Abstract
Allochronic speciation refers to a mode of sympatric speciation in which the differentiation of populations is primarily due to a phenological shift without habitat or host change. However, it has been so far rarely documented. The present paper reports on a plausible case of allochronic differentiation between sympatric populations of the pine processionary moth (PPM), Thaumetopoea pityocampa. The PPM is a Mediterranean insect with winter larval development. A phenologically atypical population with early adult activity and summer larval development was detected 10 years ago in Portugal. Mitochondrial and nuclear sequences strongly suggest that the ‘summer’ individuals are closely related to the sympatric winter population, while microsatellite data show a reduction in allelic richness, a distortion of allelic frequencies and significant genetic differentiation. Moreover, monitoring of adult flights suggests that reproductive activity does not overlap between the summer and winter populations. We postulate that the summer population appeared after a sudden phenological shift of some individuals of the sympatric winter population, leading to a founder effect and complete reproductive isolation. Given that the individuals showing this new phenology are subject to different selection pressures, the observed allochronic differentiation may rapidly lead to deeper divergence.
Keywords: allochronic isolation, microsatellite, phenology, pine processionary moth, founder effect, sequencing
1. Introduction
Speciation is the process through which one species diverges into different strains that ultimately become reproductively isolated and evolutionarily independent. ‘Allopatric speciation’ refers to any speciation process that resulted initially from spatial separation. It has been the dominant view of speciation for the past decades (Turelli et al. 2001; Via 2001; Berlocher & Feder 2002). On the contrary, sympatric speciation—which occurs without geographical separation through ecological isolation of breeding populations—has been extremely controversial. Yet, many recent empirical studies as well as mathematical models have provided evidence that sympatric speciation can occur (Turelli et al. 2001; Via 2001; Berlocher & Feder 2002). The most documented case concerns the cichlid fishes that speciated via ecological specialization and sexual selection (Kornfield & Smith 2000) and many phytophagous insects for which populations primarily diverged through host plant or habitat specialization (e.g. Wood et al. 1999; Dres & Mallet 2002). Yet, a model called ‘allochronic speciation’, in which speciation results initially from temporal separation alone (i.e. without geographical isolation or colonization of new hosts or habitats), was proposed as early as 1960 in field crickets (Alexander & Bigelow 1960), even though it was not supported in that case by further phylogenetic data (Harrison 1979). Plausible cases of allochronic speciation are still scarce in the literature and concern mainly the 13- and 17-year periodical cicadas (Ritchie 2001). Such a model of speciation has also been hypothesized for gall-forming aphids (Abbot & Withgott 2004). In both the cases, the sister species are fully separated and the mode of speciation was inferred from phylogeny and comparison of biological cycles. Very recent genetic divergence and allochronic reproductive isolation were described in a hybrid zone for swallowtail butterflies (Scriber & Ording 2005). We here report on an ongoing case of allochronic differentiation in a phytophagous insect that experienced a local and probably sudden phenological shift.
The pine processionary moth (PPM), Thaumetopoea pityocampa (Lepidoptera: Notodontidae), belongs to a genus that presents diverse phenologies, with larval development occurring either in summer or in winter depending on the species. Thaumetopoea pityocampa is native to the Mediterranean Basin and has a typical winter larval development (Démolin 1969). Adults lay eggs on pine leaves in summer and caterpillars feed on the needle-shaped leaves during autumn and winter. They pupate in the soil in late winter or early spring, and newly emerged adults disperse to reproduce during summer. In August 1997, an aberrant population with summer larval development was found in the southern region of the National Pinewood of Leiria, Portugal, in a huge outbreak situation. Larvae were mostly in the late instars of their development and pupated in September. In subsequent years, caterpillars were found to develop between mid-June and September, and pupated in the soil until May, when adults' flight and egg laying started (M. Branco 2002, personal observation). This unusual population, hereafter called the Leiria ‘summer population’ (SP), coexists in the same area with a ‘normal’ population with winter larval development, hereafter called the Leiria ‘winter population’ (WP).
The aim of our study was to determine whether reproductive isolation was in process between the summer and winter populations of T. pityocampa, and to determine the origin of the SP and the genetic consequences of this novel phenology. (i) As cases of cryptic morphological species are frequent in insects, we first reconstructed the phylogenetic relationships of the SP and the surrounding WPs using mitochondrial and nuclear sequence data to check whether they belonged to the same species and to determine the level of divergence. (ii) We also used polymorphic microsatellite markers to infer the genetic characteristics of the SP when compared with normal WPs. (iii) Finally, we monitored adult flights of both the phenological populations to assess whether adult activity overlapped between SP and WP.
2. Material and methods
(a) Sampling and DNA extraction
Samples of T. pityocampa were collected in Portugal, Spain and France between 2001 and 2005. Samples consisted of 25–35 larvae collected from different trees to prevent sampling siblings, except in Apostiça (Portugal) where adult males were sampled. Additionally, larvae of the congeneric species Thaumetopoea wilkinsoni, Thaumetopoea processionea, Thaumetopoea solitaria and Thaumetopoea pinivora were collected to infer interspecific divergence in the genus Thaumetopoea. All insects were killed and stored in absolute alcohol until DNA extraction. Collection site descriptions are available as electronic supplementary material.
DNA was extracted from the whole body of PPM larvae and from the legs and thorax of adults, using the GenElute mammalian Genomic DNA Miniprep kit (Sigma), and eluted in 200 μl of buffer.
(b) PCR amplification and sequencing of COI and ITS1
Part of the mitochondrial cytochrome oxidase I (COI) gene and the nuclear internal transcribed spacer 1 (ITS1) was amplified and sequenced, respectively, for four to six (COI) and for two (ITS1) individuals per population. PCR amplifications were performed using the primer pair C1-J-2183 (Jerry) 5′CAACATTTATTTTGATTTTTTGG3′ and TL2-N-3014 (Pat) 5′TCCAATGCACTAATCTGCCATATTA3′ for COI (Simon et al. 1994), and ITS1F 5′GCGTTCGAAATGCGATGATCAA3′ and ITS1R 5′GTAGGTGAACCTGCAGAAGG3′ for ITS1 (Vogler & DeSalle 1994). Annealing temperatures were set to 48 and 50°C for COI and ITS1, respectively. The PCR products were purified using the GenElute PCR clean-up kit (Sigma) and directly sequenced in both directions. Sequencing was performed using the BigDye terminator sequencing kit (Applied Biosystems) and carried out with an ABI 3100 automatic sequencer.
(c) Microsatellite genotyping
Five microsatellite loci were used to genotype approximately 30 individuals per population in six localities of the Iberian Peninsula. Four of these, namely MS-Thpit1, MS-Thpit3, MS-Thpit4 and MS-Thpit5, are described elsewhere (Rousselet et al. 2004). PCR primers were designed for a new microsatellite locus (MS-Thpit6) isolated from the same microsatellite library (motif (GA)16CA(GA)2, GenBank accession number EF 190999) as MS-Thpit6F 5′TCCCAAGCACTCTCGCTTTC3′ and MS-Thpit6R 5′ATAACGTGGGATGCTCAGCG3′. Amplification conditions were the same as for MS-Thpit1. Fluorescent PCR products were run and detected on an ABI 3100 automatic sequencer and product sizes were determined using the GeneScan software (Applied Biosystems).
(d) Analyses of molecular data
All the obtained sequences for COI and ITS1 were aligned using ClustalW (Thompson et al. 1994), as implemented in Bioedit v. 7.0. The best-fit model of sequence evolution was estimated for COI sequences using the Akaike information criterion (Posada & Buckley 2004) with Modeltest v. 3.7 (Posada & Crandall 1998). The chosen model of sequence evolution was applied to calculate genetic distances between haplotypes and between species with PAUP* v. 4b10 (Swofford 2003). For each of the COI and ITS1 datasets, a statistical parsimony network was computed using the TCS v. 1.21 software (Clement et al. 2000), which estimates gene genealogies from DNA sequences following the method described by Templeton et al. (1992). For the ITS1 dataset that contained both indels and substitutions, gaps were treated as a fifth base and any insertion of more than one base was considered as a single evolutionary event.
Concerning microsatellite data, allelic richness and frequencies were calculated using Genetix v. 4.04 (Belkhir et al. 1996–2004). Population structure was analysed using pairwise FST (Weir & Cockerham 1984) and calculated using Arlequin v. 3.0 (Excoffier et al. 2005), their significance being estimated with 3000 permutations. Neighbour-joining trees of populations were constructed using Cavalli-Sforza and Edwards' chord distance using Populations v. 1.2.28 (O. Langella, http://www.pge.cnrs-gif.fr/bioinfo/populations/index.php). Bootstrap values were computed by resampling loci and are given as per cent values of 2000 replications. To test for population bottleneck, we used the program Bottleneck v. 1.2 (Cornuet & Luikart 1996; Piry et al. 1999) using both the mode-shift test and the Wilcoxon test for heterozygote excess under the stepwise mutation model and the two-phase model. We chose the Wilcoxon test because it is preferable when few loci are used (Cornuet & Luikart 1996; Cornuet et al. 1999).
(e) Males' flight period
The flight of adult males was monitored in Leiria from early May to the end of September 2005. Funnel traps baited with synthetic PPM pheromone dispensers (pityolure 40 mg) were hung on trees at a reachable height. In Leiria, 15 traps were distributed in a south–north transect at 1–3 km intervals. Male activity of a WP was also monitored in Apostiça using five traps. Pheromone dispensers were replaced every six weeks. Traps were assessed weekly in Leiria and every two weeks in Apostiça.
3. Results
(a) Sequence data
(i) COI
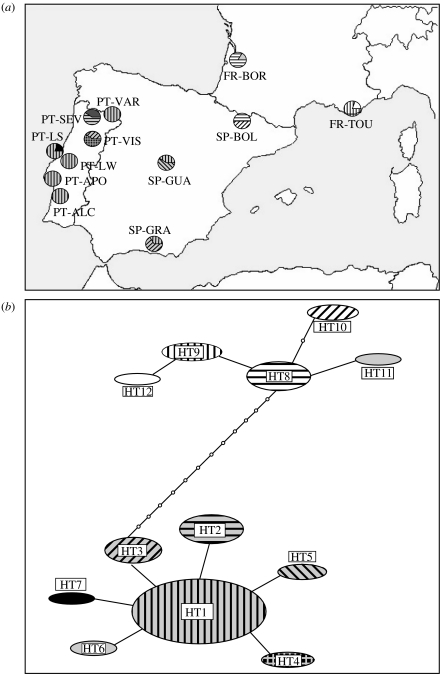
We obtained 65 sequences, 736 bp long, for T. pityocampa from Portugal, Spain and France, as well as for four T. wilkinsoni, five T. pinivora and five T. solitaria sequences. All sequences are available in GenBank under accession numbers EF 185128–EF 185146. The dataset for T. pityocampa corresponded to 12 haplotypes and 23 polymorphic sites. Figure 1 shows the distribution of the 12 haplotypes in the sampled populations, as well as the haplotype network. The pityocampa haplotypes fell in two clades separated by 12 mutation steps. One clade comprised all the haplotypes (HT8–HT12) found in France and near the Pyrenees (Boltaña, Spain), whereas the other clade grouped all other haplotypes from Spain and Portugal (HT1–HT7). Interestingly, most individuals of the Leiria SP had the same haplotype as Leiria WP and two neighbouring winter populations (HT1). Yet, one individual of the SP had a unique haplotype that differed from HT1 by one mutation.
Figure 1.
Haplotype distribution and network of T. pityocampa COI sequences. (a) Geographical distribution of the haplotypes among the 12 sampled populations. PT-VAR: Varges; PT-SEV: Sevivas; PT-VIS: Viseu; PT-LS: Leiria SP; PT-LW; Leiria WP; PT-APO: Apostica; PT-ALC: Alcacer; SP-BOL: Boltaña; SP-GUA: Guadarrama; SP-GRA: Granada; FR-BOR: Bordeaux; FR-TOU: Toulon. (b) Haplotype network of the 12 haplotypes with the corresponding shaded codes. Haplotype frequencies are represented by the circle area. Each line corresponds to a mutational step and each empty circle to a missing intermediate haplotype.
The most appropriate model of sequence evolution was the GTR+I+G model, including a proportion of invariable sites (I=0.63) and gamma distribution shape parameter (G=7.35) with unequal base frequencies (freqA=0.314; freqC=0.161; freqG=0.133; freqT=0.392) and a rate matrix [(A−C)=16 518 312; (A−G)=20 208 544; (A−T)=2745754.75; (C−G)=5.71; (C−T)=92 313 760; (G−T)=1]. Using that model, between-haplotype distances were between 0.0013 and 0.0027 within the Iberian clade, between 0.0013 and 0.0054 within the ‘French clade’ and between 0.0190 and 0.0255 between both the groups. All distances thus clearly fell in the intraspecific range for the genus Thaumetopoea, as the interspecific distances measured fell between 0.121 (T. pityocampa–T. wilkinsoni) and 0.419 (T. pinivora–T. solitaria).
(ii) ITS1
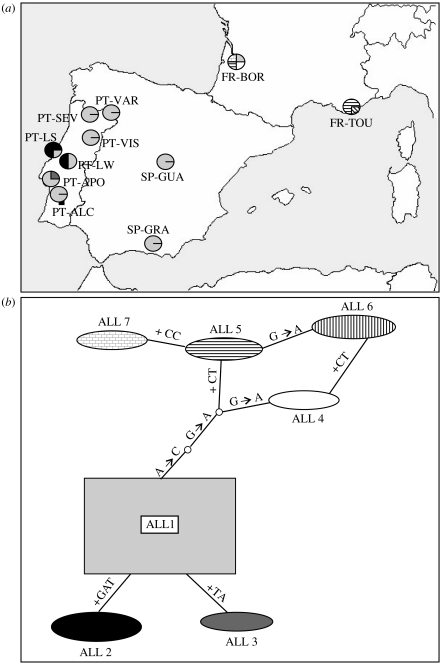
We obtained 513–518 bp long sequences for 22 individuals of T. pityocampa from France, Spain and Portugal, and 435–559 bp long sequences for two individuals of T. wilkinsoni, two T. pinivora, two T. processionea and two T. solitaria. All sequences are available in GenBank (EF 189679–EF 189687). The final alignment of all T. pityocampa was 522 bp long, including gaps. It showed seven alleles differing by both insertions and substitution events. Most of the individuals were homozygous. Both alleles of heterozygous individuals could be unambiguously identified without cloning, by direct sequencing of both strands. Three alleles (ALL1–ALL3) were mostly found in the Iberian Peninsula, while four (ALL4–ALL7) were exclusively found in the French populations (figure 2a). Interestingly, ALL2 was only found in Leiria and was shared between SP and WP. The network shows the relationships between alleles (figure 2b). We could only align the ITS1 gene between T. pityocampa and T. wilkinsoni. The differences between species were much higher than the inter-haplotype variations observed within T. pityocampa, as there were 7 indels (1–20 bp) and 17 substitutions between T. pityocampa and T. wilkinsoni. The sequences obtained from all other species were too divergent and could not be unambiguously aligned.
Figure 2.
Allelic distribution and network of T. pityocampa ITS1 sequences. (a) Geographical distribution of the alleles among the 11 sampled populations. (b) Network of the seven alleles with the corresponding shaded codes. Allelic frequencies are represented by the circle area. Each line corresponds to a mutational step (either a substitution or an indel) and each empty circle to a missing intermediate allele.
(b) Microsatellite results
We genotyped 182 individuals for the five microsatellite loci. The total number of alleles per locus ranged from 8 for MS-Thpit5 to 34 for MS-Thpit6. The mean number of alleles per population was as low as 3.4 in the SP, while it was 8.6–9 in all other populations. Distributions of allelic frequencies per population and per locus are available as electronic supplementary material. For all loci, the lowest number of alleles was found in the SP, which also had a reduced number of rare alleles. Furthermore, in several cases, allelic frequencies were distorted comparatively to the frequencies observed in the surrounding WPs. For instance, MS-Thpit1 was almost fixed for allele 165 (frequency 0.97), although this allele never exceeded a proportion of 0.61 in Portuguese WPs. Similarly, the main alleles of MS-Thpit3 and MS-Thpit6 (allele 239, frequency 0.47 and allele 166, frequency 0.75, respectively) were systematically found within frequencies below 0.08 and 0.07 (respectively) in WPs.
For the four loci, MS-Thpit1, MS-Thpit3, MS-Thpit4 and MS-Thpit5, all alleles found in the depauperate SP in Leiria were also present in the sympatric WP. For the locus MS-Thpit6, six alleles were present in the SP, of which one was found with a frequency of 0.75. Four of these alleles (including the main one) were also found in the Leiria WP and the fifth one was found in a preliminary study of the WP in caterpillars sampled in 2004 (not shown). The last allele of the SP was found only in one heterozygous individual; that particular allele was not found in the Leiria WP, but was present in the Portuguese population of Viseu.
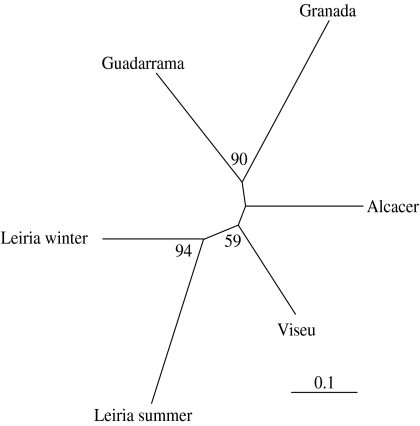
The phylogenetic tree of populations (figure 3) showed that the Leiria SP and WP were significantly grouped together (bootstrap value 92), and that the two Spanish populations also formed a well-supported clade. The pairwise FST showed that all pairs of populations were significantly structured (table 1). The SP of Leiria had the highest pairwise FST values, irrespective of the population it was compared to. No significant bottleneck effect was revealed.
Figure 3.
Neighbour-joining tree of T. pityocampa populations obtained from microsatellite data.
Table 1.
Pairwise FST estimates between the six genotyped populations. (*p<0.01.)
| Leiria summer | Leiria winter | Alcacer | Viseu | Guadarrama | Granada | |
|---|---|---|---|---|---|---|
| Leiria summer | — | 0.16* | 0.18* | 0.19* | 0.22* | 0.26* |
| Leiria winter | — | 0.07* | 0.03* | 0.12* | 0.12* | |
| Alcacer | — | 0.04* | 0.04* | 0.08* | ||
| Viseu | — | 0.05* | 0.07* | |||
| Guadarrama | — | 0.09* | ||||
| Granada | — |
(c) Males' flight period
A total of 156 males were sampled in Leiria. The first captures were recorded on 12th May, reaching a maximum of 52 individuals on 3rd June. No adult was captured between 1st July and 5th August, but six WP males were trapped in August and September. In Apostiça, the WP flight began on 17th August and extended until 4th October, the maximum being reached on 31st August. A total of 306 males were caught during this period.
4. Discussion
(a) Molecular identification and the origin of the summer population
Both mitochondrial and nuclear data suggest that the individuals sampled from the Leiria SP belong to the same species as the classical WPs sampled from Portugal, Spain and France, rather than to a cryptic Thaumetopoea species. Furthermore, the network of COI haplotypes shows a significant divergence between Iberian and French–Pyrenean haplotypes, which probably reflects the effective role of barrier to gene flow that the Pyrenean mountains played in the Late Tertiary or Quaternary, during post-glacial recolonization(s). Even though the study of the phylogeographical pattern of the PPM is beyond the scope of the present paper, this result clearly shows that the SP is genetically similar to other Iberian individuals. Based on our data, we thus cannot retain the hypothesis of a recent past introgression by a closely related species or a different clade of PPM as the origin for the phenological shift observed. The geographical distribution of the different alleles found for the nuclear ITS1 as well as their relationships is consistent with the conclusions drawn from the mitochondrial gene and also suggest that the SP belong to the Iberian clade. Most interestingly, one of the seven alleles was exclusively found in Leiria and was shared between the sympatric summer and winter populations. This strongly suggests that either the SP originated from individuals of the local WP or the recurrent gene flow occurs between the two local phenological ecotypes. As sequence data obtained from few individuals could not permit to determine which of these two hypotheses was the most plausible, we also conducted a study using polymorphic nuclear markers and a monitoring of adult moths' activity.
(b) Population genetics and adult monitoring
The five microsatellite loci could successfully be used to genotype individuals belonging to the Iberian clade (i.e. excluding the populations from the French–Pyrenean clade, see §3a(i)) to compare allelic richness and frequencies and to infer the genetic structure between populations. The pairwise FST show that the SP is significantly differentiated from all other populations, suggesting that the temporal isolation is even greater than the geographical structure of population that was already evidenced in this species (Kerdelhué et al. 2006). The Leiria SP presented the smallest number of alleles for each of the five loci studied in comparison with all other populations, having very few or no rare alleles. Furthermore, in several cases, allelic frequencies were distorted comparatively to the frequencies observed in the surrounding WPs. These characteristics strongly suggest that the SP originated from a founder effect resulting from the establishment of a small number of mutant individuals (Nei et al. 1975; Allendorf 1986). The bottleneck was not detected using the Wilcoxon test for heterozygous excess or the mode-shift test most probably because the number of loci analysed was too low (Piry et al. 1999), or a sufficient number of generations have occurred since the time of the founder effect, which permitted to reach a new equilibrium (Cornuet & Luikart 1996). Moreover, the fact that all the alleles found in the SP were also found in either the sympatric or nearby WP is consistent with a local origin of the SP through a sudden phenological shift, as was hypothesized from the sequence data. The SP was discovered approximately 10 years ago at extremely high population size, which suggests that it is actually much older than 10 years. As genetic signs of the founder effect are still detected in this population, one can confidently conclude that gene flow is highly reduced between the two phenological ecotypes. The genetic results are also consistent with the monitoring of adult activity, as the curves of pheromone trapping are clearly bimodal and non-overlapping, showing a one-month gap between the capture of the last ‘summer’ male and that of the first ‘winter’ male.
(c) Ecological consequences of the phenological shift
Leiria SP adults fly and reproduce in late May, which is during a cooler season than WP moths (August), and temperature is known to affect PPM reproductive behaviour and pheromone emission (Zhang & Paiva 1998). While eggs are also subjected to lower temperatures than the WP, larval development by contrast takes place in summer rather than winter, and consequently requires a smaller investment of metabolic energy for cold resistance, allowing for faster development (Pimentel 2004). Consistently, Leiria SP nests are looser than those of WP, implying lower energetic silk spinning costs, and contain a smaller number of larvae (M. Branco & H. Santos 2003, personal observation). The two populations are further subjected to different ecological pressures, particularly regarding natural enemies. By contrast, the SP pupal stage spent in the soil lasts up to three months longer than that of the WP.
(d) General conclusions
The results obtained on sequence, microsatellite and male monitoring all converge towards the conclusion that the SP was recently established by a reduced number of individuals with early adult emergence and consecutive rapid larval development. These individuals were ‘instantly’ reproductively isolated from the surrounding winter individuals. The phenological shift was not correlated with a change of host species, habitat or resource, as the summer larvae still feed on 1-year-old needles (like the conspecific winter caterpillar), and on the same pine trees as the sympatric winter individuals (H. Santos 2003, personal observation). This situation strictly corresponds to the definition of the incipient stage of allochronic speciation (Alexander & Bigelow 1960), in which asynchronous populations ‘become instantly isolated, even though they initially lack ecological or spatial differentiation, without shifting to a new habitat or host (Abbot & Withgott 2004). The Leiria SP thus represents a unique opportunity to study the very first steps of sympatric differentiation that could ultimately lead to sympatric, allochronic speciation. The founder effect by itself may be important in some mode of speciation, as it can cause extensive genetic changes leading to reproductive isolation (founder effect speciation; see Harrison 1991). Moreover, the SP, albeit sympatric and syntopic with the original WP, is subjected to different selection pressures that could lead to disruptive selection and accelerate the process of differentiation (Turelli et al. 2001).
Acknowledgments
We thank F. Goussard, P. Ménassieu, P. Arnaldo, T. Vasconcelos, N. Nemer, B. Frérot, J. Poirot (and his colleagues from DSF), A. Roques, S. Larsson and R. Gonzalez Ruiz for the field collection of PPM larvae and related species. J.-P. Rossi is greatly acknowledged for his help in the data analyses and graphic representations using the R-package. We are grateful to Y. Yildiz and L. Maudemain for genotyping and sequencing of some of the populations studied here. This work was partly financed by the PESSOA programme (Program for Integrated Actions, EGIDE-GRICES) and the European Project PROMOTH QLK5-CT-2002-00852.
Footnotes
This paper is dedicated to the memory of the late Daniel Lachaise, who suddenly passed away in July 2006. His work in evolution and speciation, and his enthusiasm, strongly influenced our thoughts.
Supplementary Material
Sampling localities for Thaumetopoea spp. and number of sequenced and genotyped individuals. No COI: number of individuals sequenced for the COI gene; No ITS1: number of individuals sequenced for the ITS1 domain; No microsat: number of individuals genotyped for the 5 microsatellite loci
Histrograms of allelic frequencies per population for the 5 microsatellite loci used. Allelic sizes were determined using the GeneScan software with the 400HD-ROX (Applied Biosystems) as size standard as described in Rousselet et al. (2004). Locus MS-Thpit6 was fluorescently-labelled with 6-Fam (Sigma) and sized with the size standard 500-LIZ (Applied Biosystems). The alleles found in the summer population are represented in colour
References
- Abbot P, Withgott J.H. Phylogenetic and molecular evidence for allochronic speciation in gall-forming aphids (Pemphigus) Evolution. 2004;58:539–553. doi:10.1554/03-043 [PubMed] [Google Scholar]
- Alexander R.D, Bigelow R.S. Allochronic speciation in field crickets and a new species, Acheta veletis. Evolution. 1960;14:334–346. doi:10.2307/2405976 [Google Scholar]
- Allendorf F.W. Genetic drift and the loss of alleles versus heterozygosity. Zoo. Biol. 1986;5:181–190. doi:10.1002/zoo.1430050212 [Google Scholar]
- Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N. & Bonhomme, F. 1996–2004 Genetix 4.05, logiciel sous WindowsTM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, France.
- Berlocher S.H, Feder J.L. Sympatric speciation in phytophagous insects: moving beyond controversy? Annu. Rev. Entomol. 2002;47:773–815. doi: 10.1146/annurev.ento.47.091201.145312. doi:10.1146/annurev.ento.47.091201.145312 [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cornuet J.-M, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet J.-M, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Démolin G. Bioecologica de la processionnaria del pino, Thaumetopoea pityocampa. Incidencia de los factores climaticos. Bol. Serv. Plagas For. 1969;23:9–24. [Google Scholar]
- Dres M, Mallet J. Host races in plant-feeding insects and their importance in sympatric speciation. Phil. Trans. R. Soc. B. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. doi:10.1098/rstb.2002.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Harrison R.G. Speciation in North American field crickets: evidence from electrophoretic comparisons. Evolution. 1979;33:1009–1023. doi: 10.1111/j.1558-5646.1979.tb04758.x. doi:10.2307/2407463 [DOI] [PubMed] [Google Scholar]
- Harrison R.G. Molecular changes at speciation. Annu. Rev. Ecol. Syst. 1991;22:281–308. doi:10.1146/annurev.es.22.110191.001433 [Google Scholar]
- Kerdelhué C, Magnoux E, Lieutier F, Roques A, Rousselet J. Comparative population genetic study of two oligophagous insects associated with the same hosts. Heredity. 2006;97:38–45. doi: 10.1038/sj.hdy.6800836. doi:10.1038/sj.hdy.6800836 [DOI] [PubMed] [Google Scholar]
- Kornfield I, Smith P.F. African cichlid fishes: model systems for evolutionary biology. Annu. Rev. Ecol. Syst. 2000;31:163–196. doi:10.1146/annurev.ecolsys.31.1.163 [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. doi:10.2307/2407137 [DOI] [PubMed] [Google Scholar]
- Pimentel, C. S. M. G. 2004 Pine processionary moth (Thaumetopoea pityocampa) and great tit (Parus major) in Portugal: population dynamics and interactions. Ph.D. thesis, Universidade Nova de Lisboa. Faculdade de Ciências e Tecnologia.
- Piry S, Luikart G, Cornuet J.-M. Bottleneck: a computer program for detecting recent reduction in the effective population size using allele frequency data. J. Hered. 1999;90:502–503. doi:10.1093/jhered/90.4.502 [Google Scholar]
- Posada D, Buckley T.R. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. doi:10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Ritchie M.G. Chronic speciation in periodical cicadas. Trends Ecol. Evol. 2001;16:59–61. doi: 10.1016/s0169-5347(00)02060-7. doi:10.1016/S0169-5347(00)02060-7 [DOI] [PubMed] [Google Scholar]
- Rousselet J, Magnoux E, Kerdelhué C. Characterization of five microsatellite loci in the pine processionary moth Thaumetopoea pityocampa (Lepidoptera Notodontidae Thaumetopoeinae) Mol. Ecol. Notes. 2004;4:213–214. doi:10.1111/j.1471-8286.2004.00620.x. [Google Scholar]
- Scriber J.M, Ording G.J. Ecological speciation without host plant specialization; possible origins of a recently described cryptic Papilio species. Entomol. Exp. Appl. 2005;115:247–263. doi:10.1111/j.1570-7458.2005.00285.x [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annu. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Templeton A.R, Crandall K.A, Sing C.F. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Barton N.H, Coyne J.A. Theory and speciation. Trends Ecol. Evol. 2001;16:330–343. doi: 10.1016/s0169-5347(01)02177-2. doi:10.1016/S0169-5347(01)02177-2. [DOI] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. doi:10.1016/S0169-5347(01)02188-7 [DOI] [PubMed] [Google Scholar]
- Vogler A.P, DeSalle R. Evolution and phylogenetic information content of the ITS-1 region in the tiger beetle Cicindela dorsalis. Mol. Biol. Evol. 1994;11:393–405. doi: 10.1093/oxfordjournals.molbev.a040121. [DOI] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Wood T.K, Tilmon K.J, Shantz A.B, Harris C.K, Pesek J. The role of host-plant fidelity in initiating insect race formation. Evol. Ecol. Res. 1999;1:317–332. [Google Scholar]
- Zhang Q.H, Paiva M.R. Female calling behaviour and male response to the sex pheromone in Thaumetopoea pityocampa (Den, Schiff.) (Lep., Thaumetopoeidae) J. Appl. Entomol. 1998;122:353–360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling localities for Thaumetopoea spp. and number of sequenced and genotyped individuals. No COI: number of individuals sequenced for the COI gene; No ITS1: number of individuals sequenced for the ITS1 domain; No microsat: number of individuals genotyped for the 5 microsatellite loci
Histrograms of allelic frequencies per population for the 5 microsatellite loci used. Allelic sizes were determined using the GeneScan software with the 400HD-ROX (Applied Biosystems) as size standard as described in Rousselet et al. (2004). Locus MS-Thpit6 was fluorescently-labelled with 6-Fam (Sigma) and sized with the size standard 500-LIZ (Applied Biosystems). The alleles found in the summer population are represented in colour