Abstract
Through a screen designed to isolate novel fission yeast genes required for chromosome segregation, we have identified mal3 +. The mal3-1 mutation decreased the transmission fidelity of a nonessential minichromosome and altered sensitivity to microtubule-destabilizing drugs. Sequence analysis revealed that the 35-kD Mal3 is a member of an evolutionary conserved protein family. Its human counterpart EB-1 was identified in an interaction screen with the tumour suppressor protein APC. EB-1 was able to substitute for the complete loss of the mal3 + gene product suggesting that the two proteins might have similar functions. Cells containing a mal3 null allele were viable but showed a variety of phenotypes, including impaired control of cell shape. A fusion protein of Mal3 with the Aequorea victoria green fluorescent protein led to in vivo visualization of both cytoplasmic and mitotic microtubule structures indicating association of Mal3 with microtubules. The absence of Mal3 protein led to abnormally short, often faint cytoplasmic microtubules as seen by indirect antitubulin immunofluorescence. While loss of the mal3 + gene product had no gross effect on mitotic spindle morphology, overexpression of mal3 + compromised spindle formation and function and led to severe growth inhibition and abnormal cell morphology. We propose that Mal3 plays a role in regulating the integrity of microtubules possibly by influencing their stability.
Precise duplication of the genetic material and its subsequent accurate segregation into two daughter cells are essential properties of the eucaryotic cell cycle, that ensure the faithful inheritance of genetic information. Genomic stability is vital for the survival of an organism and genetic instability has long been considered an integral characteristic of certain types of cancers (reviewed in Hartwell, 1992).
Entry into mitosis is controlled by the activation of the protein kinase complex MPF which results in chromosome condensation and the formation of the mitotic spindle. The spindle is a dynamic array of microtubules and a large number of microtubule-associated proteins (Hoyt and Geiser, 1996; reviewed in Hyman and Karsenti, 1996; Walczak and Mitchison, 1996). Alignment of chromosomes to the middle of the spindle in metaphase and the subsequent separation of the sister chromatids to opposite poles in anaphase (reviewed in Yanagida, 1995) requires interactions between kinetochores and spindle microtubules. Although substantial progress has been made in recent years in identifying components required for spindle function, such as mitotic motor proteins (Hoyt and Geiser, 1996), the complexity of the system suggests that a large number of molecules involved in ensuring the accurate segregation of the chromosomes await identification.
Mutations in genes required for chromosome transmission fidelity can be identified by scoring for the increased loss of a genetically marked, nonessential minichromosome. Using such an assay we have identified a number of new factors required for chromosome segregation (Fleig et al., 1996). Here we report the characterization of a further component mal3 +. Cloning of the wild-type mal3 + gene revealed that the 35-kD predicted product of the mal3 + gene, Mal3, belongs to an evolutionary conserved protein family. A human homologue of Mal3 is EB-1, which was identified on the basis of its ability to interact with the carboxyl terminus of the tumour-suppressor protein adenomatous polyposis coli (APC)1 (Su et al., 1995).
Germline mutations in the APC gene lead to the inherited disease familial adenomatous polyposis (FAP). FAP shows a dominant mode of inheritance occurring in 1 in 7,000 individuals and results in the development of hundreds to thousands of colorectal tumors in early adult life. Some of these will give rise to carcinomas (reviewed in Kinzler and Vogelstein, 1996). APC gene defects are also present in the majority of sporadic colorectal tumors and represent an early event in colorectal tumor development (Groden et al., 1991; Nishisho et al., 1991). The APC gene encodes a 310-kD cytoplasmic protein with no strong homologies to proteins of known function (reviewed in Kinzler and Vogelstein, 1996). APC has been implicated in a number of cellular processes including the regulation of directed cell migration (reviewed in Polakis, 1995; Näthke et al., 1996; Wong et al., 1996). The amino-terminal part of APC contains an oligomerization domain, while the central third mediates β-catenin binding (reviewed in Kinzler and Vogelstein, 1996; Pfeifer, 1996). Association of APC with β-catenin links APC to the process of cell adhesion as well as placing it into the Drosophila Wg/ mouse Wnt signal transduction pathway (reviewed in Kemler, 1993; Kinzler and Vogelstein, 1996; Pfeifer, 1996). The carboxyl-terminal end of APC is required for association with EB-1, the human homologue of the Drosophila tumor suppressor protein DLG and the microtubule cytoskeleton (Munemitsu et al., 1994; Smith et al., 1994; Matsumine et al., 1996). Over 95% of APC mutations in FAP and sporadic tumors result in truncations which delete the carboxyl-terminal third of the protein (reviewed in Polakis, 1995) suggesting that interactions in this region of APC play key roles in regulating the growth-controlling function of APC.
Here we show by the ability of EB-1 to substitute for fission yeast Mal3 function that the two proteins appear to have similar functions, implying that an analysis of fission yeast Mal3 could provide valuable insight into the function of the APC-interacting protein EB-1. Our data indicate that the microtubule-interacting mal3 + gene product plays an important role in regulating/maintaining the integrity of microtubule arrays.
Materials and Methods
Media and Strains
The Schizosaccharomyces pombe strains HM348 (h +, leu1-32, fur1, ade6-M210, tps1, Ch16 [ade6-M216]) (Niwa et al., 1986), YP10.22 (h −, leu1-32, ade6-M210, ura4-D6, Ch16 [ade6-M216]) and YP10.22a (h +, ade6-M210, ura4-D6, Ch16 (ade6-M216)), the isolation of the mal3-1 carrying strain YP30.1 and the calculation of the minichromosome loss rate have been described (Fleig et al., 1996). Strains YP246 (h −, his3-D1, leu1-32, ura4-D18, ade6-M210) and YP249 (h +, his3-D1, leu1-32, ura4-D18, ade6-M216) (Ohi et al., 1996) were used to generate diploid strain YP41. The mal3 deletion strain YP350 (h +, mal3Δ::his3 + , his3-D1, leu1-32, ura4-D18, ade6-M210) was isolated by tetrad analysis. Strains YP113 (h +, nda2-KM52, leu1-32) and YP114 (h +, nda3-KM311, leu1-32) have been described (Umesono et al., 1983; Toda et al., 1984). Strains were grown in rich (YE5S) or EMM minimal medium with appropriate supplements (Moreno et al., 1991). For low- or high-level expression from the nmt1 + promoter cells were grown in EMM with 15 μM thiamine or pregrown on EMM plus thiamine plates for 18 h at 30°C before inoculation into thiamine-less EMM liquid medium, respectively. Sensitivity to thiabendazole (TBZ) was monitored at 24 or 30°C on YE5S plates or EMM plates containing 10 μg/ml of TBZ.
Microscopy
Photomicrographs of cells were taken with a Zeiss Axioskop fitted with differential interference optics. For determination of phenotypes a minimum of 300 cells was scored microscopically. Staining with calcofluor (Sigma Chem. Co., St. Louis, MO) and processing of cells for immunofluorescence microscopy was carried out as described (Verde et al., 1995; Hagan and Hyams, 1988). For tubulin staining we used the primary monoclonal antitubulin antibody TAT1 (Woods et al., 1989) followed by FITC-conjugated goat anti–mouse antibodies (Hagan and Yanagada, 1997). Spindle pole bodies were stained using AP9.2 affinity-purified anti-sad1-primary and Cy3-conjugated secondary sheep anti–rabbit antibodies (Sigma) (Hagan and Yanagida, 1995). Immunofluorescence images were processed as described previously (Lange et al., 1995).
Identification of mal3+ ORF and Plasmid Constructions
Strain YP30.1 was transformed with an S. pombe genomic bank (Barbet et al., 1992) and Ura+ transformants were selected by incubation for 50 h at 32°C, followed by replica-plating twice onto plates containing 10 μg/ml TBZ. Plasmids were isolated (Moreno et al., 1991) from the 3 out of 24,000 transformants able to grow following this selection process, amplified in E. coli XL1-blue (Stratagene Inc., La Jolla, CA) and retransformed into YP30.1 to ascertain plasmid borne rescue. These plasmids were tested for the ability to suppress the increased minichromosome loss of the mal3-1 mutant by visual screening for the suppression of colony sectoring. Only one of the genomic DNA inserts, present in pUR3-1, was able to rescue all defects of strain YP30.1.
30 nucleotides of sequence from the 5′ (5′TCGCTTATCTGTCCCTTGAACCTACGACAA 3′) as well as the 3′ (5′GATCAGGTGGTAACTCAAAACCATCCTCAG 3′) end of the 2.9-kb genomic DNA insert of pUR3-1 were used for an homology search in the S. pombe database, identifying a region of identity on chromosome I that contained two putative ORFs SPAC18G6.13c and SPAC18G6.14. To determine which ORF rescued the mal3-1 mutant strain, pUR3-1 was cut with PacI/SmaI and re-ligated thus rendering pUR3-1a which contains only the SPAC-18G6.13c ORF. The 1.6 kb PvuI/BamHI fragment containing SPAC-18G6.14 was cloned blunt-ended into SmaI cut pUR18 (Barbet et al., 1992) resulting in plasmid pUR3-1b. Only pUR3-1b was able to rescue the defects of the mal3-1 mutant. SPAC18G6.14 was thus named mal3 +. To obtain full-length mal3 +, genomic DNA prepared from strain YP10.22 (Moreno et al., 1991) was used as a PCR template with oligonucleotides a (5′TACCAGTCGACATGTCTGAATCTCGGCAAGA 3′) and b (5′CTTAGCCCGGGTTAAAACGTGATATTCTCATC 3′). Oligonucleotides a and b are homologous to the 5′ and 3′ ends of the mal3 + ORF (sequences in italics) and contain a SalI and SmaI site, respectively (see Fig. 1 C). These restrictions sites were used to clone the 980-bp mal3 + PCR fragment into SalI/SmaI cut pREP3 (Maundrell, 1993) generating pREP3-mal3. The sequence of three independently PCR-derived mal3 + DNAs was determined.
Figure 1.
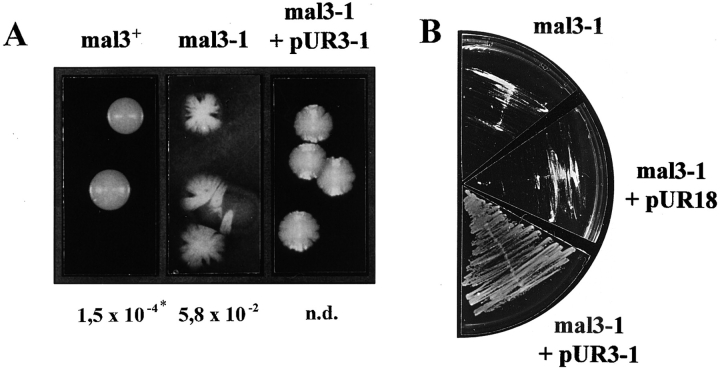
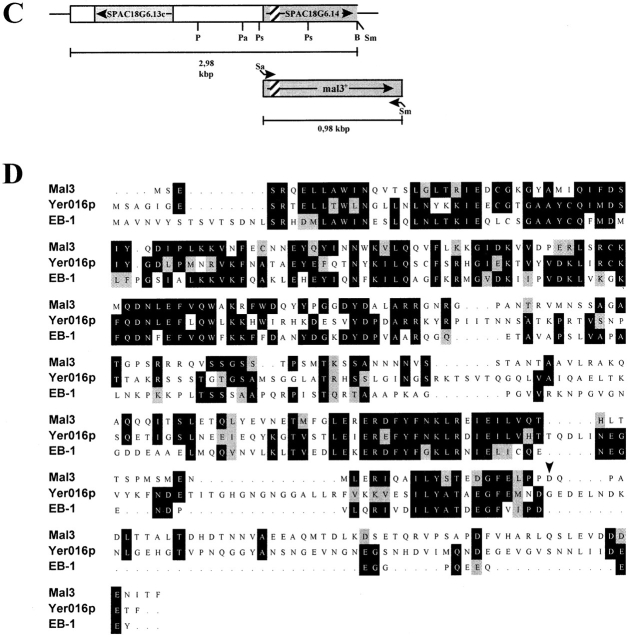
Phenotypic characterization of the mal3-1 strain, isolation of mal3 + gene and Mal3 amino acid sequence. (A) Sectoring phenotypes of mal3 +, mal3-1, and mal3-1 transformed with plasmid pUR3-1 strains grown at 24°C are shown. *Minichromosome loss rate; n. d., not determined. (B) While the mal3-1 strain or mal3-1 plus vector (pUR18) are unable to grow on TBZ containing EMM medium at 30°C, this defect is rescued by plasmid pUR3-1. (C) Diagrammatic representation of genomic DNA insert of pUR3-1 (top) and full length mal3 + gene obtained by PCR (bottom). PCR primers have been marked by arrows, ORF by grey boxes and a putative intron in SPAC18G6.14/mal3 + by a striped box. P, PvuI; Pa, PacI; Ps, PstI; B, BamHI; Sm, SmaI; Sa, SalI. (D) Amino acid sequence comparison of fission yeast Mal3, budding yeast Yer016p (GenBank accession number P40013) and human EB-1 (GenBank accession number U24166). Identical and similar amino acids are indicated by black and grey boxes, respectively. The arrow head indicates the end of the Mal3 truncated protein in pUR3-1. The mal3 + sequence data are available from EMBL/GenBank/DDBJ under accession number Y09518.
To fuse the yEGFP and mal3 + ORFs, we generated a mal3 + ORF without the TAA stop codon via PCR using pREP3-mal3 and the following oligonucleotides: 5′TACCAGTCGACTATGTCTGAATCTCGCAAGA 3′ and 5′AGTACCTCGAGAAACGTGATATTCTCATCGT 3′ and cloned it into SalI/XhoI pBluescript (Stratagene). The mal3 + DNA fragment was excised with BamHI/XhoI and cloned into BamHI/SalI cut pUG23 (Güldener, U., and J.H. Hegemann, unpublished observations), thereby fusing the mal3 + and yEGFP (Cormack et al., 1997) ORFs. This fusion was subsequently cloned into SalI cut pREP3 behind the nmt1 + promoter.
To demonstrate linkage between the cloned mal3 + gene and the mal3-1 mutant we constructed a mal3Δ/mal3-1 diploid strain. Both mal3 alleles show increased sensitivity to TBZ. Random spore analysis of this diploid strain revealed that the 400 spores tested showed increased sensitivity to TBZ indicating that mal3Δ and mal3-1 were linked and thus that the complementing DNA fragment was the mal3 + gene.
Database Searches and Cloning of Human EB-1
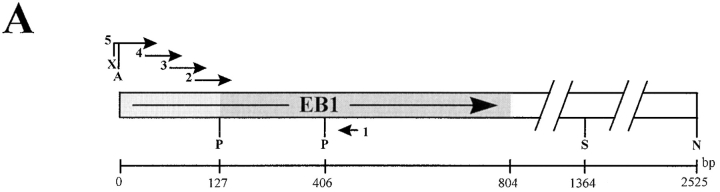
The NCBI's protein database was searched using BLASTP. The Mal3 query showed strongest homologies to human EB-1 (GenBank U24166; P = 3.7 × 10−53) and budding yeast protein Yer016p (GenBank P40013; P = 4.3 × 10−53). Sequence alignments were performed using CLUSTAL W (Thompson et al., 1994). The dbEST database was searched using BLAST. The EST clone (NCBI 184415, GenBank R13836) with the highest homology to Mal3 (P = 1.8 × 10−19) was ordered from Research Genetics Inc. (Huntsville, AL) (clone ID 26991) and sequenced revealing 100% identity to EB-1 (Su et al., 1995) albeit without the first 127 bp of the EB-1 ORF. To obtain full-length EB-1 the missing 5′ end of the cDNA was constructed via PCR using the cDNA as template and overlapping oligonucleotides (see Fig. 2 A). Oligonucleotide 1 (5′ GCTGGAGCAACAAGGGAA G 3′) was used in combination with oligonucleotides 2 (5′ AATCTGACAAAGATCGAACAGTTGTGCTCAGGGGCTGCGTATTGTCAGTTTATGGAC 3′), 3 (5′ TGACATGCTGGCCTGGATCAATGAGTCTCTGCAGTTGAATCTGACAAAGATCGAACAG 3′), 4 (5′ TCAACGTCAGTGACCAGTGATAACCTAAGTCGACATGACATGCTGGCCTGGATC 3′), and 5 (5′ TTCTGCTCTAGACTCGAGATGGCAGTGAACGTATACTCAACGTCAGTGACCAGTG 3′) in such a way that, for example, the PCR product resulting from the use of oligonucleotides 1 and 2 was used as a template for the next PCR (see Fig. 2 A). After sequence analysis of the 452-bp DNA product obtained from the last PCR reaction, the fragment was cloned as a PstI/XbaI fragment into pBluescript generating plasmid pJB17. To obtain the entire EB-1 ORF the 2-kb long PstI/NotI fragment from the original cDNA clone was cloned into pJB17 resulting in plasmid pJB23. The 1,367-bp AvaI/StuI fragment from pJB23 containing the entire EB-1 was cloned into XhoI/ SmaI cut pREP3.
Figure 2.
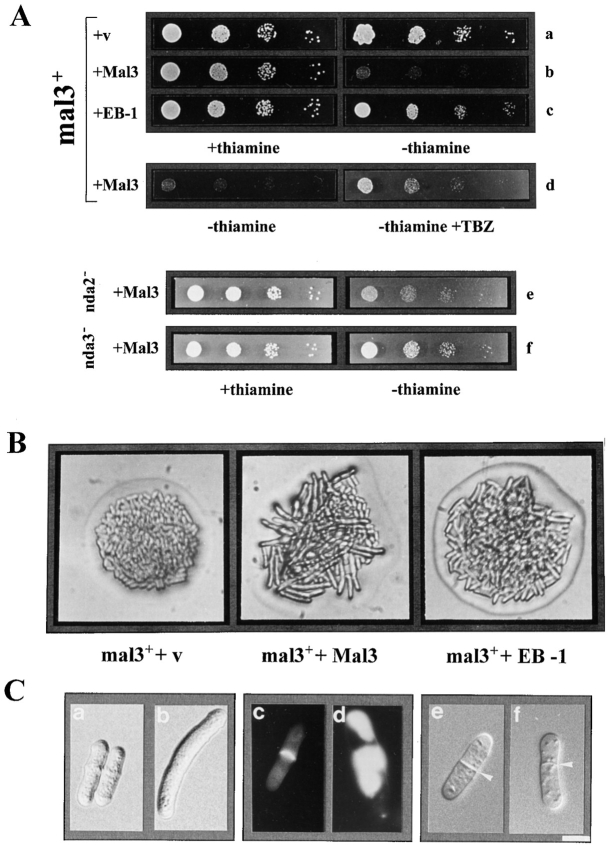
Suppression of the TBZ sensitivity of the mal3-1 and mal3Δ strains by mal3 + and EB-1. (A) Diagrammatic representation of cloning of EB-1 cDNA. The positions of the five oligonucleotides used to construct the 5′ end (lightly shaded box) of EB-1 not present in the original cDNA (dark shaded box) are indicated. X, XbaI; A, AvaI; P, PstI; S, StuI; N, NotI. (B) The mal3 + strain transformed with vector (a), the mal3-1 strain transformed with vector (b) or with plasmids expressing mal3 +(c) or EB-1 (d) and the mal3Δ strain transformed with vector (e), or with plasmids expressing mal3 + (f) or EB-1 (g) were all grown for 18 h at 30°C in thiamine-less EMM medium, diluted in water and serial dilutions (1:10) starting with 104 cells were spotted on selective EMM medium with 10 μg/ml TBZ and incubated for 3 d at 30°C.
Disruption of mal3+ Open Reading Frame
To disrupt the mal3 + gene we replaced the 0.8-kb PacI-PstI mal3 + fragment with the 1.9-kb his3 + (Ohi et al. 1996) marker in diploid strain YP41. Correct integration was verified by Southern-hybridization and PCR analysis. PCR primers used were: a (5′ GAGCCTCTTTGCGAAGGAGCATTG 3′) and b (5′ TAAAACGTGATATTCTCATCGTCA 3′) homologous to the his3 + 3′ end and the last 24 base pairs of the mal3 + ORF, respectively. Transformants with a correctly disrupted mal3 + ORF were sporulated and asci dissected by tetrad analysis.
Results
Isolation of the mal3-1 Mutant and Cloning of mal3+
Through a screen designed to isolate new fission yeast genes involved in chromosome segregation, we have identified 25 mutant strains which show an increase of loss rate of a nonessential minichromosome. A decrease in minichromosome transmission fidelity was identified by using an ade6-based colony color assay and was manifested as an increase in the number of red sectors in a white colony (Fleig et al., 1996). Strain YP30.1, which showed a 380-fold increase in minichromosome loss at 24°C (Fig. 1 A, middle) and was hypersensitive to the microtubule-destabilizing drug thiabendazole (TBZ) (Fig. 1 B), was chosen for further analysis. Crossing this mutant strain to a wild-type strain and pulling tetrads indicated that a single locus was responsible for both the increased loss of the minichromosome and the TBZ-sensitivity. We called this allele mal3-1.
The wild-type mal3 + gene was isolated by complementation of the TBZ-sensitive phenotype. 10 μg/ml of TBZ had little effect on a mal3 + strain at 30°C (Fig. 2 B, a), but severely inhibited growth of mal3-1 (Fig. 1 B). Transformation of the mal3-1 strain with an S. pombe genomic library identified three plasmids which were able to rescue the growth defect of the mal3-1 strain on TBZ medium. One of these plasmids contained a 2.9-kb genomic DNA insert which also complemented the increased loss of the minichromosome (Fig. 1 A, right).
DNA sequence analysis of either end of this 2.9-kb insert was used for a homology search in the Schizosaccharomyces pombe database (Sanger Center, Hinxton, UK). A match of 100% identity was found on chromosome I. This region contained the two putative open reading frames (ORF) SPAC18G6.13c and SPAC18G6.14. The DNA insert contained the entire SPAC18G.13c ORF and 82% of the SPAC18G6.14 ORF (Fig. 1 C, top diagram). Only SPAC- 18G6.14 rescued the mal3-1 defects (data not shown) and was thus named mal3 +. Linkage was demonstrated between the cloned mal3 + gene and the mal3-1 mutant (see Materials and Methods).
The DNA sequence of the mal3 + ORF was identical to that reported for SPAC18G6.14 with the exception of a silent change at position 210 of the ORF where the mal3 + sequence had a C and SPAC18G6.14 a T nucleotide. mal3 + encodes a protein of 308 amino acid residues with a mass of 35.1 kD and is interrupted by a 60-bp intron (Fig. 1, C and D). Database searches indicated that no previously characterized sequence motifs were present in the Mal3 amino acid sequence.
Mal3 Belongs to an Evolutionary Conserved Protein Family
A search of NCBI's nonredundant protein database identified several proteins with significant homology to Mal3, in particular the 30-kD human protein EB-1 (28.9% identity and 39.6% similarity) and the 38.3-kD hypothetical S. cerevisiae ORF Yer016p (33.1% identity and 44.1% similarity) (Su et al., 1995; Dietrich et al., 1997). Blocks of homology were distributed over the entire length of the protein with the exception of the COOH-terminal end (Fig. 1 D). Searches of the dbEST database (Boguski et al., 1993) identified 16 ESTs of human and mouse origin with significant homology to Mal3, indicating that it is a member of a conserved multiprotein family. Therefore we sought to determine whether members of this family were functionally homologous. The EST cDNA clone with the highest homology to the Mal3 sequence was sequenced revealing 100% identity to the published sequence of human EB-1 (Su et al., 1995). The first 127 bp of the EB-1 ORF not present in the cDNA fragment were generated via PCR using appropriate overlapping oligonucleotides (Fig. 2 A).
Human EB-1 Rescues the TBZ-Hypersensitivity of the mal3 Deletion Strain
The functional interchangeability of Mal3 and EB-1 proteins was tested by assessing the ability of EB-1 to suppress the TBZ-hypersensitive phenotype of mal3-1 and mal3Δ (mal3 + null allele, see next section). Both strains were transformed with plasmids expressing either mal3 + or EB-1 under the control of the thiamine-repressible nmt1 + promoter. The presence of 15 μM thiamine leads to low-level expression from this promoter, while the absence of thiamine leads to high-level expression after 12– 14 h (Fleig and Nurse, 1991; Maundrell, 1993). Transformants were pregrown in medium lacking thiamine for 18 h at 30°C and then tested for the ability to grow on TBZ-containing plates. As shown in Fig. 2 B the wild-type mal3 + strain transformed with vector only grows well on plates containing 10 μg/ml TBZ (Fig. 2 B, a), while the mal3Δ strain grows poorly (e) and mal3-1 is unable to grow (b). Plasmid-borne expression of Mal3 suppresses the growth defect on TBZ plates of both the mal3-1 and mal3Δ mutants (Fig. 2 B, c and f). Human EB-1 partially rescued the increased TBZ-sensitivity of mal3-1 (Fig. 2 B, d) and fully suppressed the TBZ sensitivity of mal3Δ (Fig. 2 B, g) suggesting that, while the human EB-1 protein is only partially able to compete with the mutant form of the Mal3 protein in the mal3-1 strain, EB-1 can substitute for the complete loss of the mal3 + gene product. Low-level expression of Mal3 and EB-1 partially rescued the TBZ-sensitivity of the mal3Δ strain (data not shown).
Deletion of the mal3+ Gene Is Viable but Leads to Altered Cell Morphology and Phenotypes Similar to those of the mal3-1 Mutant
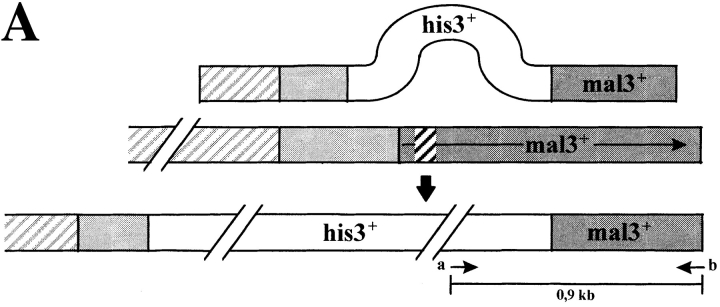
A mal3 null allele was constructed by replacing the first 152 amino acids of the mal3 + ORF and promoter region with the his3 + marker in the diploid strain YP41 (Fig. 3 A, diagram). Southern analysis identified the resultant mal3 deletion (data not shown). Diploid transformants that carried the mal3 + null allele (named mal3Δ) were sporulated and subjected to tetrad analysis. While all four spores in each tetrad were viable at 32°C histidine prototrophy segregated 2:2, indicating that mal3 + is not essential for cell proliferation. PCR analysis of the histidine prototrophic spores confirmed the correct integration of the mal3 + disruption cassette (Fig. 3 A).
Figure 3.
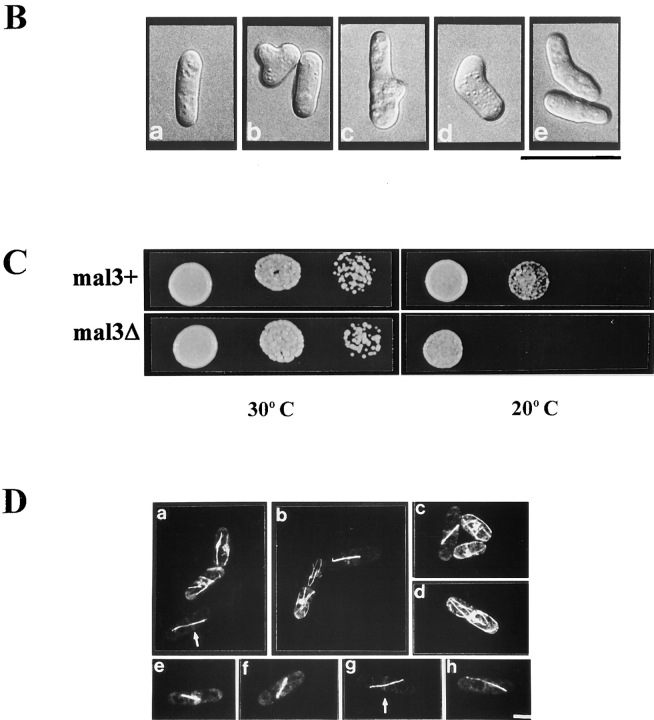
Phenotypic characterization of mal3Δ strain and in vivo localization of Mal3 protein. (A) Diagrammatic representation and PCR analysis of the mal3 + disruption. Diploid strain YP41 was transformed with a linear 3.2-kb DNA fragment where 800 bp of the mal3 + gene had been replaced by the 1.9-kb his3 + gene. Spores were grown and their DNA analysed by PCR for the presence of the mal3 + disruption. The positions of the PCR primers a and b are shown in the bottom diagram. The 900-bp DNA fragment generated by PCR can be obtained only if the linear mal3 + disruption fragment has integrated correctly, as the DNA sequence homologous to primer b is not present on the 3.2-kb DNA fragment used for replacement. a and b show the PCR product of his− and his+ spore DNA, respectively. M, DNA molecular mass markers (0.94 and 0.83 kb). (B) Photomicrographs of mal3Δ cells. A wild-type strain (a) and the mal3Δ strain (b–e) were grown logarithmically at 24°C. (C) Serial dilution patch test for cold-sensitivity of mal3 +and mal3Δ strains. Dilutions shown were tenfold. Strains were incubated at 30°C and 20°C for 3 and 5 d, respectively. (D) In vivo localization of Mal3 protein. A wild-type S. pombe strain expressing Mal3-yEGFP under the control of the nmt1 + promoter was pregrown on solid EMM thiamine medium, resuspended in liquid EMM medium with 0.05 μM thiamine, grown for 16 h at 30°C and photographed directly. Bars: (B) 20 μm; (D) 5 μm.
Further analysis of the haploid mal3Δ strain revealed phenotypes similar to those of the mal3-1 mutant strain i.e., increased sensitivity to TBZ (Fig. 2 B, e) and a decreased transmission fidelity of the minichromosome (data not shown). In addition while no difference in growth between a wild-type and a mal3Δ strain was observed at 30 or 24°C (Fig. 3 C, left; data not shown), deletion of the mal3 + gene conferred cold sensitivity (Fig. 3 C, bottom right).
Absence of the mal3 + gene product resulted in alterations in cell form and polarity as branched and curved cells were seen instead of the cylindrical shape typical of wild-type cells (Figs. 3 B and 4 A). Bent cells were often elongated. Altered cell morphology was observed at all temperatures tested, but was most prominent at 20°C, where >30% of mal3Δ cells had abnormal shapes (Fig. 4 A).
Figure 4.
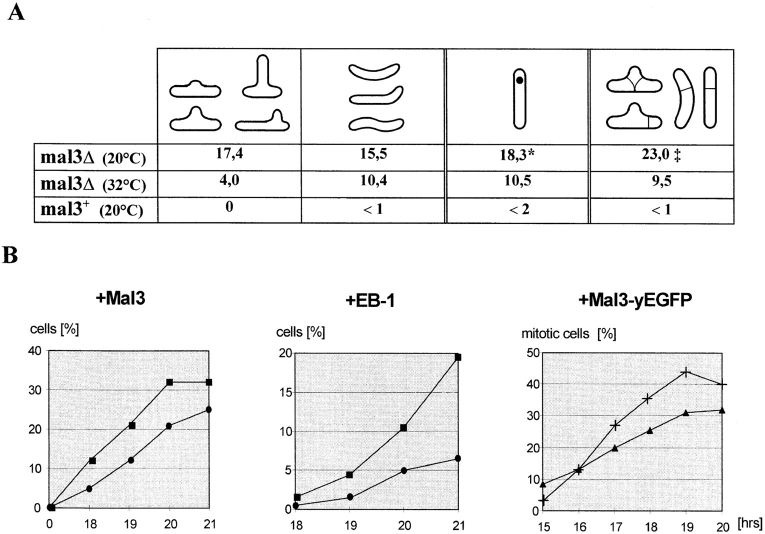
Phenotypes of the mal3Δ strain and Mal3 and EB-1 overproducing strains. (A) mal3Δ cells were grown at 20 or 32°C in liquid YE5S medium and the percentage of cells with abnormal cell morphology, a displaced nucleus and misplaced septum was determined. The mal3 + control strain was grown at 20°C. *Percentage of all interphase cells with a displaced nucleus; ‡percentage of septated cells with a misplaced septum. (B) Wild-type strains overproducing Mal3, EB-1 or Mal3-yEGFP were grown for 0–21 h at 30°C in expression inducing medium and the percentage of cells with altered cell shape (•), a displaced nucleus (▪), a mitotic spindle (▴) and aberrant spindles out of the total number of spindles (+) was determined.
The Mal3 Protein Associates with Microtubules In Vivo
To determine the intracellular localization of Mal3 in living cells, a yeast-enhanced version of the green fluorescent protein named yEGFP (Cormack et al., 1997) was fused to the COOH-terminal end of the mal3 + coding region and the fusion protein was expressed under the control of the nmt1 + promoter (see Materials and Methods). The Mal3-yEGFP fusion protein rescued the TBZ-sensitivity of mal3Δ when expressed at a high level from the nmt1 + promoter, albeit less efficient than Mal3, indicating that the fusion protein was functional (data not shown).
We observed green fluorescence when the Mal3-yEGFP protein was expressed at low or high levels in the cell. Low-level expression of the fusion protein resulted in staining along microtubule structures and some uniform green fluorescence of the cytoplasm and faint nuclear staining. However, as fluorescence was weak under these conditions, we looked for intermediate levels of expression of the fusion protein which had no effect on cell growth and cell morphology but gave better staining. Growing cells on medium containing 0.05 μM thiamine (Javerzat et al., 1996; data not shown) resulted in fluorescence that was clearly highlighting filamentous structures reminiscent of microtubules. The Mal3-yEGFP fusion protein associated with both cytoplasmic (Fig. 3 D, a–d) and mitotic spindle microtubules (Fig. 3 D, e–h). Fluorescence was distributed uniformly along the elongating spindle, with early mitotic spindles staining most intensely. Interestingly in anaphase the Mal3-yEGFP fusion protein also localized to a ring present at the cell equator (Fig. 3 D, a and g, arrows). Expression of yEGFP alone from the nmt1 + promoter led to uniform green staining of the entire yeast cell (data not shown; Fleig et al., 1996).
Loss of Mal3 Function Results in Altered Interphase Microtubule Arrays
As our data indicated that Mal3 is a microtubule interacting protein we examined the consequences of a loss of Mal3 protein upon the microtubule cytoskeleton.
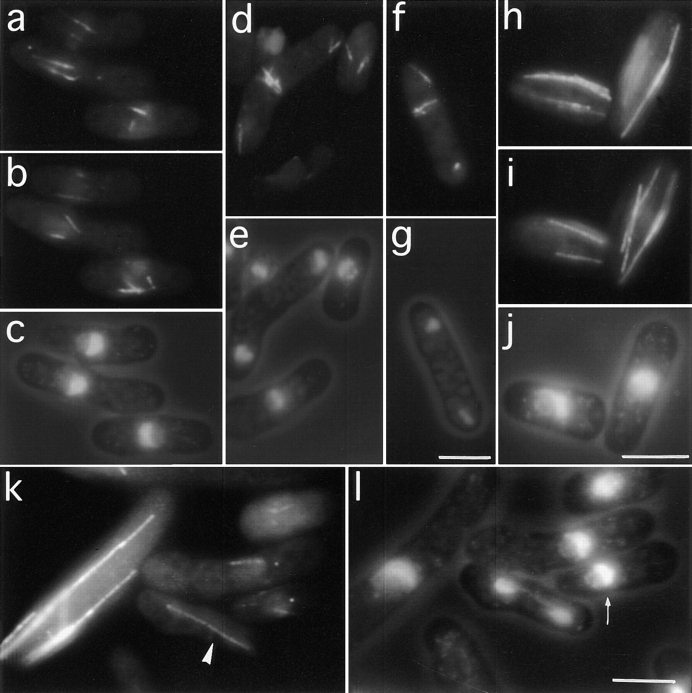
In wild-type fission yeast cells cytoplasmic microtubules are aligned along the long axis of the cell between the cell tips (Marks et al., 1986; Hagan and Hyams, 1988; Fig. 5, h and i). In contrast morphologically wild-type mal3Δ cells grown at 20°C had microtubules that were largely short (approximately one-fifth of wild-type length) and present only around the nucleus (Fig. 5, a and b). In addition, the staining in branched cells was often absent or very weak possibly indicating that less than the wild-type complement of microtubules were present in each cytoplasmic bundle (data not shown). To determine that these findings were not artefacts of fixation, a simple test was performed. At the nonpermissive temperature cdc25-22 cells arrest cell cycle progression at the G2/M boundary but continue to grow thus becoming highly elongated with cytoplasmic microtubules (Hagan and Hyams, 1988). By mixing arrested cdc25-22 cells with shorter mal3Δ cells before processing them for immunofluorescence, one can determine whether the microtubules in both strains are unable to reach the cell tips (i.e., fixation artefact) or whether the defect is present only in mal3Δ. Consistent with the latter interpretation the images in Fig. 5 k show normal microtubules in the cdc25-22 cell and short microtubules in mal3Δ cells. In contrast to the severe alteration of cytoplasmic microtubules no gross spindle abnormalities were observed in mal3Δ cells although again spindle staining was reduced. (Fig. 5 k, arrowhead). However we observed a fourfold increase in the number of cells showing condensed chromosomes in comparison to an isogenic wild-type strain indicating defects in some aspects of mitosis.
Figure 5.
Antitubulin immunofluorescence images of mal3Δ cells cultured at 20°C. a, b, d, f, h, i, and k show antitubulin staining while c, e, g, j, and l show DAPI and phase contrast images. a and b represent two different focal planes of three mal3Δ cells showing several short microtubules around the location of the nucleus shown in c. (d) Strong post-anaphase-array staining in the upper, bent cell. (f) Displaced post-anaphase-array. h–i show antitubulin immunofluorescence images of two mal3 + cells grown at 20°C at different focal planes, while j shows DAPI staining. k and l show a mixed culture of mal3Δ and elongated cdc25-22 cells. k shows antitubulin while l shows DAPI staining. cdc25-22 cells were grown at 25°C in YE5S to early log phase, shifted to the restrictive temperature for 210 min, returned to 20°C in 30 s by immersion in an ice bath and mixed at a ratio of 1:4 with the mal3Δ culture grown at 20°C inYE5S. k shows four mal3Δ cells and an elongated cdc25-22 cell. The abnormally faint spindle in the mal3Δ cell is indicated by an arrow head in k. The small arrow in l indicates a mal3Δ cell with a misplaced nucleus. Bars, 5 μm.
Consistent with the notion that the microtubule cytoskeleton plays a role in nuclear positioning (Toda et al., 1984; Hagan and Yanagida, 1997) mal3Δ cells frequently show displaced nuclei (Fig. 5 l, small arrow). At 20°C 18% of mal3Δ cells had off center premitotic nuclei (Fig. 4 A). Interestingly we also observed displaced post-anaphase-arrays (Fig. 5 f). Intriguingly the post-anaphase-arrays were the most strongly staining structures in mal3Δ cells observed by antitubulin immunofluorescence possibly indicating that these microtubules have a different composition or properties to other cytoplasmic microtubules (Fig. 5 d).
In addition to displacement of the nucleus and post-anaphase-array lack of Mal3 also led to displacement of the septum from its normal central location. Approximately 23% of mal3Δ cells had a septum not positioned in the central fifth of a cell at 20°C (Fig. 4 A).
Overexpression of mal3+ Inhibits Colony Formation and Leads to Aberrant Cell Morphology
Overproducing Mal3 by inducing mal3 + expression from the nmt1 + promoter in a wild-type strain resulted in severe inhibition of colony formation (Fig. 6 A, b) and cell elongation, indicative of a possible role of Mal3 in cell cycle regulation. Cells started to elongate following 18 h of induction and subsequently became highly elongated with phenotypes similar to those of the mal3Δ strain, i.e., aberrant cell morphologies and displaced interphase nuclei (Figs. 4 B and 6 B). The proportion of cells with altered cell shapes correlated directly with the duration of mal3 + overexpression; the most drastic phenotypes were observed after 21 h or more (Figs. 4 B and 6 C, b).
Figure 6.
Phenotypic characterization of overexpression of Mal3 and EB-1 proteins. (A) Serial dilution patch test for inhibition of colony formation by overproduction of Mal3 and EB-1. Dilutions shown were tenfold. A mal3 + strain was transformed with either the insert-less vector (a) or plasmids expressing mal3 + (b) or EB-1 (c) from the nmt1 + promoter. Strains were grown for 18 h at 30°C in thiamineless medium and then plated on EMM medium with (+) or without thiamine (−).The growth inhibition phenotype of Mal3 overexpression was suppressed by 10 μg/ml TBZ (d) or the presence of the α- and β-tubulin mutants, nda2-KM52 and nda3-KM311, respectively (e and f). Colonies were photographed after incubation for 3 d at 30 or 29°C (mutant tubulin strains). (B) Photomicrographs of colonies of wild-type cells transformed with vector control (v), or plasmids expressing mal3 + or EB-1. Strains were pregrown as in A before streaking cells on thiamineless plates and incubating them for 1.5 d at 30°C. (C) mal3 + was overexpressed in a wild-type strain for 0 (a, c, and e), 19 (f), 21 (b), or 24 (d) h at 30°C. c and d show staining of septa by calcofluor. Septa in e and f are indicated by arrowheads. Bar, 5 μm.
Cells with extra Mal3 protein also displayed mitotic defects and were unable to undergo proper cytokinesis. The cytokinesis retardation appeared to be coupled with the appearance of abnormally formed and misplaced septa. Following 21 h of mal3 + induction 7.2% of cells had a septum and in 25% of these the septa were not in the central fifth of the cell. Later (e.g., 24 h) more than 80% of cells with a septum showed highly abnormal, multiple and/or misplaced septa (Fig. 6 C, d and f). Overexpression of human EB-1 protein led to similar, albeit less severe phenotypes (Figs. 4 B and 6 A, c and B). Overexpression of either Mal3 or EB-1 protein in the mal3Δ strain led to moderate or no inhibition of colony formation, respectively (data not shown).
Overexpression of Mal3 Protein Compromises Spindle Formation and Function
Immunofluorescence staining of a wild-type strain overexpressing mal3 + for 20–22 h at 32°C showed no gross defects in microtubule integrity in interphase cells. Cytoplasmic microtubule bundles were present in a full complement and extended along the main cell axis to the cell tips. The bundles in branched cells were slightly more curved than those of wild-type cells (Fig. 7 e).
Figure 7.
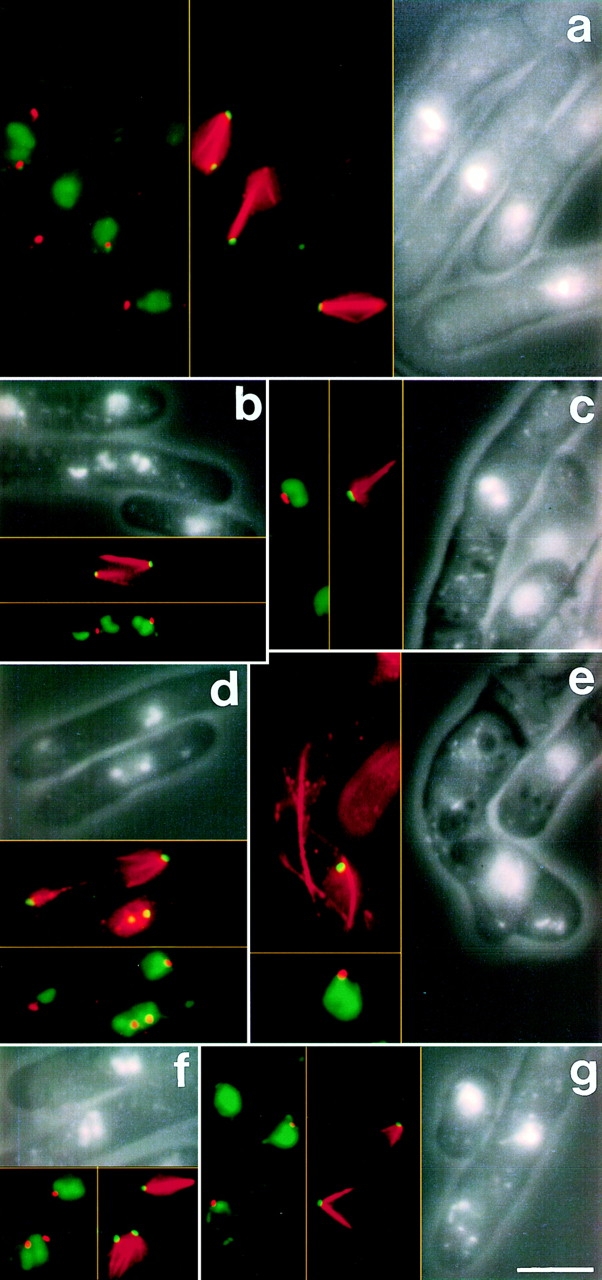
Overexpression of mal3 + results in defects in form and function of the mitotic spindle. Wild-type cells overexpressing mal3 + were grown at 32°C for 21 h in inducing conditions. Each panel shows three different images of the same cells: in the first, containing the lettering, the relative location of the chromatin and the cell outline is shown by combining DAPI fluorescence and phase contrast images; in the second the microtubules (red) and the SPBs (green) are shown; the third panel shows the position of the SPBs (red) relative to the chromatin (green). a shows three strongly staining spindles of roughly equal length. The chromosomes are highly condensed and randomly positioned along the spindle axis. In each case bars of tubulin staining emanate from the SPB away from the main body of the spindle. b, d, and g show examples of cells where the two half spindles appear to be losing their interdigitation presumably leading to the complete loss of interdigitation seen in g. In b the smaller chromosome III has wandered from the spindle axis, indicating that the microtubules are not contacting the kinetochores. e shows a bent interphase cell which has not separated from its partner during the previous cytokinesis. c and f show examples of defects during the early stages of spindle formation. Bar, 5 μm.
Excess Mal3 led to an increase in the proportion of cells which were in mitosis and strongly affected the formation and function of the spindle. After 20 h of induction 24% of cells were in mitosis and 16% had condensed chromosomes. Two predominant defective spindle phenotypes were observed: (a) elongating spindles with severe defects in chromosome segregation to the poles (Fig. 7 a), and (b) the disintegration of the spindle evidenced by spindle fraying (Fig. 7, a and b) and two separated V-shaped tubulin structures present in one cell (Fig. 7, d and g). 52% of cells with a spindle exhibited the former phenotype, while 6% of all cells displayed the latter one. Additionally, 2.5% of cells in the population were unable to form a spindle and showed V-shaped tubulin staining patterns typical of mutants with a defective spindle pole body component or mitotic motor protein (Fig. 7, c and f; Hagan and Yanagida, 1995). We interpret the significant number of cells which showed two separate V-shaped structures in one cell as being those in which the two halves of the mitotic spindle no longer interdigitate and yet the microtubules are stable and do not depolymerize.
A time course analysis of Mal3-yEGFP overexpression revealed that the increase in the number of cells in mitosis and the appearance of aberrant spindle structures was correlated directly with the duration of overexpression (Fig. 4 B, right) and peaked following induction for 19–20 h. At this time ∼32% of cells showed green fluorescing mitotic spindles with >40% of these being aberrant.
Decreasing Microtubule Stability Partially Rescues the Mal3 Overexpression Phenotype of Severe Growth Inhibition
The abnormally short cytoplasmic microtubule arrays in the mal3Δ strain suggested that Mal3 might somehow influence microtubule stability. As excess Mal3 led to defective spindles and mitotic microtubules have shorter half-lives than their interphase counterparts (Salmon et al., 1984; Saxton et al., 1984) these data also support a possible role for Mal3 in stabilizing microtubules. If Mal3 does indeed play a role in stabilizing microtubules then destabilizing microtubules by some means would be expected to alleviate the severity of the phenotypes observed upon Mal3 overexpression. We tested this idea by destabilizing microtubules in two ways and assaying colony forming potential: first, wild-type cells overexpressing Mal3 were grown in the presence of TBZ and secondly by overproducing Mal3 in strains carrying the cold-sensitive mutant α1- and β-tubulin alleles, nda2-KM52 and nda3-KM-311, respectively (Umesono et al., 1983). The inhibition of colony formation of wild-type cells expressing Mal3 at high levels was partially suppressed by growth on medium containing 10 μg/ml TBZ (Fig. 6 A, d) and was also rescued by the presence of either mutant tubulin allele (Fig. 6 A, e–f). The experiment was carried out at 29°C, which while being permissive for both mutant tubulin strains does result in 10–20% of cells having abnormal bent cell shapes (Umesono et al., 1983), indicating that the tubulin alleles do not impart a fully wild-type function. We tested three different transformants expressing extra Mal3 per strain and consistently found that while the presence of the nda3-KM311 allele almost totally suppressed the inhibition of colony formation conferred by mal3 + overexpression, the nda2-KM52 allele was only partially able to do so.
Discussion
By a screen designed to isolate new components of the chromosome transmission machinery in fission yeast we have identified and characterized the mal3 + gene. The mal3-1 mutant strain shows a ∼400-fold increase in the rate of loss of a nonessential minichromosome at 24°C but otherwise grows normally. However, as an increase in chromosome loss rate less than three orders of magnitude might not severely affect colony formation, we cannot rule out an effect of mal3-1 on the transmission of regular chromosomes. Fission yeast Mal3 and the human APC-interacting protein EB-1 belong to the same protein family. Mutations in the APC gene result in the cancer predisposition syndrome FAP and are believed to be an early event in the development of the majority of sporadic colorectal tumors (reviewed in Kinzler and Vogelstein, 1996). Recently it has been demonstrated that the genetic instability that forms an integral part of colorectal tumors can be caused by severe defects in chromosome segregation (Lengauer et al., 1997). In this context it is intriguing that we have identified the mal3 + gene product, whose loss can be rescued by human EB-1, in a screen for components influencing chromosome transmission fidelity in fission yeast.
Mal3 Is a Microtubule-associated Protein that Influences Microtubule Integrity
In vivo localization of the Mal3 fusion protein showed, in addition to faint cytoplasmic and nuclear staining, strong fluorescence associated with both cytoplasmic and mitotic microtubules (Fig. 3 D). As is the case with antitubulin immunofluorescence mitotic spindles were brighter than cytoplasmic microtubules, consistent with more microtubules in spindles as opposed to the cytoplasmic microtubule bundles (Ding et al., 1993). These data strongly indicate that Mal3 interacts with microtubules although we do not yet know if the interaction is a direct one.
Indirect antitubulin immunofluorescence of mal3Δ cells showed altered microtubule structures. Instead of the typical wild-type cytoplasmic microtubule network that extends between the cell tips (Hagan and Hyams, 1988; Fig. 5, h and i) the interphase microtubules of the mal3Δ strain were extremely short, never reached the cell tips and were closely associated with the nucleus (Fig. 5, a and b). Regrowth of microtubules after depolymerization by incubation at low temperatures shows microtubule regrowth occurs predominantly around the nucleus (cited in Verde et al., 1995; Hagan, I.M., unpublished observations). The often fainter staining of microtubules in the mal3 + deletion strain indicates either poor fixation or fewer microtubules as opposed to the bundles of three microtubules observed in wild-type S. pombe cells (Streiblova and Girbardt, 1980; Tanaka and Kanbe, 1986).
The effect on spindle morphology was less marked in mal3Δ cells with the only visible defect being weaker staining intensity (Fig. 5, k). However a mal3Δ cell population showed a significantly increased chromosome condensation index indicating a delay during mitosis suggesting that the weaker staining reflects a compromised function. This finding is complemented by the observation that deletion of the mal3 + homologue in S. cerevisiae leads to an accumulation of cells with a 2C DNA content (Fleig, U., S. Heck, and J.H. Hegemann, unpublished observation).
Overproduction of the Mal3 protein severely compromised the formation and function of the mitotic spindle. Microtubules can be altered by the association of microtubule associating proteins (MAP) and interaction with microtubule severing proteins and motors (reviewed in Hirokawa, 1994; Desai and Mitchison, 1995; Hyman and Karsenti, 1996). Mitotic microtubules have shorter half-lives than interphase microtubules and microtubule turnover plays an important part in the assembly of the mitotic spindle (reviewed in Hyman and Karsenti, 1996). In addition proteins that increase the catastrophe rate of microtubules are required for correct mitotic spindle assembly (Belmont and Mitchison, 1996; Walczak et al., 1996). We speculate that overexpression of Mal3 might hyperstabilize microtubules and thus upset the fine-tuned balance of microtubule dynamics required for mitosis. In this context it is interesting that the mal3 + overexpression phenotype of severe growth inhibition was rescued partially by growth on TBZ-containing medium or in the presence of mutant tubulin alleles (Fig. 6 A, d–f). Rescue of a number of phenotypes caused by the absence of the microtubule-destabilizing mitotic motor protein Kar3p has also been observed in the presence of microtubule-destabilizing drugs or a reduction in tubulin gene dosage (Saunders et al., 1997).
An alternative explanation for the effect of excess Mal3 on the mitotic spindle could be that the excess protein is coating the mitotic microtubules, thus blocking/hindering binding of proteins required for proper spindle function.
Absence or Overexpression of Mal3 Protein Results in Displacement of Nucleus and Septum
The interphase nucleus of mal3Δ cells was often displaced from the center of the cells (Fig. 4 A), thus reinforcing the role played by the microtubule cytoskeleton and associated motor proteins in the positioning of the nucleus (for example Oakley and Morris, 1980; Jacobs et al., 1988; Hagan and Yanagida, 1997). In S. pombe, mutations in tubulin genes or the presence of microtubule-destabilizing chemicals can lead to a displacement of the nucleus (Walker, 1982; Umesono et al., 1983; Hiraoka et al., 1984; Verde et al., 1995).
Excess Mal3 protein also had a profound effect on the morphology and position of the division septum, resulting in abnormal and/or misplaced septa (Fig. 6 C, d and f) reminiscent of loss of function mutations in the fission yeast dmf1/mid1 gene, which is required for the correct positioning of the division septum (Chang et al., 1996; Sohrmann et al., 1996). A possible connection between the actin and microtubule cytoskeleton has been demonstrated in S. pombe by the existence of the benomyl-resistant ben4 mutant, which displays an altered actin cytoskeleton and is rendered lethal by the presence of an extra actin gene in the genome (Fantes, 1989). In this context the existence of an equatorial tubulin ring present in anaphase after the formation of the actin contractile ring is particularly interesting and led to the suggestion that this microtubule ring also plays a role in the determination of the division plane (Pichova et al., 1995). Similarly rings have also been observed with the in vivo localization of the S. pombe microtubule-associating protein Dis1 (Nabeshima et al., 1995) and our Mal3-yEGFP.
Deletion or Overexpression of mal3+ Leads to Aberrant Cell Morphology
A large number of abnormal curved or branched cells were observed in the mal3Δ strain and in cells overexpressing mal3 + indicating a possible role for Mal3 in the control of directed cell growth. The altered cell shape is most likely a consequence of the defective microtubule cytoskeleton of the mal3Δ strain as disruption or alteration of the microtubule cytoskeleton by mutation of the tubulin genes or by treatment with microtubule-destabilizing chemicals also leads to misshaped cells (Walker, 1982; Umesono et al., 1983; Hiraoka et al., 1984; Verde et al., 1995). The molecular basis for the importance of the microtubular cytoskeleton in the spatial organization of fission yeast has begun to emerge with the characterization of the tea1 + gene product, whose localization at the cell poles is dependent upon an intact cytoplasmic microtubule cytoskeleton and which is required to maintain antipodal growth (Mata and Nurse, 1997).
Excess Mal3 also resulted in morphological abnormalities in the presence of seemingly normal interphase microtubule arrays. This resembles the effect of overexpression of the microtubule-interacting protein Dis1 (Nabeshima et al., 1995; Nakaseko et al., 1996). The most likely explanation for these phenotypes is that the overproduction of a MAP occupies so many binding sites on the microtubule surface that it either directly competes out or sterically blocks the binding of other MAPs and motors. Thus the microtubules appear normal but are nonfunctional.
Function of the Human APC-interacting Protein EB-1?
The tumor suppressor protein APC has been implicated in a number of cellular processes including cell migration (Kinzler and Vogelstein, 1996; reviewed in Pfeifer, 1996). Although mutations in APC lead to an accumulation of enterocytes close to the crypt-villus transition in the intestine (reviewed in Polakis, 1995), overexpression of wild-type APC protein results in disordered cell migration in the intestinal epithelium of chimeric mice (Wong et al., 1996). In the presence of an intact microtubule cytoskeleton, endogenous APC protein in epithelial cells is concentrated in puncta near the end of microtubules that protrude into actively migrating membrane structures (Näthke et al., 1996). As the APC protein can promote assembly and bundling of microtubules in vitro (Munemitsu et al., 1994), one of its functions might be the recruitment of microtubules to protruding membrane structures thereby stabilizing/determining the direction of cell migration (Näthke et al., 1996). The important role of microtubule dynamics in guiding directed movement of, for example, the motile portion of the axon terminal, the growth cone, has been well documented (Sabry et al., 1991; Tanaka and Kirschner, 1995).
The carboxyl terminus of APC is required for the association with both microtubules and EB-1 (Munemitsu et al., 1994; Smith et al., 1994; Su et al., 1995). As human EB-1 and fission yeast Mal3 appear to have similar functions and our data indicate that Mal3 is a microtubule-interacting protein involved in microtubule integrity, we suggest that APC might exert the recruitment of microtubules to actively migrating membranes via EB-1 protein. Whether EB-1 is normally distributed all over microtubules (as is the case for Mal3) and APC only interacts with a sub-population, or whether EB-1 is always complexed with APC and microtubule ends awaits further analysis. Indeed interaction with APC might be only one of multiple functions executed by EB-1.
The control of directed cell growth is vital for cell function and cellular differentiation. The cylindrical form of the wild-type fission yeast cell and the highly polarized manner of growth of this organism facilitate the identification of components required for these processes. Our results raise the exciting possibility that some of the components required for directed cell growth in fission yeast and directed cell movement in higher eucaryotes have been conserved.
Acknowledgments
We thank K. Gould for the his3-D1 strains and pAF1; O. Niwa and M. Yanagida for strain HM438; K. Gull for the TAT1 antibody; A. Carr for the genomic DNA library; G. Schlauderer, M. Sen-Gupta, and T. Fiedler for help with the sequence analysis; B. McCormack for yEGFP; A. Fischer-Heuschkel for Mal3-yEGFP; and C. Reitz for photographic artwork. U. Fleig and J.H. Hegemann are grateful to E. Fleig for his continuous support.
This work was supported by the Cancer Research Campaign (CRC) and the Wellcome Trust equipment grant (to Manchester University, School of Biological Sciences) to I.M. Hagan, and the Deutsche Forschungsgemeinschaft (SFB 272, TP A1) and the European Union Programme BIOTECH II to J.H. Hegemann.
Abbreviations used in this paper
- APC
adenomatous polyposis coli
- FAP
familial adenomatous polyposis
- GFP
green fluorescent protein
- ORF
open reading frame
- MAP
microtubule-associating protein
- TBZ
thiabendazole
Footnotes
Please address all correspondence to Dr. Ursula Fleig, Institut für Mikrobiologie und Molekularbiologie, Frankfurterstrasse 107, 35392 Giessen, Germany. Tel.: +49 641 99 35544. Fax: +49 641 99 35549. E-mail: ursula. fleig@bio.uni-giessen.de
References
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. . Gene (Amst) 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST-database for “Expressed Sequence Tags.” . Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Chang F, Wollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile ring. J Cell Sci. 1996;109:132–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. A new role for motor proteins as couplers to depolymerizing microtubules. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, F.S., J. Mulligan, K. Hennessy, M.A. Yelton, E. Allen, R. Araujo, E. Aviles, A. Berno, T. Brennan, J. Carpenter et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome V. Nature (Lond.). 387(suppl):78–81. [PMC free article] [PubMed]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. . J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes, P.A. 1989. Molecular biology of the fission yeast. Academic Press, San Diego. 127–182.
- Fleig UN, Nurse P. Expression of a dominant negative allele of cdc2 prevents activation of the endogenous p34cdc2kinase. Mol Gen Genet. 1991;226:432–440. doi: 10.1007/BF00260656. [DOI] [PubMed] [Google Scholar]
- Fleig U, Sen-Gupta M, Hegemann JH. Fission yeast mal2 +is required for chromosome segregation. Mol Cell Biol. 1996;16:6169–6177. doi: 10.1128/mcb.16.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. . J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1 +associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Evidence for cell cycle-specific, spindle pole body mediated nuclear positioning in the fission yeast Schizosaccharomyces pombe. . J Cell Sci. 1997;110:1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The nda3 gene of fission yeast encodes β-tubulin: a cold sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Mikrotubule organization and dynamics dependent on microtubule-associated proteins. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Geiser JR. Genetic analysis of the mitotic spindle. Annu Rev Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Jacobs CW, Adams AEM, Szaniszlo PJ, Pringle JR. Functions of microtubles in the Saccharomyces cerevisiaecell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javerzat J-P, Cranston G, Allshire RC. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 1996;24:4674–4683. doi: 10.1093/nar/24.23.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of the cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Lange BH, Sherwin T, Hagan IM, Gull K. The basics of immunofluorescence video-microscopy for mammalian cells and microbial systems. Trends in Cell Biol. 1995;5:328–332. doi: 10.1016/s0962-8924(00)89056-x. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature (Lond) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci(Suppl) 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Matsumine A, Ogai A, Senda T, Okumura N, Satho K, Baeg G-H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Binding of APC to the human homolog of the drosophila discs large tumor suppressor protein. Science (Wash DC) 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- Maundrell KG. Thiamine repressible expression vectors pREP and pRIP for fission yeast. Gene (Amst) 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Munemitsu S, Souza B, Müller O, Albert I, Rubinfeld B, Polakis P. The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Nabeshima K, Kinoshita K, Yanagida M. Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes to Cells. 1996;1:633–644. doi: 10.1046/j.1365-2443.1996.00253.x. [DOI] [PubMed] [Google Scholar]
- Näthke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumour suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol. 1996;134:165–197. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsonomiya J, Baba S, Hedge P. Mutation of chromosome 5q21genes in FAP and colorectal cancer patients. Science (Wash DC) 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet. 1986;203:397–405. [Google Scholar]
- Oakley BR, Morris NR. Nuclear movement is β-tubulin dependent in Aspergillus nidulans. Cell. 1980;19:255–262. doi: 10.1016/0092-8674(80)90407-9. [DOI] [PubMed] [Google Scholar]
- Ohi R, Feoktistova A, Gould KL. Construction of vectors and a genomic library for use with his3-deficient strains of Schizosaccharomyces pombe. . Gene (Amst) 1996;174:315–318. doi: 10.1016/0378-1119(96)00085-6. [DOI] [PubMed] [Google Scholar]
- Pfeifer M. Regulating cell proliferation: as easy as APC. Science (Wash DC) 1996;272:974–975. doi: 10.1126/science.272.5264.974. [DOI] [PubMed] [Google Scholar]
- Pichova A, Kohlwein SD, Yamamoto M. New arrays of cytoplasmic microtubules in the fission yeast Schizosaccharomyces pombe. . Protoplasma. 1995;188:252–257. [Google Scholar]
- Polakis P. Mutations in the APC gene and their implications for protein structure and function. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Sabry JH, O'Connor TP, Evans L, Toroian-Raymond A, Kirschner M, Bentley D. Microtubule behavior during guidance of pioneer neuron growth cones in sito. J Cell Biol. 1991;115:381–395. doi: 10.1083/jcb.115.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Leslie RJ, Karow WM, McIntosh JR, Saxton WM. Spindle microtubule dynamics in sea urchin embryos: analysis using fluoresence-labeled tubulin and measurements of fluoresence distribution after photobleaching. J Cell Biol. 1984;99:2165–2186. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Frankhauser C, Brodbeck C, Simanis V. The dmf1/ mid1gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Smith KJ, Levy DB, Maupin P, Pollard TD, Vogelstein B, Kinzler KW. Wild-type but not mutant APC associates with the microtubule cytoskeleton. Cancer Res. 1994;54:3627–3675. [PubMed] [Google Scholar]
- Streiblova E, Girbardt M. Microfilaments and microtubules in cell division cycle mutants of Schizosaccharomyces pombe. . Can J Microbiol. 1980;26:250–261. doi: 10.1139/m80-038. [DOI] [PubMed] [Google Scholar]
- Su L-K, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler KW. APC binds to the novel protein EB-1. Cancer Res. 1995;55:2972–2977. [PubMed] [Google Scholar]
- Tanaka E, Kirschner MW. The role of microtubule dynamics in growth cone motility and axonal growth. J Cell Biol. 1995;128:139–155. doi: 10.1083/jcb.128.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kanbe T. Mitosis in the fission yeast Schizosaccharomyces pombeas revealed by freeze-substitution electron microscopy. J Cell Sci. 1986;80:253–268. doi: 10.1242/jcs.80.1.253. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell division gene NDA2 as one of the two different α-tubulin genes in Schizosaccharomyces pombe. . Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M. Two cell division cycle genes NDA2 and NDA3 of the fission yeast Schizosaccharomyces pombecontrol microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J Mol Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GM. Cell cycle specificity of certain anti-microtubular drugs in Schizosaccharomyces pombe. . J Gen Micro. 1982;128:61–71. doi: 10.1099/00221287-128-1-61. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ. Kinesin-related proteins at mitotic spindle poles. Function and regulation. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]
- Wong MH, Hermiston ML, Syder AJ, Gordon JI. Forced expression of the tumour suppressor adenomatosis polyposis protein induces disordered cell migration in the intestinal epithelium. Proc Natl Acad Sci USA. 1996;93:9588–9593. doi: 10.1073/pnas.93.18.9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sassa R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal anitbodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Frontier questions about sister chromatid separation in anaphase. Bioessays. 1995;17:519–526. doi: 10.1002/bies.950170608. [DOI] [PubMed] [Google Scholar]