Abstract
Site-directed N-ethylmaleimide labeling was studied with Glu-126 and/or Arg-144 mutants in lactose permease containing a single, native Cys residue at position 148 in the substrate-binding site. Replacement of either Glu-126 or Arg-144 with Ala markedly decreases Cys-148 reactivity, whereas interchanging the residues, double-Ala replacement, or replacement of Arg-144 with Lys or His does not alter reactivity, indicating that Glu-126 and Arg-144 are charge-paired. Importantly, although alkylation of Cys-148 is blocked by ligand in wild-type permease, no protection whatsoever is observed with any of the Glu-126 or Arg-144 mutants. Site-directed fluorescence with 2-(4-maleimidoanilino)-naphthalene-6-sulfonic acid (MIANS) in mutant Val-331 → Cys was also studied. In marked contrast to Val-331 → Cys permease, ligand does not alter MIANS reactivity in mutant Glu-126 → Ala/Val-331 → Cys, Arg-144 → Ala/Val-331 → Cys, or Arg-144 → Lys/Val-331 → Cys and does not cause either quenching or a shift in the emission maximum of the MIANS-labeled mutants. However, mutation Glu-126 → Ala or Arg-144 → Ala and, to a lesser extent, Arg-144 → Lys cause a red-shift in the emission spectrum and render the fluorophore more accessible to I−. The results demonstrate that Glu-126 and Arg-144 are irreplaceable for substrate binding and suggest a model for the substrate-binding site in the permease. In addition, the findings are consistent with the notion that alterations in the substrate translocation pathway at the interface between helices IV and V are transmitted conformationally to the H+ translocation pathway at the interface between helices IX and X.
Keywords: bioenergetics/transport/membrane proteins/site-directed thiol labeling/charge-pair interaction
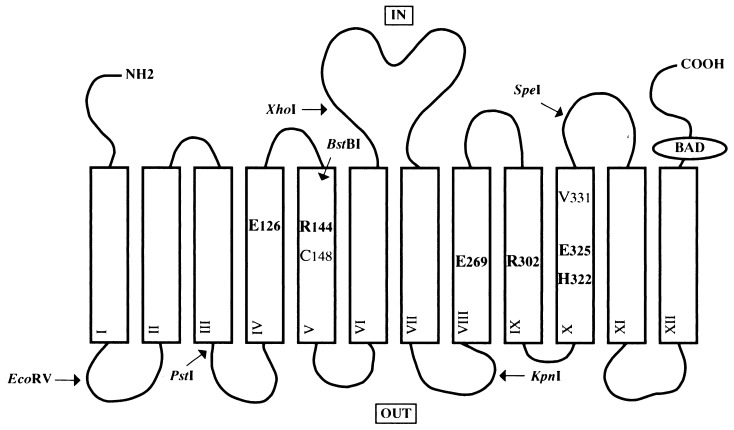
The lactose permease (lac permease) of Escherichia coli is representative of secondary active transport proteins that convert free energy stored in electrochemical ion gradients into work in the form of a concentration gradient (reviewed in ref. 1). This hydrophobic, polytopic, cytoplasmic membrane protein catalyzes the coupled stoichiometric translocation of β-galactosides and H+. The lacY gene that encodes the permease has been cloned and sequenced, and the product of the lacY gene has been solubilized, purified, reconstituted into proteoliposomes, and shown to be solely responsible for β-galactoside transport as a monomer (see ref. 2). All available evidence indicates that the permease consists of 12 hydrophobic, membrane-spanning, α-helical domains connected by hydrophilic loops with both the N and C termini on the cytoplasmic face of the membrane (Fig. 1) (reviewed in refs. 3 and 4).
Figure 1.
Secondary structure model of lac permease. Putative transmembrane helices are shown in boxes. The positions of the six irreplaceable residues (Glu-126, Arg-144, Glu-269, Arg-302, His-322, and Glu-325), as well as Cys-148 and Val-331, are indicated. Also shown are the restriction endonuclease sites used for constructing the mutants and the biotin acceptor domain (BAD). The topology of helices IV and V have been modified as described in the text.
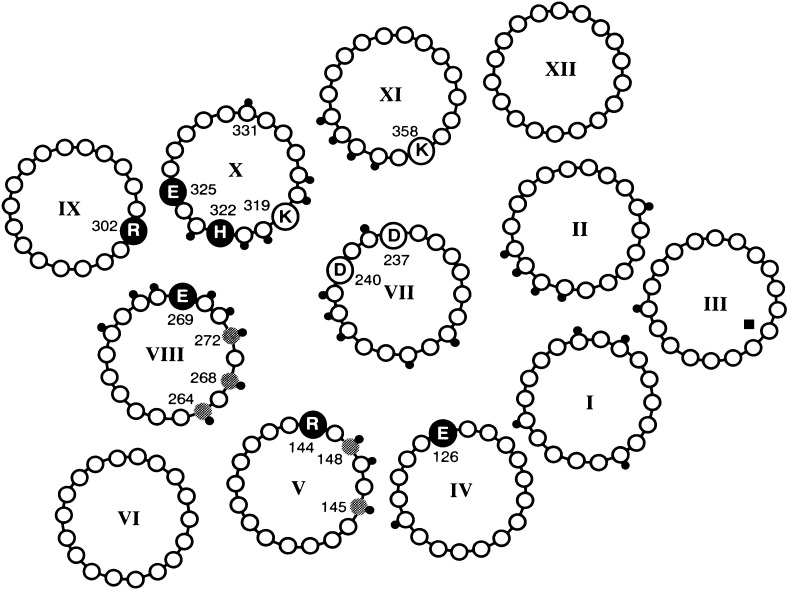
Site-directed mutagenesis of wild-type permease and Cys-scanning mutagenesis of a functional mutant devoid of Cys residues (C-less permease) reveals that 4 of 417 residues in the protein are irreplaceable with respect to coupling between lactose and H+ translocation—Glu-269 (helix VIII), Arg-302 (helix IX), His-322 (helix X), and Glu-325 (helix X) (reviewed in refs. 3–5). Although the permease has not been crystallized, application of a battery of site-directed biochemical and biophysical techniques that include second-site suppressor analysis, excimer fluorescence, engineered divalent metal binding sites, chemical cleavage, electron paramagnetic resonance, thiol crosslinking, and identification of discontinuous mAb epitopes has enabled the formulation of a helix packing model (Fig. 2) (see refs. 4–6). Experimental observations from structural and extensive mutational analysis have led to a proposed mechanism for energy coupling between sugar and H+ transport (5, 7).
Figure 2.
Helix packing model of lac permease viewed from the cytoplasmic surface. The positions of the six irreplaceable residues (Glu-126, Arg-144, Glu-269, Arg-302, His-322, and Glu-325) are indicated by filled, enlarged circles. The two interacting pairs of Asp-Lys residues (Asp-237/Lys-358 and Asp-240/Lys-319) are highlighted by open, enlarged circles. The gray circles represent residues where the NEM reactivity of Cys replacement mutants is blocked by substrate. The positions of NEM-sensitive Cys replacements are indicated with a small, black dot.
Lac permease is inactivated irreversibly by N-ethylmaleimide (NEM) and protection is afforded by substrate (8) which led to the postulate that a Cys residue is at or near the binding site, and the substrate-protectable residue was shown later to be Cys-148 (9). Although Cys-148 is not essential for transport, the size and polarity of the side chain at this position can modify transport activity and substrate specificity (10). Small hydrophobic side chains (Ala and Val) increase apparent affinity for substrate, hydrophilic side chains (Ser, Thr, and Asp) decrease apparent affinity, and bulky or positively charged side chains (Phe and Lys) virtually abolish activity. Single Cys-148 permease reacts rapidly with 2-(4-maleimidoanilino)naphthalene-6-sulfonic acid (MIANS), a fluorophore whose quantum yield increases dramatically upon reaction with a thiol (11). Ligands of the permease block the reaction, and the concentration dependence correlates with the affinity of ligand [i.e., β-d-galactopyranosyl 1-thio-β-d-galactopyranoside (TDG) ≫ lactose > galactose]. However, neither glucose, which differs from galactose in only the stereochemistry of the C4-OH, nor sucrose has any effect. Met-145 is located on the same face of helix V as Cys-148, and M145C permease reacts with MIANS in a similar fashion, but the effect of ligand is far less dramatic. The fluorescence of MIANS-labeled Cys-148 or Cys-145 permease is quenched by I−, indicating that this face of helix V is solvent accessible. Moreover, I− quenching of MIANS-labeled M145C permease is attenuated by TDG. These observations and others (12) indicate that Cys-148 and Met-145 are components of the substrate translocation pathway, Cys-148 interacting weakly and hydrophobically with the galactosyl moiety of substrate and Met-145 interacting even more weakly with the glucosyl moiety.
Fluorescence studies (13) with purified single-Cys V331C permease show that concentrations of TDG ranging from 1 to 10 mM increase the MIANS reactivity of V331C permease and cause quenching and a blue-shift in the emission spectrum; over a range of TDG concentrations from 0 to 0.5 mM, only fluorescence quenching is observed. These results and others (14) are consistent with the presence of two binding sites, occupation of the high affinity site facing the periplasmic side (Kd ≅ 0.12 mM) resulting in quenching and occupation of the low affinity site facing the cytoplasmic side (Kd ≅ 3−5 mM) causing position 331 to move into a more hydrophobic environment.
Among the last residues in the permease to be mutagenized (15)—Glu-126 and Arg-144 in the cytoplasmic halves of helices IV and V, respectively—are irreplaceable for active transport. Initially, the two residues were placed at the membrane– water interface at the cytoplasmic ends of helices IV and V, respectively (16). However, studies using single amino acid deletions (C. Wolin and H.R.K., unpublished observations), nitroxide-scanning and accessibility measurements (M. Zhao, J. Hernandez-Borrell, W. L. Hubbell, and H.R.K., unpublished observations), and lac permease fusions with the NG domain of FtsY (E. Bibi, personal communication) indicate that loop IV/V is smaller than indicated by hydropathy profiling, extending only from about Val-132 to Phe-138 (Fig. 1). Wild-type or C-less permease with a neutral replacement for Glu-126 or Arg-144 is totally inactive, even with respect to downhill lactose translocation, in contrast to mutations in residues involved in H+ translocation and coupling (reviewed in refs. 4, 5, and 7). Activity is not rescued by double-neutral replacements or by interchanging the residues. The only mutations tested that retain partial activity are E126D and R144K. E126D permease catalyzes lactose accumulation to a steady-state level comparable to wild type, but the apparent Km is higher with no change in Vmax (M. Sahin-Tóth and H.R.K., unpublished observations). R144K permease transports lactose at a very slow rate to only about 25% of the wild-type steady-state (Fig. 2).
In this communication, site-directed labeling of Cys-148 and Val-331 was used to study the role of Glu-126 and Arg-144. The results indicate that both residues are irreplaceable for substrate binding, and a model for the binding site is formulated.
MATERIALS AND METHODS
Materials.
Deoxyoligonucleotides were synthesized on an Applied Biosystems 391 DNA synthesizer. Restriction endonucleases and T4 DNA ligase were from New England Biolabs. Taq DNA polymerase was from Promega. N-[1-14C]ethylmaleimide (40 mCi/mmol) was purchased from DuPont/NEN. 125I-labeled protein A was from Amersham. Immobilized monomeric avidin was from Pierce. Site-directed rabbit polyclonal antiserum against a dodecapeptide corresponding to the C terminus was prepared as described (17). MIANS was from Molecular Probes. All other materials were reagent grade and obtained from commercial sources.
Plasmid Construction.
Mutant permease R144A/C148, R144K/C148, R144H/C148, or E126R/R144E/C148 was constructed by introducing mutation R144A, R144K, R144H, or E126R/R144E into plasmid pT7–5/cassette lacY encoding single-Cys-148 by oligonucleotide-directed, site-specific mutagenesis using either two- (18) or one-step PCR. The PCR products were digested by using restriction sites PstI, BstBI, XhoI, or KpnI and ligated to similarly treated plasmid pKR35/C-less lacY-CXB (encoding Cys-less permease with a biotin acceptor domain at the C terminus) (19). Mutant E126A/C148 was constructed by restriction fragment replacement of the DNA fragment encoding E126A (15) into pKR35/Cys-148-CXB using the EcoRV and BstBI restriction sites. Mutant E126A/R144A/C148 was constructed by restriction fragment replacement of the DNA fragment encoding R144A/C148 into pKR35/C-less lacY-CXB encoding E126A using the BstBI and SpeI restriction sites. Mutants E126A/V331C, R144A/V331C, or R144K/V331C were constructed by restriction fragment replacement of the DNA fragment encoding V331C (20) into plasmid pKR35/C-less lacY-CXB encoding E126A, R144A, or R144K using the KpnI and SpeI restriction sites. Mutations were verified by sequencing the length of the PCR-generated or replacement segment through the ligation junctions by using dideoxynucleotide termination (21).
Growth of Bacteria.
E. coli T184 (lacY−Z−) transformed with each plasmid was grown aerobically at 37°C in Luria– Bertani broth containing streptomycin (10 μg/ml) and ampicillin (100 μg/ml). Fully grown cultures were diluted 10-fold and grown for 2 h more before induction with 0.5 mM iso-propyl 1-thio-β-d-galactopyranoside. After additional growth for 2 h, cells were harvested and used for membrane preparations.
Labeling with [14C]NEM.
Crude membranes were prepared by osmotic lysis and sonification as described (22), and in situ labeling of Cys-148 with [1-14C]NEM was performed (14).
Protein Determinations.
Protein was assayed by using a Micro BCA protein determination kit (Pierce).
Purification of Mutant Lac Permease for Fluorescence Studies.
Crude membranes were prepared by disruption of cells in a French pressure cell, followed by centrifugation. Membrane proteins were solubilized with 2% n-dodecyl β-d-maltopyranoside (DM), and permease was purified by affinity chromatography on immobilized monomeric avidin (11).
Labeling of Permease Mutants with MIANS and Fluorescence Measurements.
Fluorescence was measured at 22°C with an SLM 8000C spectrofluorometer (SLM–Aminco, Urbana, IL). Emission spectra were recorded by using an excitation wavelength of 330 nm and 4-nm slits for excitation and emission. The rate of MIANS labeling of V331C, fluorescence emission spectra of MIANS-labeled mutants, and quenching by I− were measured as described (13). Sucrose, which does not bind (11), was used as a control for all the measurements.
RESULTS
Site-Directed Sulfhydryl Modification and Protection by TDG in Situ.
Reactivity of single-Cys-148 permease with many sulfhydryl reagents is blocked by ligand (14). To study the effect of mutation E126A, R144A, R144K, R144H, E126R/R144E, or E126A/R144A on substrate binding, each mutation was introduced into single-Cys-148 permease, and reactivity of Cys-148 with [14C]NEM was determined in the absence or presence of the high-affinity ligand TDG.
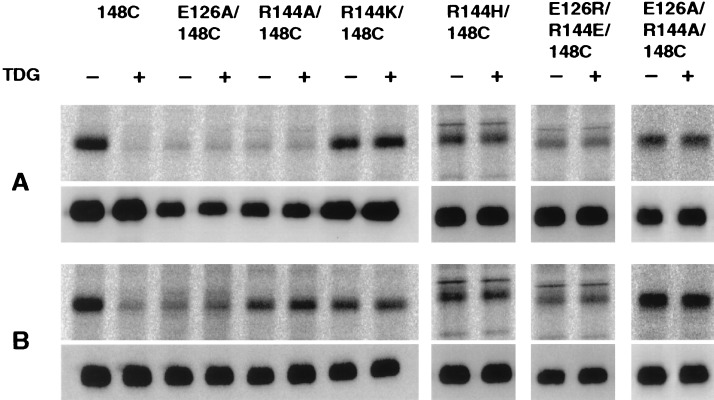
Single-Cys-148 permease is readily alkylated by NEM in 10 min (Fig. 3A) (14). Under the same conditions, E126A/C148 or R144A/C148 hardly react. In contrast, normal reactivity is observed in mutants R144K/C148 and R144H/C148. Furthermore, when the charges are interchanged or neutralized in mutant E126R/R144E/C148 or E126A/R144A/C148, respectively, essentially normal reactivity also is observed. After incubation with NEM for 30 min, the extent of labeling increases in all the mutants (Fig. 3B).
Figure 3.
Reactivity of single-Cys permease mutants with [1-14C]NEM and the effect of TDG. Membranes [0.2–0.4 mg of protein in 50 μl of 100 mM KPi (pH 7.5)/10 mM MgSO4] prepared from E. coli T184 transformed with pKR35/lacY-CXB encoding single-Cys mutants E126A/C148, R144A/C148, R144K/C148, R144H/C148, E126R/R144E/C148, or E126A/R144A/C148, as indicated, were incubated with [1-14C]NEM (40 mCi/mmol; 0.4 mM final concentration) for 10 min (A) or 30 min (B) in the absence or presence of 10 mM TDG at 25°C. The reactions were terminated by the addition of DTT to a final concentration of 15 mM, and biotinylated permease was solubilized and purified as described (14). Aliquots containing 5 μg of protein were separated by sodium dodecyl sulfate/12% PAGE, and the labeled protein was visualized by autoradiography (Upper, A and B). A fraction of the eluted protein (0.5 μg) was analyzed by Western blotting with anti-C-terminal antibody (Lower, A and B).
The reactivity of single-Cys-148 permease with NEM is almost completely blocked by TDG (Fig. 3 A and B). Strikingly, with each of the other mutants tested, TDG affords no protection whatsoever against NEM labeling. Thus, a guanidino group at position 144 and a carboxylate at position 126 appear to be absolutely required for ligand binding.
Site-Directed Fluorescence.
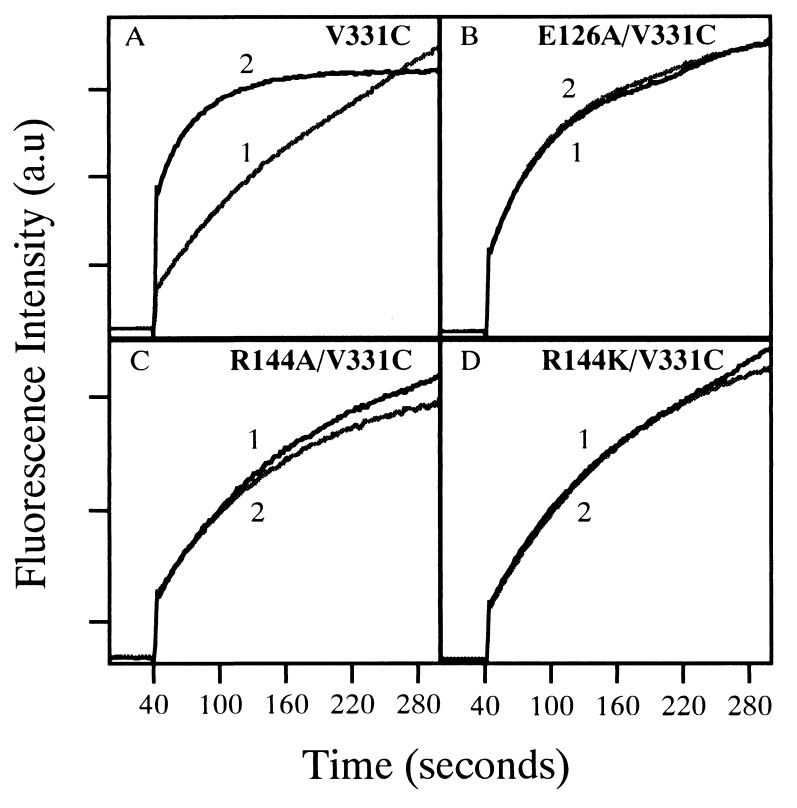
To determine whether the Glu-126 or Arg-144 mutants are defective in either the high- or low-affinity site or both, the E126A, R144A, or R144K mutations were transferred into V331C permease. As shown (13), TDG enhances the reactivity of V331C permease with MIANS (Fig. 4A). In contrast, the substrate analog fails to have any effect on the reactivity of mutant E126A/V331C, R144A/V331C, or R144K/V331C (Fig. 4 B–D).
Figure 4.
Reaction of MIANS with purified permease mutants and effect of TDG. Affinity-purified single-Cys permease mutant V331C (A), E126A/V331C (B), R144A/V331C (C), or R144K/V331C (D) [40 μg of pure protein in 0.5 ml of 50 mM KPi (pH 7.5)/150 mM NaCl/0.02% DM] was preincubated with a given concentration of ligand. MIANS labeling was initiated by addition of MIANS to a final concentration of 1 μM, and fluorescence increase was recorded continuously at 415 nm (excitation 330 nm) as a function of time as described (13). Ligand was added as follows: curve 1, no addition or 10 mM sucrose; curve 2, 10 mM TDG.
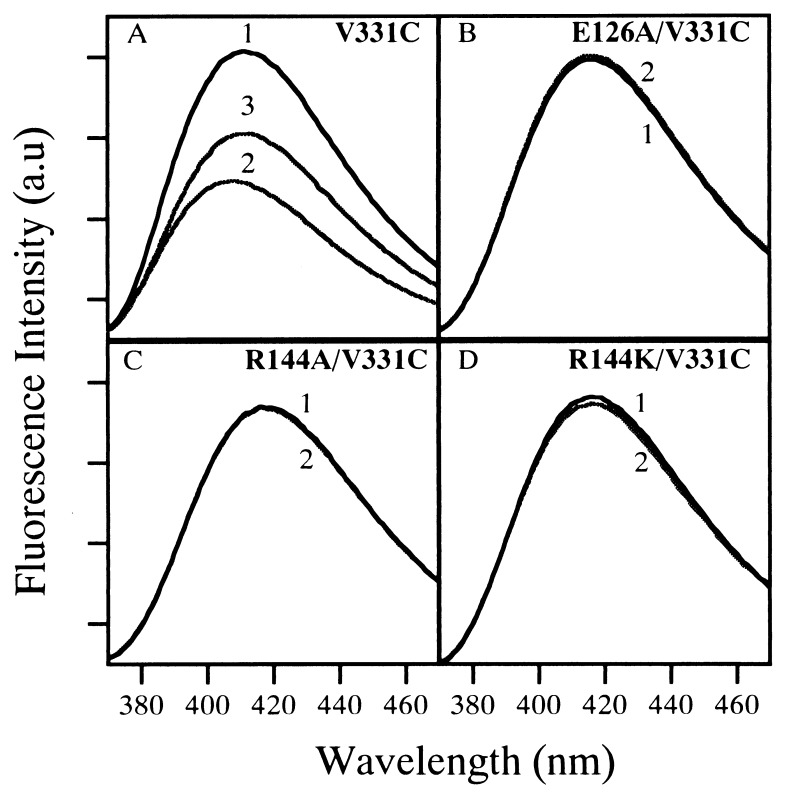
The emission spectrum of MIANS-labeled V331C permease exhibits a maximum at 411 nm in the absence of ligand, and 10 mM TDG causes 40% quenching and a 6-nm blue-shift (Fig. 5A). Interestingly, the emission maximum of MIANS-labeled E126A/V331C, R144A/V331C, or R144K/V331C permease is red-shifted by about 5–7 nm, indicating that the fluorophore is in a relatively more polar environment (Fig. 5 B–D). In dramatic contrast to V331C permease, TDG has no effect on the emission spectrum of MIANS-labeled E126A/V331C, R144A/V331C, or R144K/V331C permease. Taken together with the NEM-labeling results, the observations demonstrate that E126A, R144A, and R144K permease are completely unable to bind substrate.
Figure 5.
Fluorescence emission spectra of MIANS-labeled permease mutants and effect of TDG. Each single-Cys permease mutant was labeled with MIANS while bound to avidin-Sepharose. Reactions were terminated with DTT, and the excess reagent was removed by extensive washing before elution of the labeled protein from the affinity column. Emission spectra of MIANS-labeled permease V331C (A), E126A/V331C (B), R144A/V331C (C), and R144K/V331C (D) [40 μg of pure protein in 0.5 ml of 50 mM KPi (pH 7.5)/150 mM NaCl/0.02% DM] were measured (excitation 330 nm) at 22°C after preincubation with a given concentration of ligand as follows: curve 1, 10 mM sucrose; curve 2, 10 mM TDG; curve 3, 0.2 mM TDG.
Information regarding the polarity of the environment in the vicinity of MIANS can be obtained by using I−, a collisional quencher (23). The Stern–Volmer quenching constant (KSV) is 10-fold higher for MIANS-labeled E126A/V331C and R144A/V331C permease relative to V331C permease (Table 1), thereby confirming the observation that the fluorophore is more solvent-accessible in the mutants. Interestingly, KSV is only about 4-fold higher with R144K/V331C permease, where a charge-pair between positions 126 and 144 should be maintained.
Table 1.
Stern–Volmer quenching constants for I− with MIANS-labeled permease mutants
| Mutant | KSV, M−1 |
|---|---|
| V331C | 0.06 |
| E126A/V331C | 0.62 |
| R144A/V331C | 0.58 |
| R144K/V331C | 0.24 |
MIANS labeling of each single-Cys permease mutant was performed with the protein bound to avidin-sepharose. Reactions were terminated with DTT, and excess reagent was removed by extensive washing before elution of the labeled protein from the affinity column. Fluorescence emission maxima (see Fig. 5) then were recorded (excitation, 330 nm) in the presence of concentrations of KI ranging from 0.025 M to 0.4 M, and the spectra were corrected for identical measurements carried out in the presence of the same concentrations of KCl. The data were fit to the Stern–Volmer equation by using a linear regression program as described (13).
DISCUSSION
By far, the most important result presented here is that TDG does not protect Cys-148 from alkylation by NEM in any of the mutants tested. Thus, a guanidino group at position 144 and a carboxylate at position 126 are absolutely required for substrate binding. In addition to Glu-126 and Arg-144, there are two single-Cys replacement mutants, K131C and F140C, in this region of the protein that exhibit low transport activity (15). Replacement of either one of these residues with Ala has no effect on the reactivity of Cys-148 to NEM or on substrate protection of Cys-148 from alkylation, which emphasizes the uniqueness of Glu-126 and Arg-144 (K. C. Zen and H.R.K., unpublished observations).
In addition, in situ labeling demonstrates that replacement of Glu-126 or Arg-144 with Ala causes a dramatic decrease in the reactivity of Cys-148. Importantly, replacement of Arg-144 with Lys, in particular, or His restores Cys-148 reactivity, which suggests that a Lys or His side chain can partially mimic Arg, a conclusion supported by I−-quenching studies (Table 1). Recent site-directed thiol crosslinking studies (C. Wolin and H.R.K., unpublished observations) with paired Cys replacements indicate that the middle portions of transmembrane helices IV and V are in close proximity. Therefore, it is likely that Glu-126 and Arg-144 interact by charge-pairing.
The NEM reactivity of Cys-148 in the Glu-126 and Arg-144 mutants is highly analogous to the transport properties of mutants in Asp-237 (helix VII) and Lys-358 (helix XI), which are charge-paired (reviewed in ref. 6). Neutral replacement of Asp-237 or Lys-358 resulting in an unpaired charge abolishes active lactose transport, whereas simultaneous neutral replacement or reversal of the charged residues yields active permease. Furthermore, D237H/K358H permease binds Mn2+ (24), providing direct evidence that the two positions are in close proximity. Similarly, normal reactivity of Cys-148 is observed in the charge-reversal mutant E126R/R144E/C148 and the double-neutral mutant E126A/R144A/C148, in marked contrast to the low reactivity in the single mutants E126A/C148 and R144A/C148. The findings provide strong support for charge-pairing between Glu-126 and Arg-144. However, unlike Asp-237 and Lys-358, Glu-126 and Arg-144 are essential for transport, which precludes functional studies.
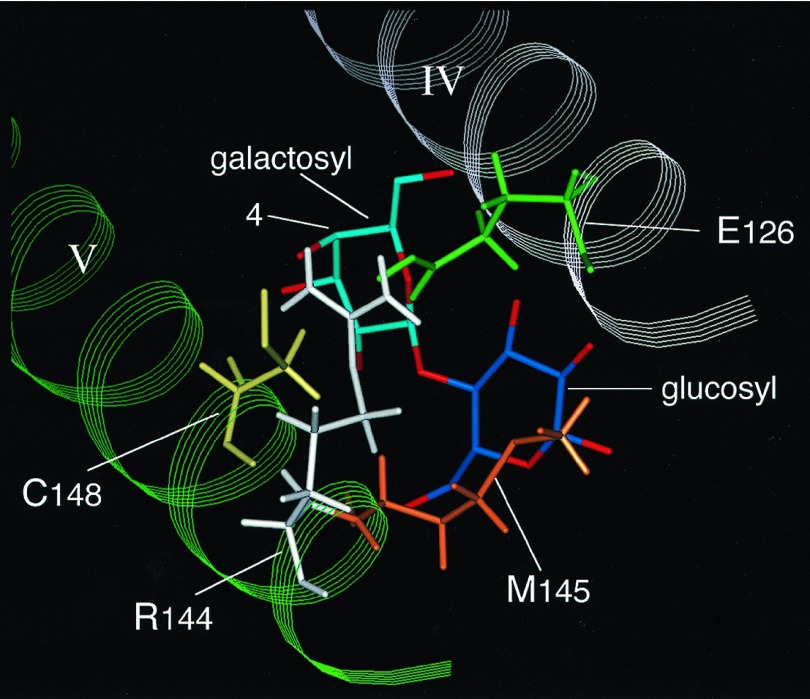
Taken together with the observations regarding the roles of Cys-148 and Met-145 in substrate binding (10, 11), the results lead to a model for the substrate-binding site depicted in Fig. 6, the chief features of which are the following. (i) One of the guanidino NH2 groups of Arg-144 H-bond(s) to the OH group at the C4 and/or C3 positions of the galactosyl moiety of the substrate, an interaction(s) that plays a key role in the substrate specificity of the permease. Galactose binds with low affinity, but gulose and glucose, which differ from galactose only in the stereochemistry of the OH group at the C3 and C4 position, respectively, have no affinity for the permease (11, 25). (ii) The other guanidino NH2 group of Arg-144 forms a salt-bridge with Glu-126, and the interaction holds Arg-144 and Cys-148 in an orientation that allows specific interaction with the galactosyl moiety. One of the oxygen atoms of the carboxylate at position 126 also could act as an H-bond acceptor from the C6-OH of the galactosyl moiety (26). (iii) Cys-148 interacts weakly and hydrophobically with the galactosyl end of lactose and other galactosides (10, 11). (iv) Met-145 interacts even more weakly with the glucosyl part of lactose (11).
Figure 6.
Putative substrate-binding site in lac permease. Helices IV (white) and V (green) are shown with Glu-126 (green), Arg-144 (white), Met-145 (orange), and Cys-148 (yellow). The galactosyl and glucosyl moieties of lactose are in light and dark blue, respectively. As indicated, one of the guanidino NH2 groups of Arg-144 interacts with the hydroxyl group at the C4 and/or C3 positions of the galactosyl moiety, and the other guanidino NH2 group interacts with Glu-126, which may also interact with the C6-OH of the galactosyl moiety. Cys-148 interacts weakly and hydrophobically with the galactosyl moiety, and Met-145 interacts even more weakly with the glucosyl part of lactose.
Kinetic measurements, binding and site-directed labeling studies indicate that lac permease has two binding sites (13, 14, 27–29). The site-directed fluorescence studies reported here demonstrate that mutants E126A/V331C, R144A/V331C, or R144K/V331C are defective in both sites. Moreover, there is a red shift in the fluorescence of MIANS-labeled E126A/V331C, R144A/V331C, and R144K/V331C permease and increased accessibility to I−, suggesting that the E126A and R144A mutations and, to a lesser extent, the R144K mutation place position 331 in a more solvent-accessible environment. Therefore, mutations at positions 126 or 144 may produce conformational alterations in helix X, which contains two residues essential for H+ translocation and coupling, Glu-325 and His-322. Although it is not surprising that Ala replacement for Glu-126 or Arg-144 alters the environment of Cys-148, a residue proximal in sequence and space, long-range conformational changes at position 331, which is distal in sequence and space, are consistent with a high degree of flexibility in the permease (reviewed in refs. 4 and 5). It is noteworthy in this context that recent site-directed thiol-crosslinking studies with coexpressed N- and C-terminal halves of the permease indicate that cytoplasmic loop IV/V is in close proximity to loops VIII/IX and X/XI (I. Kwaw and H.R.K., unpublished observations). Hence, as postulated (4, 5, 7) conformational changes at the interface between helices IV/V could alter the conformation of helix X and other helices in the C-terminal half of the permease.
In summary, it is clear that Glu-126 and Arg-144 are indispensable for substrate binding. Moreover, the contrast between the properties of the charge-reversal mutant E126R/R144E, the double-neutral substitution mutant E126A/R144A/C148, and mutant R144K/C148 with the single-neutral mutants E126A and R144A argues strongly that the two residues interact by charge-pairing. The finding that permease mutants E126A, R144A, R144K, R144H, E126R/R144E, or E126A/R144A do not bind ligand raises two possibilities that may not be mutually exclusive: (i) Glu-126 and Arg-144 are directly involved in substrate recognition, as discussed, or (ii) Glu-126 and Arg-144 play a crucial part in maintaining the conformation of the substrate translocation pathway because mutations at the two positions obliterate both high- and low-affinity binding on either side of the membrane. Furthermore, long-range conformational alterations caused by such mutations could encompass helical interfaces involved in coupling H+ and substrate translocation.
Finally, interactions between conserved Arg and Glu residues on the cytosolic face of the mammalian glucose facilitator GLUT4 have been found to be important for the integrity of the outward- and inward-facing conformations and for oscillation between these two states (30). Specifically, two conserved residues, Glu-146 and Arg-153 in putative loop IV/V, which are analogous to Glu-126 and Arg-144 in lac permease, appear to be required for all conformational states of the transporter, as mutation of either residue impairs ligand binding and transport.
Acknowledgments
We are indebted to John Voss for the molecular graphics shown in Fig. 6 and Alberto Gonzalez for constructing mutants R144H/C148 and E126R/R144E/C148. P.V. is the recipient of National Institutes of Health National Research Service Award Postdoctoral Fellowship 5 F32 DK09287. The work was supported in part by National Institutes of Health Grant DK51131 to H.R.K.
ABBREVIATIONS
- lac permease
lactose permease
- C-less permease
functional lac permease devoid of Cys residues
- TDG
β-d-galactopyranosyl 1-thio-β-d-galactopyranoside
- DM
n-dodecyl β-d-maltopyranoside
- NEM
N-ethylmaleimide
- KPi
potassium phosphate
- MIANS
2-(4-maleimidoanilino)naphthalene-6-sulfonic acid
References
- 1.Kaback H R. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 2.Sahin-Tóth M, Lawrence M C, Kaback H R. Proc Natl Acad Sci USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaback H R. In: Handbook of Biological Physics: Transport Processes in Eukaryotic and Prokaryotic Organisms. Konings W N, Kaback H R, Lolkema J S, editors. II. Amsterdam: Elsevier; 1996. pp. 203–227. [Google Scholar]
- 4.Kaback, H. R. & Wu, J. (1998) Q. Rev. Biophys., in press. [DOI] [PubMed]
- 5.Frillingos, S., Sahin-Tóth, M., Wu, J. & Kaback, H. R. (1998) FASEB J., in press. [DOI] [PubMed]
- 6.Kaback H R, Voss J, Wu J. Curr Opin Struct Biol. 1997;7:537–542. doi: 10.1016/s0959-440x(97)80119-4. [DOI] [PubMed] [Google Scholar]
- 7.Kaback H R. Proc Natl Acad Sci USA. 1997;94:5539–5543. doi: 10.1073/pnas.94.11.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox C F, Kennedy E P. Proc Natl Acad Sci USA. 1965;54:891–899. doi: 10.1073/pnas.54.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyreuther K, Bieseler B, Ehring R, Müller-Hill B. In: Methods in Protein Sequence Analysis. Elzinga M, editor. Clifton, NJ: Humana; 1981. pp. 139–148. [Google Scholar]
- 10.Jung H, Jung K, Kaback H R. Biochemistry. 1994;33:12160–12165. doi: 10.1021/bi00206a019. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Kaback H R. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 12.Weitzman C, Kaback H R. Biochemistry. 1995;4:2310–2318. doi: 10.1002/pro.5560041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Frillingos S, Voss J, Kaback H R. Protein Sci. 1994;3:2294–2301. doi: 10.1002/pro.5560031214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frillingos S, Kaback H R. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 15.Frillingos S, Gonzalez A, Kaback H R. Biochemistry. 1997;47:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 16.Foster D L, Boublik M, Kaback H R. J Biol Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- 17.Carrasco N, Herzlinger D, Mitchell R, DeChiara S, Danho W, Gabriel T F, Kaback H R. Proc Natl Acad Sci USA. 1984;81:4672–4676. doi: 10.1073/pnas.81.15.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Consler T G, Persson B L, Jung H, Zen K H, Jung K, Prive G G, Verner G E, Kaback H R. Proc Natl Acad Sci USA. 1993;90:6934–6938. doi: 10.1073/pnas.90.15.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahin-Tóth M, Kaback H R. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulsen A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frillingos S, Sahin-Toth M, Persson B, Kaback H R. Biochemistry. 1994;33:8074–8081. doi: 10.1021/bi00192a012. [DOI] [PubMed] [Google Scholar]
- 23.Eftink M R, Gryczynski I, Wiczk W, Laczko G, Lakowicz J R. Biochemistry. 1991;30:8945–8953. doi: 10.1021/bi00101a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He M M, Voss J, Hubbell W L, Kaback H R. Biochemistry. 1995;34:15661–15666. doi: 10.1021/bi00048a009. [DOI] [PubMed] [Google Scholar]
- 25.Olsen S G, Brooker R J. J Biol Chem. 1989;264:15982–15987. [PubMed] [Google Scholar]
- 26.Sandermann H., Jr Eur J Biochem. 1977;80:507–515. doi: 10.1111/j.1432-1033.1977.tb11906.x. [DOI] [PubMed] [Google Scholar]
- 27.Lolkema J S, Walz D. Biochemistry. 1990;29:11180–11188. doi: 10.1021/bi00503a005. [DOI] [PubMed] [Google Scholar]
- 28.Lolkema J S, Carrasco N, Kaback H R. Biochemistry. 1991;30:1284–1290. doi: 10.1021/bi00219a018. [DOI] [PubMed] [Google Scholar]
- 29.van Iwaarden P R, Driessen A J, Lolkema J S, Kaback H R, Konings W N. Biochemistry. 1993;32:5419–5424. doi: 10.1021/bi00071a017. [DOI] [PubMed] [Google Scholar]
- 30.Schürmann A, Doege H, Ohnimus H, Monser V, Buchs A, Joos H-G. Biochemistry. 1997;36:12897–12902. doi: 10.1021/bi971173c. [DOI] [PubMed] [Google Scholar]