) method (Sanderson 2002) as implemented in r8s v. 1.6 for Unix (Sanderson 2003). PL is a ‘relaxed’ molecular clock method that allows for different rates of evolution across the tree, but minimizes hypothesized changes in rates. Tropical bolitoglossines presently are not represented in the fossil record. Therefore, we used an estimate for the age of the root of the tropical bolitoglossines based on an analysis of divergence dates across all salamanders using PL analyses of the RAG-1 gene and 11 fossil calibration points (Wiens in press). Methods and results were generally similar to those of Chippindale et al. (2004). These analyses identified three possible dates for the root of tropical bolitoglossines (depending upon the root age used for all salamanders) of 39, 49 and 55 Myr ago. However, use of these different ages has little impact on our results, and we only present those using 49 Myr ago.
PL analyses were implemented using the truncated Newton algorithm. We used cross-validated assessment to select the best-fitting smoothing parameter, with values from 100 to 104 in exponential increments of 0.5. For each root age, we performed 10 replicate optimizations. To assess confidence in the estimated ages of select clades, we sampled 280 post-burn-in trees from the Bayesian analyses (1 tree every 50 000 generations), reran the PL analysis on each tree, and obtained the standard deviation among the estimated ages using the ‘profile’ command in r8s. However, this assessment is potentially compromised for some clades that do not appear in every tree.
(d) Diversification rates and elevational ranges of clades
We tested for a possible relationship between elevation and changes in diversification rate using two general approaches. First, we calculated diversification rates and elevational midpoints of clades and looked for a possible association between them. By making minimal assumptions about the fit between the taxonomy and phylogeny (i.e. unsampled species can be assigned to taxa that appear monophyletic based on the sampled species), these analyses incorporated all described tropical bolitoglossine species, regardless of whether they were included in our phylogeny. However, this approach may not detect shifts in diversification rates and elevation within these clades. As an alternate approach, we tested for significant departures from constant rates of speciation and extinction across all of the tropical bolitoglossine species sampled in our phylogeny using the relative cladogenesis statistic (Pk) of Nee et al. (1994), as implemented in the program End-Epi v. 1.0 (Rambaut et al. 1997). We assumed that our sampling of species (approx. 60% of described species) was adequate to detect major shifts in diversification rates.
For the first approach, diversification rates were calculated using the method-of-moments estimator for stem groups, where the diversification rate is
when t is the age of the clade; n is the number of species in the clade; and ϵ is the relative extinction rate (Magallón & Sanderson 2001). The relative extinction rate (ϵ) is the extinction rate divided by the speciation rate (Magallón & Sanderson 2001).
The method-of-moments estimator is advantageous relative to the maximum likelihood estimator (i.e. ln[n]/t) because the latter assumes the extinction rate is negligible. Given that the relative extinction rate (ϵ) is unknown, we used two extreme values. We used a value of 0.90 as an upper limit and 0 as a lower limit (for justification see Magallón & Sanderson 2001), and both gave similar results.
The stem group is the most inclusive monophyletic group containing the extant species of a clade (but no other extant species), whereas the crown group is the least inclusive monophyletic group that includes all extant members of a clade (Magallón & Sanderson 2001). Use of the crown group assumes that the deepest divergence within each clade has been included, whereas use of the stem group does not. We used the stem ages of clades given that our sampling of bolitoglossine genera is complete but our sampling of species is not (137 out of 228 currently recognized species included). However, analyses using the crown-group estimator gave similar results (J. J. Wiens 2006, unpublished data).
In order to maximize the statistical power of these analyses, we used the largest number of clades that could be reasonably justified based on our phylogenetic results. Assignment of unsampled species to genera is relatively unambiguous (and almost all genera were supported as monophyletic), whereas assignment of unsampled species to subclades within most genera is not (except for Bolitoglossa). Therefore, we used all currently recognized genera as units, but with the following modifications: (i) monotypic genera (or genera represented by a single species in our sampling) were combined with their closest relatives, given that it would be difficult to estimate diversification rates for monotypic clades, (ii) Bolitoglossa was divided into 12 clades corresponding to subgenera or species groups recognized by Parra-Olea et al. (2004), each of which was strongly supported in our results, and (iii) the clade including Ixalotriton, Lineatriton, Parvimolge and Pseudoeurycea was treated as a single unit, given that the relationships within this clade are somewhat uncertain based on our results and that the placement of unsampled species within subclades of Pseudoeurycea is uncertain. In total, 19 clades were used: (i) Cryptotriton, (ii) Dendrotriton and Nyctanolis, (iii) Nototriton, (iv) Bradytriton and Oedipina, (v) Thorius, (vi) Chiropterotriton, (vii) Ixalotriton, Lineatriton, Parvimolge and Pseudoeurycea, and the following clades within Bolitoglossa (viii) subgenus Oaxakia, (ix) subgenus Pachymandra, (x) dunni group (subgenus Magnadigita), (xi) franklini group (subgenus Magnadigita), (xii) rostrata group (subgenus Magnadigita), (xiii) subgenus Bolitoglossa, (xiv) subgenus Nanotriton, (xv) subgenus Mayamandra, (xvi) adspersa group (subgenus Eladinea), (xvii) epimela group (subgenus Eladinea), (xviii) schizodactyla group (subgenus Eladinea), (xix) subpalmata group (subgenus Eladinea).
We tested for a relationship between diversification rates of clades and their generalized elevational ranges, where the latter is the average of the elevational midpoint of the ranges of all described species within that clade. Elevational data were compiled as described above. We acknowledge that use of average elevational midpoints for clades cannot reflect all of the elevational variation within species or clades. Nevertheless, despite some loss of information, the elevational midpoints for clades reflect that various clades occur predominately at lower elevations (less than 1000 m), higher elevations (greater than 2000 m) and intermediate elevations (1000–2000 m). Overall, the elevational ranges of tropical bolitoglossine species appear to be relatively narrow (mean elevational range=583.22 m; n=228; range=0–2400 m; but note that many species are known from few localities). Analyses were also performed using the average minimum elevation of species within a clade, the average maximum elevation, and the absolute minimum elevation and maximum elevation of the species within a clade. All four analyses gave results similar to those using the average elevational midpoint of species (i.e. no significant relationship between diversification rate and elevation of clades), and are not reported.
Given that there may be phylogenetic effects on elevational ranges and diversification rates, a set of analyses were conducted using independent contrasts (Felsenstein 1985). The phylogeny and branch lengths were based on the chronogram estimated from the PL analyses. Contrasts assuming equal branch lengths also were used. A set of non-phylogenetic analyses were also performed. Independent contrasts were calculated using Compare v. 4.6b (Martins 2004). Regression analyses were performed using Statview, forcing the origin through zero for contrasts. Given that Compare retains only two decimal places, diversification rates were multiplied by 10 000 to maintain precision. Using linear regression to test for a potentially nonlinear (hump-shaped) relationship could be problematic, but use of independent contrasts tests for a simple relationship between changes in elevation and changes in diversification rate.
To test for an association between changes in elevational distribution and changes in diversification rate across all the sampled species, we identified branches with significant departures from constant diversification rates using the Pk statistic applied to the chronogram (root age of 49 Myr; use of 39 and 55 Myr gives identical results for this test). We then estimated the elevational changes on each branch by reconstructing the elevational midpoint of the range of each species on the chronogram as a continuous trait using linear generalized least squares (GLS; Martins & Hansen 1997) as implemented in Compare. Following the general approach of McPeek (1995), we performed a t-test comparing the reconstructed changes in elevational distribution on those branches having significant changes in diversification rate with elevational changes on those branches lacking significant shifts in diversification rates.
New species of tropical bolitoglossines continue to be described, and our estimates of species numbers within clades (and within elevational bands) should be considered approximate. Although failure to include all species in all clades may be a source of random error, it seems unlikely that failure to include all species will create a systematic bias in the estimated relationship between diversification rate and elevation.
(e) Relative timing of colonization and species richness of elevational bands
We performed regression of the timing of colonization of each 500 m elevational band versus the number of species in that band (with both variables natural log-transformed). Rough estimates of colonization times were based on reconstructing the elevational midpoint of the range of each species on the phylogeny using linear generalized least squares in Compare (but some analyses also were conducted using the minimum elevation of each species). The phylogeny and branch lengths were based on the chronogram from the PL analyses. We then identified the approximate timing of the first colonization of each elevational band by identifying the divergence date (crown-group age) of the oldest clade occurring in that band (based on the PL analysis). We also performed a set of analyses in which we used the summed ages of each inferred colonization of each elevational band, in order to accommodate multiple colonizations of a single band. All other things being equal, the number of species within a clade is thought to increase exponentially over time, suggesting that this variable should be log-transformed for statistical analysis (Magallón & Sanderson 2001). Furthermore, the relationship between time and the natural log of species richness appeared to be nonlinear, and thus time was also natural log-transformed. We emphasize that we are interested in the relative timing of colonization of different climatic zones and habitats that presently occur at the elevations observed today (i.e. habitats have probably shifted in elevation over time due to climate change), rather than the absolute dates of colonization of specific elevational bands.
Some colonizations of an elevational band appear to involve a single species. In these cases, we assumed that the elevational band was colonized sometime between the initial splitting of that species and the present day, and we arbitrarily used the midpoint of these two ages in the analyses. The three highest elevational bands (3501–5000 m) are occupied by a single species (Pseudoeurycea gadovii). We excluded the two highest elevational bands (4001–4500 and 4501–5000 m) to reduce non-independent data points (i.e. dispersal of this species to elevations above 4501 m is not independent of its presence at elevations above 4001 m).
We acknowledge that there is considerable uncertainty in estimating the phylogeny, ancestral trait values, divergence dates and species numbers within elevational bands. Furthermore, the elevational midpoint of a species does not necessarily reflect when a given elevation was actually colonized, given that the elevational range of a species can differ from the midpoint (although elevational ranges of species average less than 600 m, see above). Clearly, there are many sources of error that could prevent us from finding a significant relationship between the timing of colonization and species richness. Nevertheless, the overall pattern is relatively simple and straightforward (i.e. clades occurring at low and high elevations are relatively derived within the tree) and the relationship between age of colonization and species richness is strong.
We also performed a Bayesian reconstruction of ancestral states (Huelsenbeck et al. 2003) in order to address the robustness of our major result (i.e. that intermediate elevations were colonized first and that lower and higher elevations were colonized subsequently) to a different method of ancestral trait reconstruction and to errors in estimating the topology and branch lengths. In current versions of MrBayes, this requires constraining a given clade to be present in all topologies, adding the trait of interest as another character to the data matrix, and rerunning the phylogenetic analysis (here using two replicate searches with 6 million generations each). We constrained the clade containing all tropical bolitoglossines to be present in all estimated trees and evaluated the posterior probabilities of different states being present at this very strongly supported node. We added elevation to the combined DNA data as an ordered multistate character with five states (the maximum allowed in current versions of MrBayes), with each state encompassing a 600 m elevational band (0, 0–600 m; 1, 601–1200; 2, 1201–1800; 3, 1801–2400; 4, 2400 or higher; very few taxa have elevational midpoints greater than 3000 m), and applied to this character the generalized likelihood model of Lewis (2001). Use of eight unordered states (using 500 m elevational bands) gave similar results. Overall, this general approach provides an estimate of the posterior probability (Pp) of each state being reconstructed at that branch, given the uncertainty in trait reconstructions on each tree and the variability in tree topologies and branch lengths among the post-burn-in trees.
3. Results
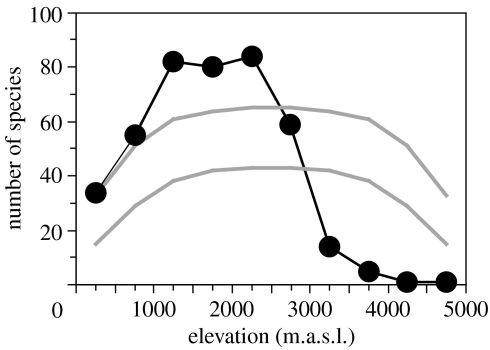
Tropical bolitoglossines in MesoAmerica show the widespread hump-shaped pattern of elevational species richness (figure 1), with the largest number of species at intermediate elevations and fewer species at the lowest and highest elevations. Almost all of the observed species richness values are outside the upper and lower 95% CIs expected under a null model of stochastic placement of elevational range midpoints, and the fit between the expected and observed values is not significant (r2=0.235, p=0.155). The elevational ranges of species and clades appear to be more concentrated at mid-elevations than expected under the mid-domain model (Colwell & Hurtt 1994).
Figure 1.
Hump-shaped pattern in species richness of tropical bolitoglossine salamanders (supergenus Bolitoglossa) in MesoAmerica. Thick grey lines indicate the upper and lower 95% CIs for the number of species at each 500 m elevational band expected from the mid-domain model.
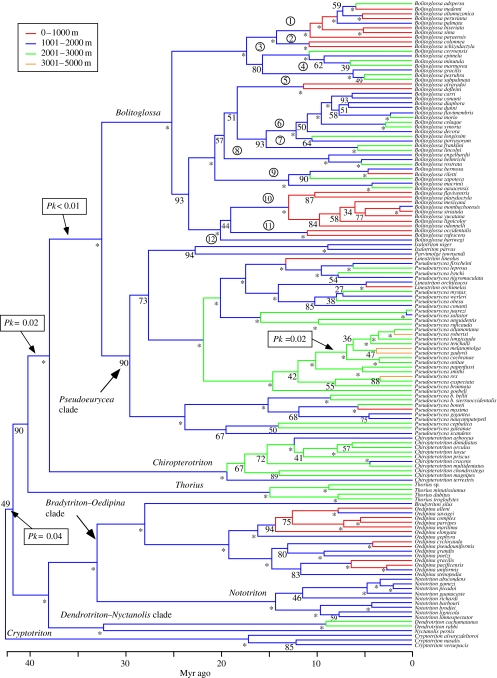
Combined, partitioned Bayesian analysis of two mitochondrial genes (cytochrome b and 16S) reveals strong support for many relationships within bolitoglossines (figure 2). Contrary to hypotheses based on morphology (Elias & Wake 1983), Nyctanolis does not appear to be the sister taxon of all other tropical bolitoglossines. Instead, Nyctanolis is the sister taxon of Dendrotriton, and these two clades are the sister group of a clade containing Nototriton and Oedipina+Bradytriton. There is strong support for a clade containing the species-rich genera Bolitoglossa and Pseudoeurycea, along with several small genera nested inside of Pseudoeurycea (Lineatriton, Ixalotriton, Parvimolge). There is also strong support for placing Chiropterotriton as sister taxon of this large clade, and moderate support for placing Thorius as sister taxon of that clade. Cryptotriton is weakly placed as sister taxon of all other tropical bolitoglossines.
Figure 2.
Estimated phylogeny and divergence times for tropical bolitoglossine salamanders (outgroup taxa not shown). Numbers adjacent to clades indicate Bayesian posterior probabilities (×100), with asterisks showing clades with Pp≧0.95. Results using parsimony (not shown) are similar. Colouring of branches shows broad-scale elevational distributions of species (midpoint of elevational range) and reconstructed ancestors (based on GLS reconstruction as a continuous trait); colours indicate 1000 m intervals containing the continuous values for species and ancestors, not character states used in ancestral reconstructions. Also shown are branches with significant deviations from constant rates of speciation and extinction (using the relative cladogenesis statistic, Pk). Branches with circled numbers correspond to 12 clades within Bolitoglossa used in the analyses of diversification rates: 1, adspersa group (subgenus Eladinea); 2, schizodactyla group (subgenus Eladinea); 3, epimela group (subgenus Eladinea); 4, subpalmata group (subgenus Eladinea); 5, subgenus Pachymandra; 6, dunni group (subgenus Magnadigita); 7, franklini group (subgenus Magnadigita); 8, rostrata group (subgenus Magnadigita); 9, subgenus Oaxakia; 10, subgenus Bolitoglossa; 11, subgenus Nanotriton; and 12, subgenus Mayamandra.
A recent analysis of amphibian phylogeny (Frost et al. 2006) included 10 out of 13 tropical bolitoglossine genera. Their tree concurs with ours in the placement of Thorius as sister group to a clade including Bolitoglossa and Pseudoeurycea and relatives (Ixalotriton, Lineatriton). Our hypotheses also agree in placing Oedipina, Nototriton and Dendrotriton in a basal clade. Our trees differ only in the placement of Parvimolge (with Pseudoeurycea versus with Bolitoglossa) and Cryptotriton (sister taxon of all other tropical bolitoglossines versus sister taxon of Dendrotriton). However, their analysis included only a single representative of each genus, and excluded three genera.
Mapping elevational distributions onto our phylogeny (figure 2) using generalized least squares shows that mid-elevations (approx. 1000–2000 m) were colonized relatively early in the history of bolitoglossines, and that there have been subsequent invasions of low elevation habitats (in Oedipina and Bolitoglossa). Major invasions of the highest elevations have occurred within Pseudoeurycea, but some other taxa (e.g. Bolitoglossa) also extend into very high elevations. The Bayesian reconstruction for the ancestor of tropical bolitoglossines also suggests that mid-elevations were colonized early, and lower and higher elevations subsequently (Pp=0.931 for elevational band from 1201 to 1800 m), and shows that this general result is robust to an alternate reconstruction method and to uncertainty in the topology and branch lengths.
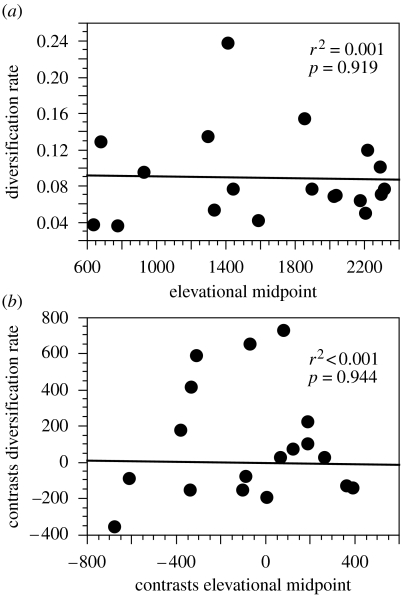
Using 19 clades that together include all known tropical bolitoglossine species, we find no relationship between rates of diversification of clades and elevational distribution of clades, regardless of whether we analyse the raw data (figure 3a) or phylogenetically independent contrasts (figure 3b). Results also are similar (no significant relationship) using different ages for Bolitoglossinae, using equal branch lengths for independent contrasts, and assuming a high extinction rate when estimating diversification rates (results not shown). Note that one clade (adspersa group of Bolitoglossa) has an unusually high diversification rate, but this clade is found primarily in South America (26 out of 31 species) and has relatively little impact on overall species richness patterns in MesoAmerica (deleting this clade gives similar results; r2=0.005, p=0.780). Data on species numbers, estimated age (including assessment of confidence), diversification rate and elevational distribution of each clade are provided in electronic supplementary material 3.
Figure 3.
Rate of diversification of clades versus average elevational midpoint of clades for tropical bolitoglossine salamanders, using (a) raw data and (b) phylogenetically independent contrasts (using branch lengths from figure 2). The results shown assume a low extinction rate, but results are similar using a high extinction rate.
We identified four branches on which there are significant shifts in diversification rates, based on the 138 sampled species (figure 2). However, changes in elevational ranges on these branches are not significantly different from those on other branches (mean difference=−5.478, d.f.=270, p=0.9721), further confirming that there is no significant association between changes in elevational range and diversification rate.
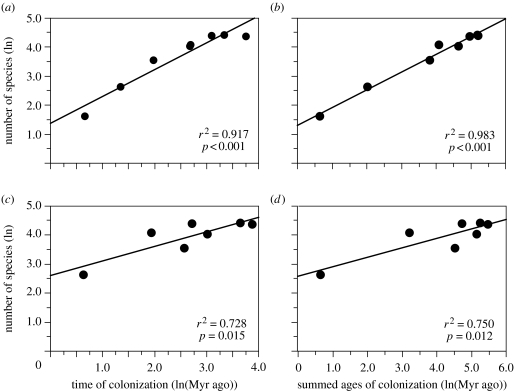
We found a strong relationship between the estimated timing of colonization of elevational bands and the present species richness of these bands, regardless of whether we use the age of the first colonization of each band (figure 4a) or the summed ages of each colonization (figure 4b). Results also are similar using the lowest elevation of occurrence of each species rather than the midpoint of their elevational ranges (figure 4c,d).
Figure 4.
The species richness of 500 m elevational bands versus the estimated date (Myr ago) when the elevational band was first colonized, for tropical bolitoglossine salamanders in MesoAmerica, using (a,c) the oldest colonization of each elevational band, (b,d) the summed ages of each colonization of each band, (a,b) the reconstructed elevational midpoint of each species, and (c,d) the reconstructed minimum elevation of each species. Both variables are natural log-transformed.
4. Discussion
What explains the widespread hump-shaped pattern of elevational species richness? Our results from tropical bolitoglossine salamanders suggest that, contrary to the species-pump model, diversification does not appear to be any more rapid in montane regions than in lowland regions. Instead, the hump-shaped pattern seemingly is related to the relative timing of colonization of different elevational zones. Specifically, intermediate elevation habitats were colonized early in the phylogenetic history of the group, and the lowest and highest elevations were colonized more recently. Thus, given that rates of species origination and extinction are similar at different elevations, it appears that there are more species at intermediate elevations simply because bolitoglossines have been present, speciating and accumulating in those habitats for longer periods of time than in lower-or higher-elevation habitats. Although there is some uncertainty in parts of our phylogenetic hypothesis, the relatively derived placements of the low and high elevation clades are strongly supported (figure 2) and our Bayesian reconstruction of a mid-elevation ancestor is robust to uncertainty in the topology and branch lengths. Our results support the idea that the relative timing of colonization (i.e. time-for-speciation effect; Stephens & Wiens 2003) may strongly influence patterns of species richness in different habitats within a region, in addition to influencing patterns at regional, continental and global scales. Our results do not directly address local-scale patterns of elevational diversity (e.g. on a single mountain slope), but it seems probable that the regional-scale patterns we address strongly influence patterns at smaller spatial scales.
Our analyses suggest that the relative timing of biogeographic dispersal is critical for explaining elevational patterns of species richness in this group, but our study does not directly address the underlying ecological and evolutionary processes that explain the biogeographic patterns. We hypothesize that limited climatic tolerances (i.e. niche conservatism; Ricklefs & Latham 1992; Peterson et al. 1999; Wiens & Graham 2005) might restrict dispersal of bolitoglossine lineages between different elevational zones. Climatic zonation is thought to be particularly important in limiting dispersal between elevations in tropical montane regions (Janzen 1967), and there is some support for this hypothesis from ecophysiological studies of tropical and temperate salamanders (e.g. Feder & Lynch 1982). Previous authors have noted that species and clades of tropical bolitoglossines tend to occur in narrow elevational bands (generally less than 1300 m; Elias 1984). Our analyses of the mid-domain effect for bolitoglossine clades separately show that each clade has a more restricted or concentrated elevational range than expected under a null model of random range placement (results not shown), which may reflect limited climatic tolerances.
Niche conservatism may also explain why bolitoglossines colonized montane mid-elevations in MesoAmerica before colonizing the tropical lowlands. Throughout their ca 200 Myr of evolutionary history, salamanders have shown only limited colonization of tropical regions (Milner 2000; Zug et al. 2001). Recent plethodontid phylogenies (Chippindale et al. 2004; Mueller et al. 2004; Min et al. 2005) suggest that bolitoglossines colonized MesoAmerica from temperate North America. In fact, all other plethodontid subfamilies occur primarily in temperate North America, as do the two successive outgroups to Plethodontidae (Amphiumidae and Rhyacotritonidae; Wiens et al. 2005a). The colonization of cooler montane regions may be the easiest way for a temperate group to initially colonize a tropical region (e.g. Smith et al. 2005), providing a climatic and ecophysiological ‘path of least resistance’. Indeed, field body temperatures of tropical bolitoglossines are extremely similar to those of terrestrial plethodontids in temperate North America (average for 43 species of tropical bolitoglossines=14.2°C; average for 28 species of temperate plethodontids=13.5°C; Feder et al. 1982). Furthermore, many montane regions that presently harbour high bolitoglossine diversity (e.g. Sierra Madre Oriental, Sierra Madre del Sur, nuclear Central American highlands) are known to have been present since the Laramide Revolution of the Late Cretaceous to Early Tertiary (Campbell 1999), which is consistent with these montane regions having been in place before the origin of the tropical bolitoglossines. Other potential explanations, which are not mutually exclusive, are that biotic interactions with the lowland fauna limit dispersal (e.g. competition) or that some lineages of bolitoglossines colonized lowland habitats in the more distant past but have now gone extinct.
The results of our study may have important conservation implications. In MesoAmerica, mid-elevation habitats not only harbour more species of bolitoglossines than lowlands, but also seem likely to harbour more phylogenetic diversity among species, given the same number of species (i.e. many older montane clades; figure 2). If the pattern found in bolitoglossines characterizes other groups as well, it would suggest even greater urgency for protecting montane habitats in MesoAmerica (and possibly other regions) than indicated by the number of species alone.
These analyses must be extended to other groups and regions to test the generality of this hypothesized explanation for the hump-shaped diversity pattern. Analyses in MesoAmerican hylid frogs (tribe Hylini) suggest that similar factors explain the hump-shaped pattern in that clade as well (Smith et al. in press). Intriguingly, the same two factors that seem to underlie elevational diversity patterns in tropical bolitoglossines (i.e. niche conservatism and the time-for-speciation effect) may also be important evolutionary factors contributing to the latitudinal diversity gradient (e.g. Wiens & Donoghue 2004; Wiens et al. 2006). Thus, these two factors may help explain a variety of latitudinal and elevational patterns of biodiversity.
Acknowledgments
Research was supported by NSF grants DEB-0334923 and DEB-0331747 (to J.J.W.), EF-0334939 (to D.B.W.), and Semarnat-Conacyt 2002-C01-0015 and PAPIIT-UNAM IN226605 (to G.P.O.). We thank K. Kozak, C. McCain, D. Moen and anonymous reviewers for their comments on the manuscript and L. Márquez (Laboratorio de Biología Molecular, I-Biología, UNAM) for technical assistance and support.
Supplementary Material
Elevational distributions of tropical bolitoglossine species
GenBank numbers for DNA sequence data
Estimated elevational distribution, diversity, age, and diversification rate for clades
References
- AmphibiaWeb: Information on amphibian biology and conservation. [web application]. 2006 Berkeley, CA: AmphibiaWeb. Available: http://amphibiaweb.org/ (Last Accessed: February 26, 2006).
- Brandley M.C, Schmitz A, Reeder T.W. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 2005;54:373–390. doi: 10.1080/10635150590946808. doi:10.1080/10635150590946808 [DOI] [PubMed] [Google Scholar]
- Campbell J.A. Distribution patterns of amphibians in Middle America. In: Duellman W.E, editor. Patterns of distribution of amphibians. A global perspective. Johns-Hopkins University Press; Baltimore, MD: 1999. pp. 111–210. [Google Scholar]
- Chippindale P.T, Bonett R.M, Baldwin A.S, Wiens J.J. Phylogenetic evidence for a major reversal in life history evolution in plethodontid salamanders. Evolution. 2004;58:2809–2822. doi: 10.1111/j.0014-3820.2004.tb01632.x. doi:10.1554/04-185R [DOI] [PubMed] [Google Scholar]
- Colwell R.K, Hurtt G.C. Nonbiological gradients in species richness and a spurious Rapoport effect. Am. Nat. 1994;144:570–595. doi:10.1086/285695 [Google Scholar]
- Elias P. Salamanders of the northwestern highlands of Guatemala. Contrib. Sci. Nat. Hist. Mus. Los Angeles Co. 1984;348:1–20. [Google Scholar]
- Elias P, Wake D.B. Nyctanolis pernix, a new genus and species of plethodontid salamander from northwestern Guatemala and Chiapas, Mexico. In: Rhodin G.J, Miyata K, editors. Advances in herpetology and evolutionary biology: essays in honor of Ernest E. Williams. Museum of Comparative Zoology, Harvard; Cambridge, MA: 1983. pp. 1–12. [Google Scholar]
- Feder M.E, Lynch J.F. Effects of latitude, season, elevation, and microhabitat on field body temperatures of neotropical and temperate zone salamanders. Ecology. 1982;63:1657–1664. doi:10.2307/1940107 [Google Scholar]
- Feder M.E, Lynch J.F, Shaffer H.B, Wake D.B. Field body temperatures of tropical and temperate zone salamanders. Smiths Herpetol. Inf. Serv. 1982;52:1–23. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. doi: 10.1086/703055. doi:10.1086/284325 [DOI] [PubMed] [Google Scholar]
- Frost D.R, et al. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 2006;297:1–370. doi:10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2 [Google Scholar]
- García-París M, Wake D.B. Molecular phylogenetic analysis of relationships of the tropical salamander genera Oedipina and Nototriton, with descriptions of a new genus and three new species. Copeia. 2000;2000:42–70. [Google Scholar]
- García-París M, Good D.A, Parra-Olea G, Wake D.B. Biodiversity of Costa Rican salamanders: implications of high levels of genetic differentiation and phylogeographic structure for species formation. Proc. Natl Acad. Sci. USA. 2000;97:1640–1647. doi: 10.1073/pnas.97.4.1640. doi:10.1073/pnas.97.4.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K.J, Blackburn T.M. Blackwell; Oxford, UK: 2000. Pattern and process in macroecology. [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., Nielsen, R. & Bollback, J. P. 2003 Stochastic mapping of morphological characters. Syst. Biol.52, 131–158. (doi:10.1080/10635150390192780) [DOI] [PubMed]
- IUCN, Conservation International, and NatureServe. 2004 Global Amphibian Assessment. www.globalamphibians.org
- Janzen D.H. Why mountain passes are higher in the tropics. Am. Nat. 1967;101:233–249. doi:10.1086/282487 [Google Scholar]
- Lewis P.O. A likelihood approach to estimating phylogeny from discrete morphological characters. Syst. Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. doi:10.1080/106351501753462876 [DOI] [PubMed] [Google Scholar]
- Lomolino M.V, Riddle B.R, Brown J.H. 3rd edn. Sinauer; Sunderland, MA: 2005. Biogeography. [Google Scholar]
- Maddison W.P. Gene trees in species trees. Syst. Biol. 1997;46:523–536. doi:10.2307/2413694 [Google Scholar]
- Magallón S, Sanderson M.J. Absolute diversification rates of angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Martins, E. P. 2004 Compare, version 4.6. Computer programs for the statistical analysis of comparative data. Distributed by the author at http://compare.bio.indiana.edu/ Department of Biology, Indiana University, Bloomington IN.
- Martins E.P, Hansen T.F. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 1997;149:646–667. doi:10.1086/286013 [Google Scholar]
- McCain C.M. The mid-domain effect applied to elevational gradients: species richness of small mammals in Costa Rica. J. Biogeogr. 2004;31:19–31. [Google Scholar]
- McCain C.M. Elevational gradients in diversity of small mammals. Ecology. 2005;86:366–372. [Google Scholar]
- McPeek M.A. Testing hypotheses about evolutionary change on single branches of a phylogeny using evolutionary contrasts. Am. Nat. 1995;145:686–703. doi:10.1086/285763 [Google Scholar]
- Milner, A. R. 2000 Mesozoic and Tertiary Caudata and Albanerpetontidae. In Amphibian biology. vol. 4. Paleontology. (eds H. Heatwole & R. L. Carroll), pp. 31–108. Chipping Norton, Australia: Surrey Beatty.
- Min M.S, Yang S.Y, Bonett R.M, Vieites D.R, Brandon R.A, Wake D.B. Discovery of the first Asian plethodontid salamander. Nature. 2005;435:87–90. doi: 10.1038/nature03474. doi:10.1038/nature03474 [DOI] [PubMed] [Google Scholar]
- Moritz C, Patton J.L, Schneider C.J, Smith T.B. Diversification of rainforest faunas: an integrated molecular approach. Annu. Rev. Ecol. Syst. 2000;31:533–563. doi:10.1146/annurev.ecolsys.31.1.533 [Google Scholar]
- Mueller R.L, Macey J.R, Jaekel M, Wake D.B, Boore J.L. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc. Natl Acad. Sci. USA. 2004;101:13 820–13 825. doi: 10.1073/pnas.0405785101. doi:10.1073/pnas.0405785101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S, May R.M, Harvey P.H. The reconstructed evolutionary process. Phil. Trans. R. Soc. B. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. doi:10.1098/rstb.1994.0068 [DOI] [PubMed] [Google Scholar]
- Nylander, J. A. A. 2004 MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. (http://www.ebc.uu.se/systzoo/staff/nylander.html).
- Nylander J.A.A, Ronquist F, Huelsenbeck J.P, Nieves-Aldrey J.L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. doi:10.1080/10635150490264699 [DOI] [PubMed] [Google Scholar]
- Oomen M.A, Shanker K. Elevational species richness patterns emerge from multiple local mechanisms in Himalayan woody plants. Ecology. 2005;86:3039–3047. [Google Scholar]
- Parra-Olea G. Molecular phylogenetic relationships of neotropical salamanders of the genus Pseudoeurycea. Mol. Phylogenet. Evol. 2002;22:234–246. doi: 10.1006/mpev.2001.1048. doi:10.1006/mpev.2001.1048 [DOI] [PubMed] [Google Scholar]
- Parra-Olea G. Phylogenetic relationships of the genus Chiropterotriton (Caudata: Plethodontidae) based on 16S ribosomal mtDNA. Can. J. Zool. 2003;81:2048–2060. doi:10.1139/z03-155 [Google Scholar]
- Parra-Olea G, Wake D.B. Extreme morphological and ecological homoplasy in tropical salamanders. Proc. Natl Acad. Sci. USA. 2001;98:7888–7891. doi: 10.1073/pnas.131203598. doi:10.1073/pnas.131203598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Olea G, García-París M, Wake D.B. Phylogenetic relationships among the salamanders of the Bolitoglossa macrinii species group (Amphibia: Plethodontidae), with descriptions of two new species from Oaxaca (México) J. Herpetol. 2002;36:356–366. [Google Scholar]
- Parra-Olea G, García-París M, Wake D.B. Molecular diversification of salamanders of the tropical American genus Bolitoglossa (Caudata: Plethodontidae) and its evolutionary and biogeographical implications. Biol. J. Linn. Soc. 2004;81:325–346. doi:10.1111/j.1095-8312.2003.00303.x [Google Scholar]
- Parra-Olea G, García-París M, Papenfuss T.J, Wake D.B. Systematics of the Pseudoeurycea bellii (Caudata: Plethodontidae) species complex. Herpetologica. 2005;61:145–158. doi:10.1655/03-02 [Google Scholar]
- Peterson A.T, Soberon J, Sanchez-Cordero V. Conservatism of ecological niches in evolutionary time. Science. 1999;285:1265–1267. doi: 10.1126/science.285.5431.1265. doi:10.1126/science.285.5431.1265 [DOI] [PubMed] [Google Scholar]
- Rahbek C. The elevational gradient of species richness: a uniform pattern? Ecography. 1995;18:200–205. doi:10.1111/j.1600-0587.1995.tb00341.x [Google Scholar]
- Rahbek C. The relationship among area, elevation, and regional species richness in neotropical birds. Am. Nat. 1997;149:875–902. doi: 10.1086/286028. doi:10.1086/286028 [DOI] [PubMed] [Google Scholar]
- Rahbek C, Graves G.R. Multiscale assessment of patterns of avian species richness. Proc. Natl Acad. Sci. USA. 2001;98:4534–4539. doi: 10.1073/pnas.071034898. doi:10.1073/pnas.071034898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Harvey P.H, Nee S. End-Epi: an application for reconstructing phylogenetic and population processes from molecular sequences. Comput. Appl. Biosci. 1997;13:303–306. doi: 10.1093/bioinformatics/13.3.303. [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E. A comprehensive framework for global patterns in biodiversity. Ecol. Lett. 2004;7:1–15. doi:10.1046/j.1461-0248.2003.00554.x [Google Scholar]
- Ricklefs R.E, Latham R.E. Intercontinental correlation of geographical ranges suggests stasis in ecological traits of relict genera of temperate perennial herbs. Am. Nat. 1992;139:1305–1321. doi:10.1086/285388 [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. doi:10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Smith S.A, Stephens P.R, Wiens J.J. Replicate patterns of species richness, historical biogeography, and phylogeny in holarctic treefrogs. Evolution. 2005;59:2433–2450. doi:10.1554/05-284.1 [PubMed] [Google Scholar]
- Smith, S. A., Nieto Montes de Oca, A., Reeder, T. W. & Wiens, J. J. In press. A phylogenetic perspective on elevational species richness patterns in Middle American treefrogs: why so few species in lowland tropical forests. Evolution [DOI] [PubMed]
- Stephens P.R, Wiens J.J. Explaining species richness from continents to communities: the time-for-speciation effect in emydid turtles. Am. Nat. 2003;161:112–128. doi: 10.1086/345091. doi:10.1086/345091 [DOI] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake D.B, Lynch J.F. The distribution, ecology, and evolutionary history of plethodontid salamanders in tropical America. Sci. Bull. Nat. Hist. Mus. Los Angeles Co. 1976;25:1–65. [Google Scholar]
- Wiens J.J. Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst. Biol. 1998;47:625–640. doi: 10.1080/106351598260635. doi:10.1080/106351598260635 [DOI] [PubMed] [Google Scholar]
- Wiens, J. J. In press. Global patterns of species richness and diversification in amphibians. Am. Nat [DOI] [PubMed]
- Wiens J.J, Donoghue M.J. Historical biogeography, ecology, and species richness. Trends Ecol. Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. doi:10.1016/j.tree.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Graham C.H. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Evol. Syst. 2005;36:519–539. doi:10.1146/annurev.ecolsys.36.102803.095431 [Google Scholar]
- Wiens J.J, Bonett R.M, Chippindale P.T. Ontogeny discombobulates phylogeny: paedomorphosis and higher-level salamander relationships. Syst. Biol. 2005a;54:91–110. doi: 10.1080/10635150590906037. doi:10.1080/10635150590906037 [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Fetzner J.W, Parkinson C.L, Reeder T.W. Hylid frog phylogeny and sampling strategies for speciose clades. Syst. Biol. 2005b;54:719–748. doi: 10.1080/10635150500234625. doi:10.1080/10635150500234625 [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Graham C.H, Moen D.S, Smith S.A, Reeder T.W. Evolutionary and ecological causes of the latitudinal diversity gradient in hylid frogs: treefrog trees unearth the roots of high tropical diversity. Am. Nat. 2006;168:579–596. doi: 10.1086/507882. doi:10.1086/507882 [DOI] [PubMed] [Google Scholar]
- Willig M.R, Kaufman D.M, Stevens R.D. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 2003;34:273–309. doi:10.1146/annurev.ecolsys.34.012103.144032 [Google Scholar]
- Zug G.R, Vitt L.J, Caldwell J.P. An introductory biology of amphibians and reptiles. 2nd edn. Academic Press; San Diego, CA: 2001. Herpetology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Elevational distributions of tropical bolitoglossine species
GenBank numbers for DNA sequence data
Estimated elevational distribution, diversity, age, and diversification rate for clades