Abstract
The dynamic rearrangement of cell–cell junctions such as tight junctions and adherens junctions is a critical step in various cellular processes, including establishment of epithelial cell polarity and developmental patterning. Tight junctions are mediated by molecules such as occludin and its associated ZO-1 and ZO-2, and adherens junctions are mediated by adhesion molecules such as cadherin and its associated catenins. The transformation of epithelial cells by activated Ras results in the perturbation of cell–cell contacts. We previously identified the ALL-1 fusion partner from chromosome 6 (AF-6) as a Ras target. AF-6 has the PDZ domain, which is thought to localize AF-6 at the specialized sites of plasma membranes such as cell–cell contact sites. We investigated roles of Ras and AF-6 in the regulation of cell–cell contacts and found that AF-6 accumulated at the cell–cell contact sites of polarized MDCKII epithelial cells and had a distribution similar to that of ZO-1 but somewhat different from those of catenins. Immunoelectron microscopy revealed a close association between AF-6 and ZO-1 at the tight junctions of MDCKII cells. Native and recombinant AF-6 interacted with ZO-1 in vitro. ZO-1 interacted with the Ras-binding domain of AF-6, and this interaction was inhibited by activated Ras. AF-6 accumulated with ZO-1 at the cell–cell contact sites in cells lacking tight junctions such as Rat1 fibroblasts and PC12 rat pheochromocytoma cells. The overexpression of activated Ras in Rat1 cells resulted in the perturbation of cell–cell contacts, followed by a decrease of the accumulation of AF-6 and ZO-1 at the cell surface. These results indicate that AF-6 serves as one of the peripheral components of tight junctions in epithelial cells and cell–cell adhesions in nonepithelial cells, and that AF-6 may participate in the regulation of cell–cell contacts, including tight junctions, via direct interaction with ZO-1 downstream of Ras.
Ras (Ha-Ras, Ki-Ras, N-Ras) is a signal-transducing, guanine nucleotide-binding protein for various membrane receptors including tyrosine kinase receptors. Ras participates in the regulation of cell proliferation, differentiation, and morphology (for reviews see Satoh et al., 1992; McCormick, 1994). Activated ras oncogenes have been identified in various forms of human cancers including epithelial carcinomas of the lung, colon, and pancreas (Barbacid, 1987). These cancer cells, as well as those that have been experimentally transformed by the activated ras gene, exhibit morphological changes and alterations of cell adhesions. Ras is thought to affect cell adhesions and cytoskeletons via alterations of the signaling pathways downstream of Ras. Ras has GDP-bound inactive and GTP-bound active forms, the latter of which interact with targets (Marshall, 1995b ). The Raf kinase family, consisting of c-Raf-1 (for reviews see Blenis, 1993; Daum et al., 1994), A-Raf (Vojtek et al., 1993), and B-Raf (Catling et al., 1994; Jaiswal et al., 1994; Moodie et al., 1994; Yamamori et al., 1995), is one of the direct targets for Ras. The activated Raf phosphorylates mitogen-activated protein (MAP)1 kinase kinase and activates it. Consequently the activated MAP kinase kinase activates MAP kinase, leading to the expression of certain genes such as c-fos (for reviews see Cano and Mahadevan, 1995; Marshall, 1995a ). Several molecules interacting with activated Ras in addition to Raf have been identified as Ras targets in mammals. These include phosphatidylinositol-3-OH kinase (Rodriguez-Viciana et al., 1994), Ral guanine nucleotide dissociation stimulator (Kikuchi et al., 1994; Spaargaren and Bischoff, 1994), and Rin1 (Han and Colicelli, 1995). We previously identified the ALL-1 fusion partner from chromosome 6 (AF-6) as a novel Ras target (Kuriyama et al., 1996). AF-6 was identified as the fusion partner of the acute lymphoblastic leukemia-1 (ALL-1) protein (Prasad et al., 1993). The ALL-1/ AF-6 chimeric protein is the critical product of the t(6;11) abnormality associated with some human leukemia. AF-6 has a PDZ (GLGF/DHR) domain that is found in a number of other proteins, including postsynaptic density protein 95 (PSD-95) (Cho et al., 1992), Drosophila discs-large tumor suppressor gene product (Dlg) (Woods and Bryant, 1991), and a tight junction-associated protein, ZO-1 (Itoh et al., 1993; Willott et al., 1993). The PDZ domain is thought to localize these proteins at the specialized sites of cell–cell contact by forming a complex with specific proteins such as the NMDA receptor and the K+ channel (Kim et al., 1995; Kornau et al., 1995).
The dynamic rearrangement of cell–cell contacts is a critical step in various cellular processes including the establishment of epithelial cell polarity and developmental patterning (for review see Gumbiner, 1996). Cell–cell junctions are categorized into at least three groups: tight, adherens, and gap junctions (for reviews see Farquhar and Palade, 1963; Tsukita et al., 1993). Tight junctions, the most apical components of the junctional complex, form a diffusion barrier that regulates the flux of ions and hydrophilic molecules through the paracellular pathway (for reviews see Diamond, 1977; Gumbiner, 1987). Tight junctions are mediated by molecules such as occludin and its associated ZO-1 and -2 (Anderson et al., 1988; Furuse et al., 1994; for review see Anderson, 1996). Adherens junctions are characterized by a well-developed plaque structure in which actin filaments are densely associated. Adherens junctions are mediated by adhesion molecules such as cadherin and its associated catenins (for reviews see Takeichi, 1990; Hülsken et al., 1994). These junctions undergo dynamic remodeling when cells move or enter the mitotic phase. Although these junctions are suspected to be regulated by certain intracellular signaling pathways, little is known at present about how they are actually regulated. Accumulating evidence suggests that small GTPases Ras and Rho family members including Rho, Rac, and Cdc42 participate in the regulation of cell adhesions (for review see Hall, 1994).
In light of these observations, we investigated roles of Ras and its target AF-6 in the regulation of cell–cell contacts. We found that AF-6 accumulated at cell–cell contact sites of MDCKII epithelial cells and had a distribution similar to that of ZO-1. AF-6 interacted with ZO-1, and this interaction was inhibited by activated Ras.
Materials and Methods
Materials and Chemicals
MDCKII cell, mouse anti–ZO-1 antibody and rat anti–α-catenin antibody were kindly provided by Dr. S. Tsukita (Kyoto University, Kyoto, Japan; Itoh et al., 1991). Rat1 and Rat1 RasVal A1 cells were kindly provided by Dr. Y. Kaziro (Tokyo Institute of Technology, Yokohama, Japan). Rabbit polyclonal antibody against AF-6 (1,130–1,612 amino acids) was generated as described previously (Harlow and Lane, 1988). FITC-conjugated anti–rabbit IgG, Texas red-conjugated anti–mouse IgG, and [35S]methionine were purchased from Amersham Intl. (Arlington Heights, IL). The expression plasmids of glutathione-S-transferase (GST)-mouse ZO-1 (1–862 amino acids), GST-mouse ZO-2 (1–400 amino acids), GST-mouse occludin (264–521 amino acids), GST–E-cadherin cytoplasmic domain, GST-mouse α-catenin and GST–β-catenin were kindly provided by Dr. S. Tsukita (Kyoto University). GST fusion proteins were expressed in Escherichia coli BL21(DE3) and purified according to the manufacturer's instructions. Bovine brain membrane fraction was prepared as described previously (Kuriyama et al., 1996) and preabsorbed to remove the native GST with glutathione Sepharose 4B (Pharmacia Biotech Inc., Grand Island, NY). To obtain recombinant HA-AF-6 (36–1,608 amino acids), the baculovirus expression plasmid pAcYM1-HA-AF-6 (36–1,608 amino acids) was constructed as follows. The cDNA fragment encoding AF-6 (36–1,608 amino acids) was amplified by polymerase chain reaction from the full length AF-6 cDNA in pBluescript and was subcloned into the KpnI site of pAcYM1-HA (Matsuura et al., 1987). Sf9 cells infected with baculovirus carrying the cDNA of HA-AF-6 (36–1,608 amino acids) were suspended in buffer A (20 mM Tris/HCl at pH 7.5, 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 10% sucrose, 10 μM [p-amidino-phenyl] methanesulfonyl fluoride, 10 μg/ml leupeptin). The suspension was sonicated and centrifuged at 100,000 g for 60 min at 4°C. The supernatant was used for affinity column chromatography. Other materials and chemicals were obtained from commercial sources.
Cell Culture
MDCKII and Rat1 cells were grown in DME containing 10% FCS, penicillin, and streptomycin in an air-5% CO2 atmosphere at constant humidity. Rat1 RasVal A1 cells were grown in DME containing 10% FCS, 1 mg/ml Geneticin, 0.4 mg/ml Hygromycin B, penicillin, and streptomycin in an air-5% CO2 atmosphere at constant humidity. PC12 cells were grown in DME containing 10% FCS, 5% horse serum, penicillin, and streptomycin in an air-5% CO2 atmosphere at constant humidity. For Ca2+ switch assay, subconfluent MDCKII cells were grown in normal growth media and transferred to low Ca2+ medium (growth media containing 4 mM EGTA) for 6 h and were then transferred back to the normal Ca2+ medium. To examine the distribution of AF-6 in Rat1 RasVal A1 cells, Rat1 RasVal A1 cells plated on 13-mm-round glass coverslips were incubated either with or without 5 mM isopropyl–β-d-thiogalactoside (IPTG) for 24 h.
Immunofluorescence and Laser Scanning Confocal Microscopy
MDCKII, PC12, Rat1, and Rat1 RasVal A1 cells plated on 13-mm-round glass coverslips were fixed in 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min. The fixed cells were incubated with primary antibodies for 1 h at room temperature and washed three times for 10 min with PBS. After the first labeling, the cells were incubated for 1 h with secondary antibodies and washed three times for 10 min with PBS. The distributions of AF-6, ZO-1, and α-catenin were examined with a laser scanning confocal microscope (Carl Zeiss, Inc., Thornwood, NY) equipped with an argon laser and a helium-neon laser for double fluorescence at 488 and 543 nm (emission filter; BP510-525 and LP590). 20 horizontal confocal sections were obtained for MDCKII cells and used to generate three-dimensional images.
Immunoelectron Microscopy
Immunoelectron microscopy was carried out as described previously (Burry et al., 1992; Jongens et al., 1994; Uchida et al., 1996). MDCKII cells were cultured on tissue culture-treated polycarbonate filters (Transwell; Coaster Corp., Cambridge, MA) with a pore size of 0.4 μm and a diameter of 6.5 mm, and maintained for 2 wk after being confluent. The samples were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, rinsed with PBS for 10 min, and permeabilized with PBS containing 5% BSA and 0.02% saponin for 10 min. The fixed cells were incubated with rabbit anti–AF-6 (1:100) in PBS containing 0.005% saponin or a hybridoma culture medium containing mouse anti–ZO-1 antibody for 2 h, followed by three washes with PBS containing 0.005% saponin for 20 min in each step. The cells were incubated with an anti–rabbit antibody or an anti–mouse antibody coupled with 1.4-nm gold particles (Nanoprobes, Inc., Stony Brook, NY) for 2 h, followed by three washes with PBS containing 0.005% saponin. For the double staining study, the samples were incubated with a hybridoma culture medium containing mouse anti–ZO-1 antibody and 0.005% saponin for 2 h and washed three times with PBS containing 0.005% saponin. The cells were then incubated with HRP-conjugated anti–mouse antibody (1:1,000; Jackson ImmunoResearch Lab., Inc., West Grove, PA) followed by three washes with PBS containing 0.005% saponin. The samples were incubated twice with 0.1 M PB (0.1 M sodium phosphate buffer at pH 7.4) for 5 min, fixed with 0.1% glutaraldehyde in 0.1 M PB for 10 min, and rinsed with 0.1 M PB for 5 min. The samples were then incubated three times with 50 mM Hepes, at pH 5.8, for 10 min. The signals were silver enhanced with the use of HQ silver (Nanoprobes) for 8 min at 18°C in the dark, followed by three washes with 0.1 M PB. The samples were incubated with a DAB solution (50 mM Hepes at pH 7.4, 0.05% 3,3′-diaminobenzidine tetrahydrochloride) for 30 min and then incubated with the DAB solution containing 0.01% H2O2 for 20 min. The samples were postfixed with 0.5% OsO4 in 0.1 M PB for 60 min at 4°C, washed twice with 0.1 M PB for 5 min, dehydrated by passage through a graded series of ethanol (50, 60, 70, 80, 90, 95, and 100%) and propylene oxide, and then embedded in epoxy resin (Polysciences, Inc., Warrington, PA). Ultrathin sections were cut, stained with lead citrate, and observed with an immunoelectron microscope (JEM-1200; JEOL, Tokyo, Japan).
Immunocytochemistry of Mouse Intestinal Epithelium
Mice were anesthetized with halothane and fixed by transcardiac perfusion with PBS containing 2% paraformaldehyde, 1 mM CaCl2 and 8% sucrose. The intestine was removed, immersed in the same fixative, and cryoprotected through a range of increasing sucrose concentrations (10, 15, 20, and 25%) in 50 mM Tris-buffered saline. The intestine was embedded in OCT compound, quick-frozen, and cut into 8-μm-thick sections in a cryostat. The frozen sections, mounted on slides, were then washed with PBS, fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized with 0.2% Triton X-100 in PBS for 10 min. The samples were subjected to immunostaining and examined as described above.
Affinity Column Chromatography
GST fusion proteins (each 6 nmol) were immobilized on 200 μl of glutathione Sepharose 4B packed into columns. Bovine membrane fraction or crude Sf-9 cell lysate expressing HA-AF-6 was loaded onto the affinity columns. The columns were washed with 2 ml (10 vol) of buffer B (20 mM Tris/HCl at pH 7.5, 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 10 μM [p-amidino-phenyl] methanesulfonyl fluoride, 10 μg/ml leupeptin), followed by washing with 2 ml (10 vol) buffer B containing 50 mM NaCl. The proteins bound to the affinity columns were eluted three times by the addition of 660 μl (3.3 vol) of buffer B containing 10 mM reduced glutathione. The eluates were subjected to SDS-PAGE and transferred to PVDF membranes. The eluted native AF-6 and recombinant HA-AF-6 were immunodetected with the anti–AF-6 antibody.
In Vitro Binding Assay (In Vitro–Translated AF-6)
The in vitro translation of pRSET-AF-6 (36–494 amino acids), AF-6 (495– 909 amino acids), AF-6 (914–1,129 amino acids), and AF-6 (1,130–1,612 amino acids) were performed using the TNT T7-coupled reticulocyte lysate system (Promega, Madison, WI) under the conditions described in the manufacturer's instruction manual. GST fusion proteins were immobilized onto 31 μl of glutathione Sepharose 4B beads and washed with 310 μl (10 vol) of buffer B. The immobilized beads were added to 40 μl of the in vitro–translated products labeled with [35S]methionine containing 1 mg/ml BSA and incubated for 1 h at 4°C with gentle mixing. The beads were washed three times with 102 μl (3.3 vol) of buffer B, and the bound proteins were eluted with GST fusion proteins three times by the addition of 102 μl (3.3 vol) of buffer B containing 10 mM reduced glutathione. The eluates were subjected to SDS-PAGE and vacuum dried. The 35S-labeled bands corresponding to in vitro–translated AF-6 were visualized with an image analyzer (BAS-2000; Fuji, Tokyo, Japan).
In Vitro Binding Assay (MBP-AF-6)
The expression of maltose-binding protein (MBP)-AF-6 (36–206 amino acids) in E. coli was performed as described previously (Kuriyama et al., 1996). GST-ZO-1 (0.1 nmol) was immobilized on 100 μl of glutathione Sepharose 4B packed into columns. E. coli BL21(DE3) producing MBP-AF-6 (36–206 amino acids) was suspended in buffer A. The suspension was sonicated and centrifuged at 100,000 g for 60 min at 4°C. The supernatants containing various concentrations of MBP-AF-6 (36–206 amino acids) were then loaded onto the GST-ZO-1 columns. The columns were washed with 1 ml (10 vol) of buffer B, followed by washing with 1 ml (10 vol) buffer B containing 50 mM NaCl. The proteins bound to GST-ZO-1 columns were eluted with GST-ZO-1 three times by the addition of 330 μl (3.3 vol) of buffer B containing 10 mM reduced glutathione. The eluted MBP-AF-6 was immunodetected with the anti-MBP antibody. The immunodetected MBP-AF-6 was visualized and estimated with a densitograph (ATTO, Tokyo, Japan).
Dissociation of AF-6 from ZO-1 by Activated Ras
GST-Ha-Ras and GST-Rac were produced in E. coli DH5α and cleaved with thrombin according to the manufacturer's instructions. The guanine nucleotide-bound forms of Ha-Ras and Rac were made by incubating Ha-Ras or Rac (5 nmol) for 1 h at 30°C with 50 μM GDP or guanosine 5′- (3-O-thio)-triphosphate (GTPγS) in 1 ml of a reaction mixture (20 mM Tris/HCl at pH 7.5, 10 mM EDTA, 1 mM DTT, 5 mM MgCl2). The binding of MBP-AF-6 (36–206 amino acids) to GST-ZO-1 columns was carried out as described above. The proteins bound to GST-ZO-1 columns were eluted three times by the addition of 330 μl (3.3 vol) of 2 μM GDP/Ha-Ras, GTPγS/Ha-Ras, or GTPγS/Rac. The eluted MBP-AF-6 was immunodetected with the anti-MBP antibody.
Other Procedures
SDS-PAGE was performed as described (Laemmli, 1970). Protein concentrations were determined with BSA as the reference protein as described (Bradford, 1976). Immunoblot analysis was carried out as described (Harlow and Lane, 1988).
Results
Colocalization of AF-6 with ZO-1 But Not with α-Catenin
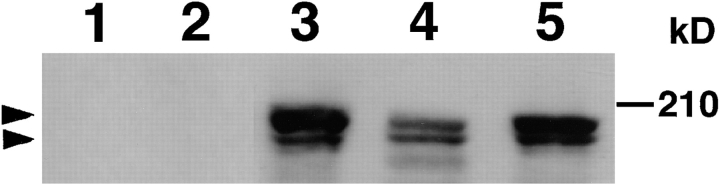
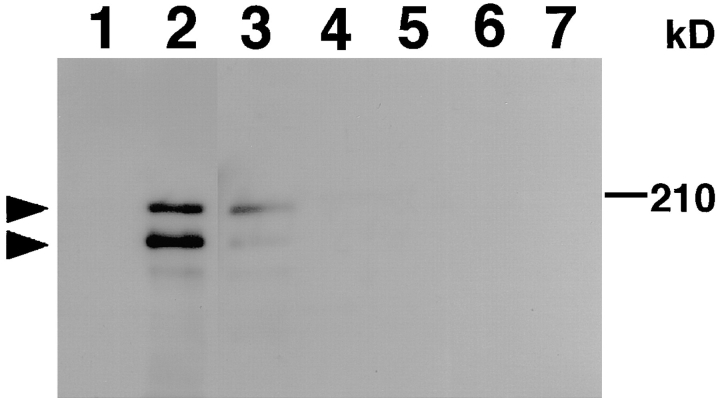
To clarify the functions of AF-6, we examined its intracellular distribution in MDCKII epithelial cells. First, immunoblot analysis was performed on cell lysates from MDCKII cells, using a polyclonal anti–AF-6 antibody. The anti–AF-6 antibody recognized two isoforms of AF-6 with molecular masses of ∼180 and 195 kD in the sample obtained from MDCKII cells (Fig. 1) as described for bovine brain extract (Kuriyama et al., 1996). Preincubation of the antibody with the recombinant AF-6 abolished the immunoreactivity, indicating the expression of AF-6 in MDCKII cells.
Figure 1.
Immunoblot analysis of MDCKII, Rat1, and PC12 cell lysates with anti–AF-6 antibody. Lane 1, MDCKII cell lysate with preimmuneserum; lane 2, MDCKII cell lysate with anti–AF-6 antibody preincubated with recombinant AF-6; lane 3, MDCKII cell lysate with anti–AF-6 antibody; lane 4, Rat1 cell lysate with anti– AF-6 antibody; lane 5, PC12 cell lysate with anti–AF-6 antibody. The results shown are representative of three independent experiments. The arrowheads denote the position of AF-6.
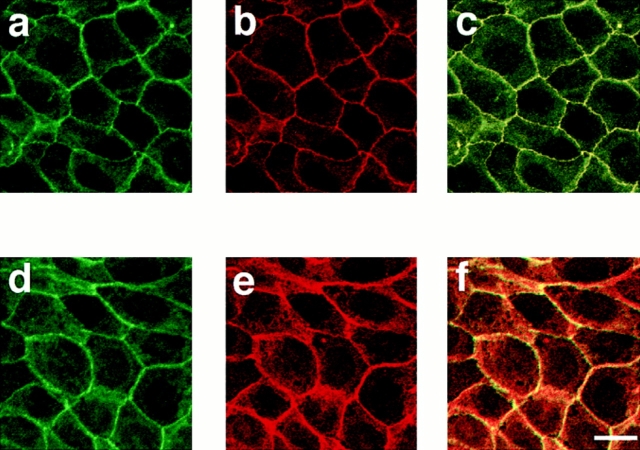
We then examined the AF-6 localization in confluent MDCKII cells, which show characteristics of polarized epithelial cells and form the junctional complex, including the tight junctions and adherens junctions, at cell–cell contact sites (Gonzalez-Mariscal et al., 1985). The immunoreactivity of AF-6 was specifically localized at sites where a cell contacted a neighbor cell, but not at the free ends, and showed a belt-like pattern of staining along the cell–cell contact sites (Fig. 2, a and d). The cytosol exhibited a relatively low level of the anti–AF-6 staining. To examine whether AF-6 exists in the tight junctions or adherens junctions, the cellular distribution of AF-6 was compared with those of ZO-1 and α-catenin, which are marker proteins of tight junctions and adherens junctions, respectively, by laser scanning confocal microscopy. Three-dimensional images of confluent MDCKII cells, doubly stained with antibodies against AF-6 and ZO-1 or α-catenin, were generated from serial optical sections obtained by laser scanning confocal microscopy. En face views showed that the localization of AF-6 overlapped with that of ZO-1 (Fig. 2, a–c), but was somewhat different from that of α-catenin (Fig. 2, d–f). To confirm that AF-6 is localized at apical lateral membranes, five serial optical sections (one section every 0.8 μm) were shown for each staining (Fig. 2, g–j). These images showed that AF-6 and ZO-1 were concentrated at the apical sections, whereas α-catenin was found at more basal sections. When the three-dimensional images were rotated latitudinally by 90°, the rotated images showed that AF-6 was colocalized with ZO-1 at the apical cell borders but not with α-catenin (Fig. 2, k and l). Thus, it is likely that AF-6 is colocalized with ZO-1 at the tight junctions rather than adherens junctions of the confluent MDCKII cells.
Figure 2.
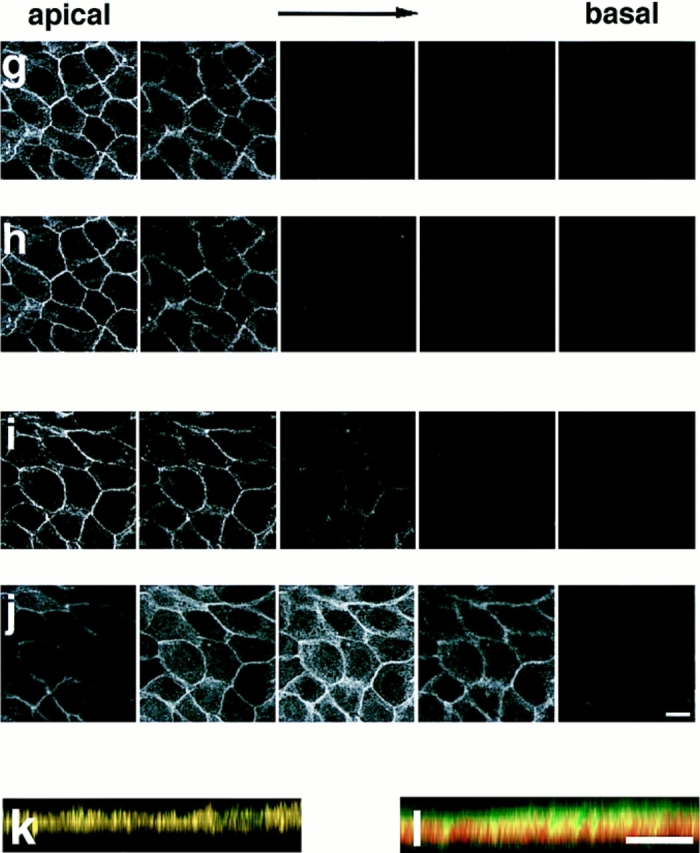
Confocal microscope images of confluent MDCKII cells showing the distributions of AF-6, ZO-1, and α-catenin. Confluent MDCKII cells were doubly stained with a rabbit polyclonal antibody against AF-6 and a mouse monoclonal antibody against ZO-1 (a–c, h, and k) or a rat monoclonal antibody against α-catenin (d–f, j, and l), followed by FITC-conjugated anti–rabbit IgG and Texas red-conjugated anti–mouse IgG or Texas red-conjugated anti–rat IgG antibodies. 20 serial optical sections were obtained at 0.8-μm intervals, and three-dimensional images were generated. In images a, c, d, f, k, and l, AF-6 is shown in green, and ZO-1 (b, c, and k) or α-catenin (e, f, and l) is shown in red. The yellow area indicates the colocalization of AF-6 and ZO-1 (c and k) or α-catenin (f and l). Images a–f are unrotated en face view. In images k and l, images c and f are rotated latitudinally by 90°, respectively. Images g–j show five confocal sections for each staining. Images g and i show the distribution of AF-6, and images h and j show that of ZO-1 and of α-catenin, respectively. Subsequent sections from apical to basal were shown from left to right. The results shown are representative of three independent experiments. Bars, 10 μm.
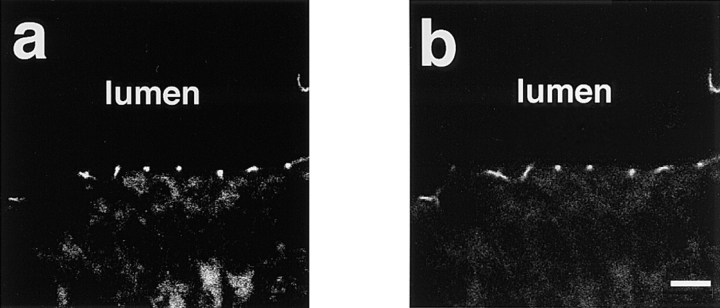
Immunoelectron Microscopic Localization of AF-6 and ZO-1 in Confluent MDCKII Cells
The ultrastructural localization of AF-6 and that of ZO-1 in MDCKII cells was further analyzed by the pre-embedding procedure. The gold signals immunoreactive for AF-6 were identified on the cytoplasmic surface of the plasma membranes in the junctional complex region (Fig. 3 a). The area immunoreactive for AF-6 was 400–500 nm long in cross-section and usually started from the apical-most point where the apical-adjacent plasma membranes converged (Fig. 3 a). A few gold particles were noted along the lateral plasma membranes outside the junctional complex region. The gold signals for ZO-1 were also identified to be almost exclusively concentrated on the cytoplasmic surface of the plasma membranes in the junctional complex region (Fig. 3 b). The area immunoreactive for ZO-1 was 300–500 nm long and always started from the apical-most point of the junctional complex region. When this junctional complex region was double labeled with anti–AF-6 polyclonal antibody and anti–ZO-1 monoclonal antibody, AF-6 immunoreactivity and that of ZO-1 was colocalized in the main part of the junctional complex region (Fig. 3, c and d). In Fig. 3 d, a cross-section of a junctional complex is seen in the center, and an oblique section of another junctional complex is seen in the right side of the photograph. Taken together, these results indicate that AF-6 is colocalized with ZO-1 in the junctional complex.
Figure 3.
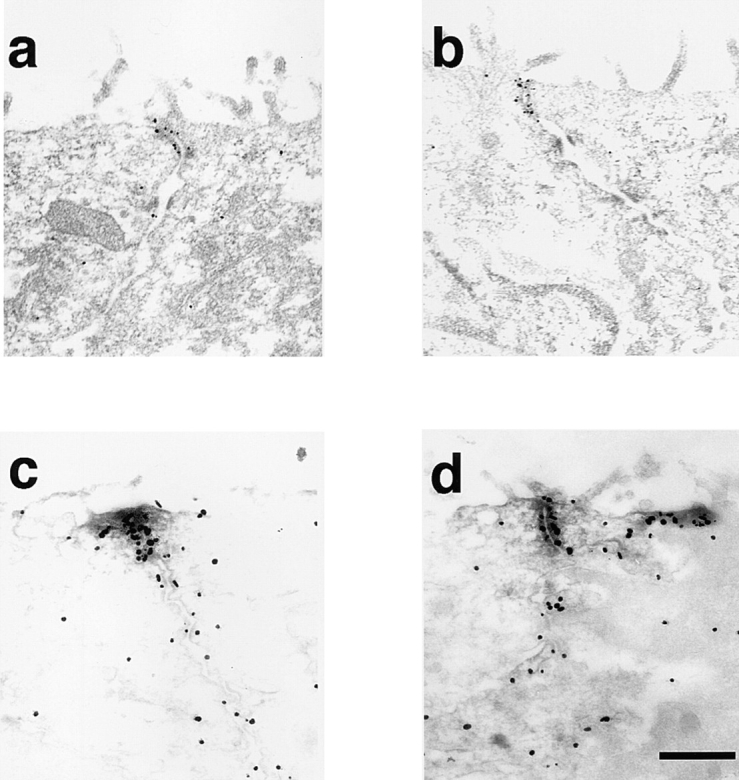
The ultrastructural localization of AF-6 and that of ZO-1 in confluent MDCKII cells. Immunoelectron micrographs of the junctional complex region in MDCKII cells stained with a rabbit polyclonal antibody against AF-6 (a), with a mouse monoclonal antibody against ZO-1 (b), or doubly stained with both anti–AF-6 polyclonal antibody (gold particles) and anti–ZO-1 monoclonal antibody (HRP reaction products; c and d). (a) The gold particles for AF-6 are accumulated on the cytoplasmic surface of the plasma membranes in the junctional complex region. (b) ZO-1 labeling is exclusively concentrated in the junctional complex region. (c and d) AF-6 immunoreactivity (gold particles) and ZO-1 immunoreactivity (HRP reaction products) are intermixed. The results shown are representative of three independent experiments. Bar, 500 nm.
Ca2+-dependent Distribution of AF-6 and ZO-1 at Cell–Cell Contact Sites of MDCKII Cells
We examined the localization of AF-6 in MDCKII cells during the formation and disappearance of cell–cell contacts. The formation of cell–cell contacts and the junctional complex can be reversed by transferring cells between low Ca2+ and normal Ca2+ media (Ca2+ switch experiment). When the MDCKII cells were cultured in low Ca2+ media for 6 h, cell–cell contacts were disrupted and the cells became rounded up, as described previously (Fig. 4 d; Gonzalez-Mariscal et al., 1985). Under such conditions, AF-6 and ZO-1 showed cytoplasmic staining without specific localization at the cell surface (Fig. 4, e and f). When the cells were transferred to normal media, they formed cell–cell contacts (Fig. 4 g), and AF-6 and ZO-1 had similar distributions at the cell–cell contact sites (Fig. 4, h and i), suggesting that AF-6 and ZO-1 show similar dynamic behavior during the formation and disappearance of cell–cell contacts.
Figure 4.
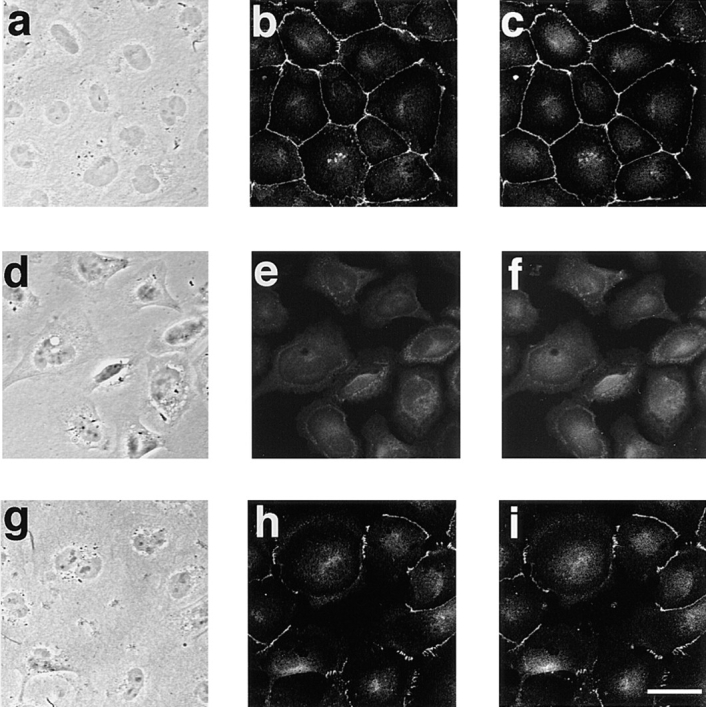
Immunofluorescence localization of AF-6 and that of ZO-1 in Ca2+ switch experiments with MDCKII cells. Subconfluent MDCKII cells were grown in normal growth media (a–c) and transferred to low Ca2+ media (growth media containing 4 mM EGTA) for 6 h (d–f) and then transferred back to the normal Ca2+ medium for 2 h (g–i). The cells were doubly stained with a rabbit polyclonal antibody against AF-6 (b, e, and h) and a mouse monoclonal antibody against ZO-1 (c, f, and i), followed by FITC-conjugated anti–rabbit IgG and Texas red-conjugated anti– mouse IgG antibodies, and examined using laser scanning confocal microscopy. Images a, d, and g are phase contrast images. The results shown are representative of three independent experiments. Bar, 10 μm.
Localization of AF-6 in Mouse Intestinal Epithelial Cells
Because intestinal epithelial cells have tight junctions at the apical cell borders, we examined the distribution of AF-6 in intestinal epithelial cells. Cryosections of mouse intestine were double labeled with AF-6 and ZO-1 antibodies and examined by laser scanning confocal microscopy. AF-6 and ZO-1 were colocalized at the apical cell borders (Fig. 5). These results indicate that AF-6 is colocalized with ZO-1 at tight junctions of intestinal epithelial cells.
Figure 5.
Immunofluorescence localization of AF-6 and that of ZO-1 in the frozen mouse intestinal epithelium. Cryosections of mouse intestine were doubly stained with a rabbit polyclonal antibody against AF-6 (a) and a mouse monoclonal antibody against ZO-1 (b), followed by FITC-conjugated anti–rabbit IgG and Texas red-conjugated anti–mouse IgG antibodies, and examined using laser scanning confocal microscopy. The results shown are representative of three independent experiments. Bar, 5 μm.
Localization of AF-6 in Cells Lacking Tight Junctions
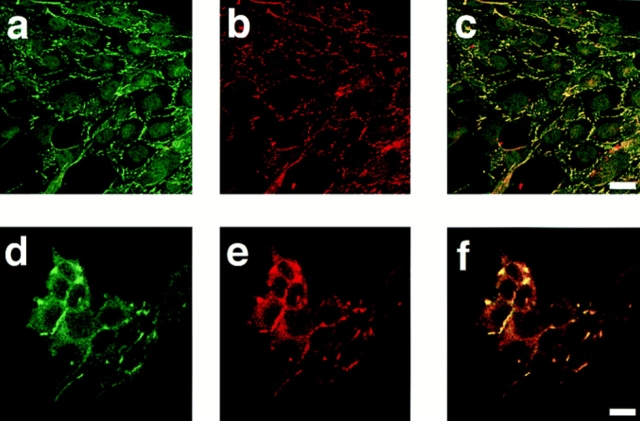
In cells lacking tight junctions such as nonepithelial cells, ZO-1 accumulates at cell–cell contact sites with cadherin (Howarth et al., 1992; Itoh et al., 1993). A recent study showed that ZO-1 interacts with catenins during the early stages of the assembly of tight junctions (Rajasekaran et al., 1996). We examined whether AF-6 accumulates at cell– cell contact sites in cells lacking tight junctions such as Rat1 fibroblasts and PC12 rat pheochromocytoma cells. First, immunoblot analysis was performed on cell lysates from Rat1 and PC12 cells. The anti–AF-6 antibody recognized two bands in the samples from Rat1 and PC12 cells as described for MDCKII cells (Fig. 1). We next examined the distribution of AF-6 in Rat1 and PC12 cells. AF-6 accumulated at the cell–cell contact sites and was colocalized with ZO-1 in both the Rat1 fibroblasts and the PC12 rat pheochromocytoma cells (Fig. 6). In the Rat1 fibroblasts, the accumulation of AF-6 was discontinuous rather than a belt-like accumulation as observed in the MDCKII epithelial cells. These results indicate that AF-6 is colocalized with ZO-1 in cell–cell contact sites of cells lacking tight junctions.
Figure 6.
Colocalization of AF-6 with ZO-1 in Rat1 and PC12 cells. Subconfluent Rat1 (a–c) and PC12 (d–f) cells were doubly stained with a rabbit polyclonal antibody against AF-6 (a, c, d, and f) and a mouse monoclonal antibody against ZO-1 (b, c, e, and f), followed by FITC-conjugated anti–rabbit IgG and Texas red-conjugated anti–mouse IgG antibodies, and examined using laser scanning confocal microscopy. AF-6 is shown in green (a, c, d, and f) and ZO-1 (b, c, e, and f) is shown in red. In images c and f, the yellow area indicates the colocalization of AF-6 and ZO-1. The results shown are representative of three independent experiments. Bars: (a–c) 10 μm; (d–f) 5 μm.
Interaction of Bovine AF-6 with ZO-1 and Occludin
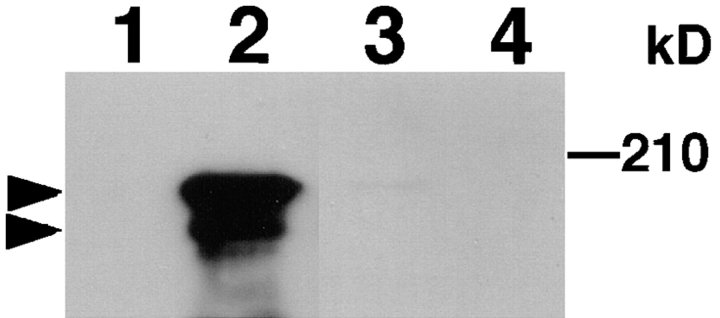
To examine the complex formation of AF-6 with tight junction components including ZO-1 and occludin, bovine brain membrane fraction was loaded onto the affinity columns immobilized with GST, GST-ZO-1, GST-occludin, GST– E-cadherin, GST–α-catenin, GST–β-catenin, and GST-CD44. The proteins bound to the affinity columns were eluted with the GST fusion proteins by the addition of glutathione, and the eluted AF-6 was detected with an anti–AF-6 antibody. AF-6 was detected in the eluate of the GST-ZO-1 affinity column and weakly in that of the GST-occludin affinity column, but not in those of the GST, GST–E-cadherin, GST–α-catenin, GST–β-catenin, or GST-CD44 affinity columns (Fig. 7). These results indicate that native AF-6 directly or indirectly interacts with ZO-1 and occludin.
Figure 7.
Interaction of bovine AF-6 with ZO-1. Bovine brain membrane fraction was loaded onto affinity columns immobilized with GST (lane 1), GST-ZO-1 (lane 2), GST-occludin (lane 3), GST–E-cadherin (lane 4), GST–α-catenin (lane 5), GST–β-catenin (lane 6), and GST-CD44 (lane 7). The interacting proteins were eluted with GST proteins by the addition of glutathione. The eluates were subjected to SDS-PAGE, followed by immunoblot analysis with anti–AF-6 antibody. The arrowheads denote the position of bovine AF-6. The results shown are representative of three independent experiments.
Interaction of Recombinant AF-6 with ZO-1
To determine whether recombinant AF-6 interacts with ZO-1 and occludin, crude lysates of Sf-9 cells infected with baculovirus carrying the cDNA of HA-AF-6 (36–1,608 amino acids) were loaded onto the GST, GST-ZO-1, GST-occludin, and GST-CD44 affinity columns. The proteins bound to the affinity columns were eluted with GST fusion proteins by the addition of glutathione, and the eluted HA-AF-6 was detected with the anti–AF-6 antibody. HA-AF-6 was detected in the eluate of the GST-ZO-1 affinity column and weakly in that of GST-occludin affinity column, but not in those of the GST or GST-CD44 affinity columns (Fig. 8). These results indicate that recombinant AF-6 interacts with ZO-1. The reason for the weak interaction of recombinant AF-6 and occludin is not clear at present. The recombinant AF-6 produced from Sf-9 cells may have a lower affinity for occludin than that of native AF-6, or the interaction of AF-6 and occludin is indirect and this interaction may be mediated by ZO-1. When the same samples were examined with HA antibody, smaller size bands were detected in addition to full size AF-6 in the eluate of the GST-ZO-1 affinity column (data not shown). Since the AF-6 used in this experiment was tagged with HA in its NH2 terminus, these smaller bands are thought to represent the products degraded from the COOH terminus. Since the size of the major smaller band was ∼50 kD, the interacting region of AF-6 with ZO-1 may contain the Ras-binding domain, which is identified as 36–206 amino acids (Kuriyama et al., 1996).
Figure 8.
Interaction of recombinant AF-6 with ZO-1. Crude lysates of Sf-9 cells infected with baculovirus carrying HA-AF-6 cDNA were loaded onto affinity columns immobilized with GST (lane 1), GST-ZO-1 (lane 2), GST-occludin (lane 3), and GST-CD44 (lane 4). The interacting proteins were eluted with GST fusion proteins by the addition of glutathione. The eluates were subjected to SDS-PAGE and followed by immunoblot analysis with anti–AF-6 antibody. The arrowheads denote the positions of HA-AF-6. The results shown are representative of three independent experiments.
We further examined which domain of AF-6 interacts with ZO-1, using in vitro–translated AF-6 such as AF-6 (36–494 amino acids), AF-6 (495–909 amino acids), AF-6 (914–1,129 amino acids), and AF-6 (1,130–1,612 amino acids) (Fig. 9 a). Affinity beads immobilized with GST-ZO-1 were mixed with the in vitro–translated AF-6 (36–494 amino acids), AF-6 (495–909 amino acids), AF-6 (914–1,129 amino acids), and AF-6 (1,130–1,612 amino acids), and the interacting proteins were then eluted with GST-ZO-1 by the addition of glutathione. As shown in Fig. 9, b–e, AF-6 (36–494 amino acids) bound to GST-ZO-1 and weakly to GST-occludin, but not to GST or GST-CD44, whereas the other domains slightly bound to GST-ZO-1 and not to GST, GST-occludin, or GST-CD44. These results together with the above observations indicate that mainly the NH2-terminal domain of AF-6 is responsible for the binding of AF-6 to ZO-1.
Figure 9.
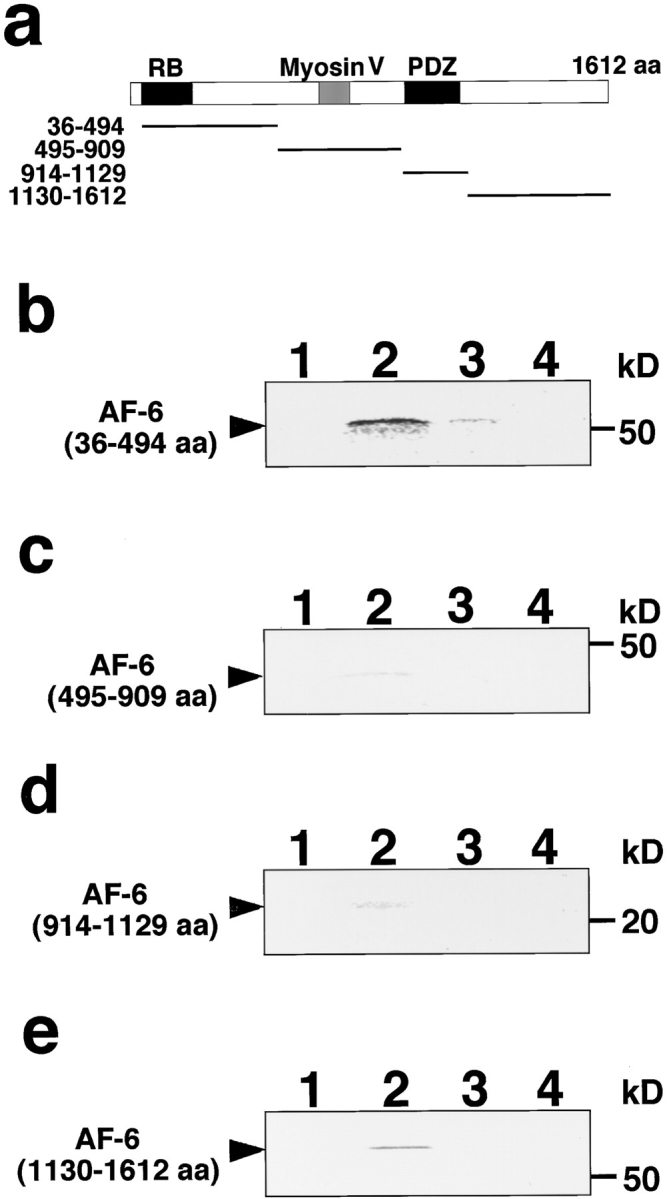
Interaction of in vitro–translated AF-6 (36–494 amino acids) with ZO-1. (a) Domain diagram of AF-6 and the recombinant fragments used for the in vitro binding assay. RB, Ras-binding domain; Myosin V, Myosin V–like domain; PDZ, PDZ domain. Bold lines show the recombinant fragments used for the in vitro binding assay. (b–e) In vitro–translated AF-6 (36–494 amino acids; b), AF-6 (495–909 amino acids; c), AF-6 (914–1,129 amino acids; d), and AF-6 (1,130–1,612 amino acids; e) were mixed with GST (lane 1), GST-ZO-1 (lane 2), GST-occludin (lane 3), and GST-CD44 (lane 4) immobilized to glutathione Sepharose 4B beads. The interacting proteins were eluted with GST fusion proteins by the addition of glutathione. The eluates were subjected to SDS-PAGE and vacuum dried. The in vitro–translated AF-6 fragments were visualized with an image analyzer. The arrowheads denote the position of in vitro–translated AF-6 fragments. The results shown are representative of three independent experiments.
Dissociation of AF-6 from ZO-1 by Activated Ras
Since the Ras-binding domain and ZO-1–binding domain are very close as noted above, we speculated that activated Ras inhibited the interaction of AF-6 and ZO-1. To test this possibility we first examined whether ZO-1 could bind to MBP-AF-6 (36–206 amino acids), which binds to activated Ras as previously described (Kuriyama et al., 1996). E. coli lysate containing MBP-AF-6 (36–206 amino acids) was loaded onto affinity columns immobilized with GST, GST-ZO-1, GDP/GST-Ha-Ras, GTPγS/GST-Ha-Ras, and GST-CD44. The proteins bound to the affinity columns were then eluted with the GST fusion proteins by the addition of glutathione, and the eluted MBP-AF-6 (36–206 amino acids) was detected with an anti-MBP antibody. MBP-AF-6 (36–206 amino acids) was detected in the eluates of the GST-ZO-1 or GTPγS/GST-Ha-Ras affinity columns, but only slightly in those of the GST, GDP/GST-Ha-Ras, or GST-CD44 affinity columns (Fig. 10 a). The weak binding of MBP-AF-6 (36–206 amino acids) to GST, GDP/GST-Ha-Ras, or GST-CD44 affinity columns appears to be background. Similar observations were obtained when MBP-AF-6 (36–494 amino acids) was used instead of MBP-AF-6 (36–206 amino acids; data not shown). To assure the specificity of the binding, we carried out the kinetic study on the binding of AF-6 to ZO-1. As shown in Fig. 10 b, MBP-AF-6 (36–206 amino acids) bound to GST-ZO-1 in a dose-dependent manner, and this binding was saturable when the amounts of MBP-AF-6 (36–206 amino acids) were increased. The apparent Kd value for the binding of MBP-AF-6 (36–206 amino acids) to GST-ZO-1 was estimated to be ∼260 nM under the conditions. Next, to examine whether activated Ras dissociates AF-6 from ZO-1, we applied GTPγS/Ras onto a GST-ZO-1 column, which retained MBP-AF-6 (36–206 amino acids). As shown in Fig. 10 c, the bound MBP-AF-6 (36–206 amino acids) was eluted with GTPγS/Ras, but was weakly eluted with GDP alone, GTPγS alone, GDP/Ras, or GTPγS/Rac. It may be noted that two bands corresponding to MBP-AF-6 were eluted from the GST-ZO-1 or GTPγS/Ras affinity columns by the addition of glutathione, whereas the upper band was eluted more efficiently than the lower band by the addition of GTPγS/Ras. The smaller size of MBP-AF-6 was probably the degradation product. We can not give the precise reasons for this complexity, but it is probable that the smaller size of MBP-AF-6 binds strongly to ZO-1 and hardly dissociates from ZO-1 by the addition of GTPγS/ Ras. Taken together, these results indicate that ZO-1 interacts with the Ras-binding domain of AF-6 (36–206 amino acids) and that this interaction may be inhibited specifically by activated Ras.
Figure 10.
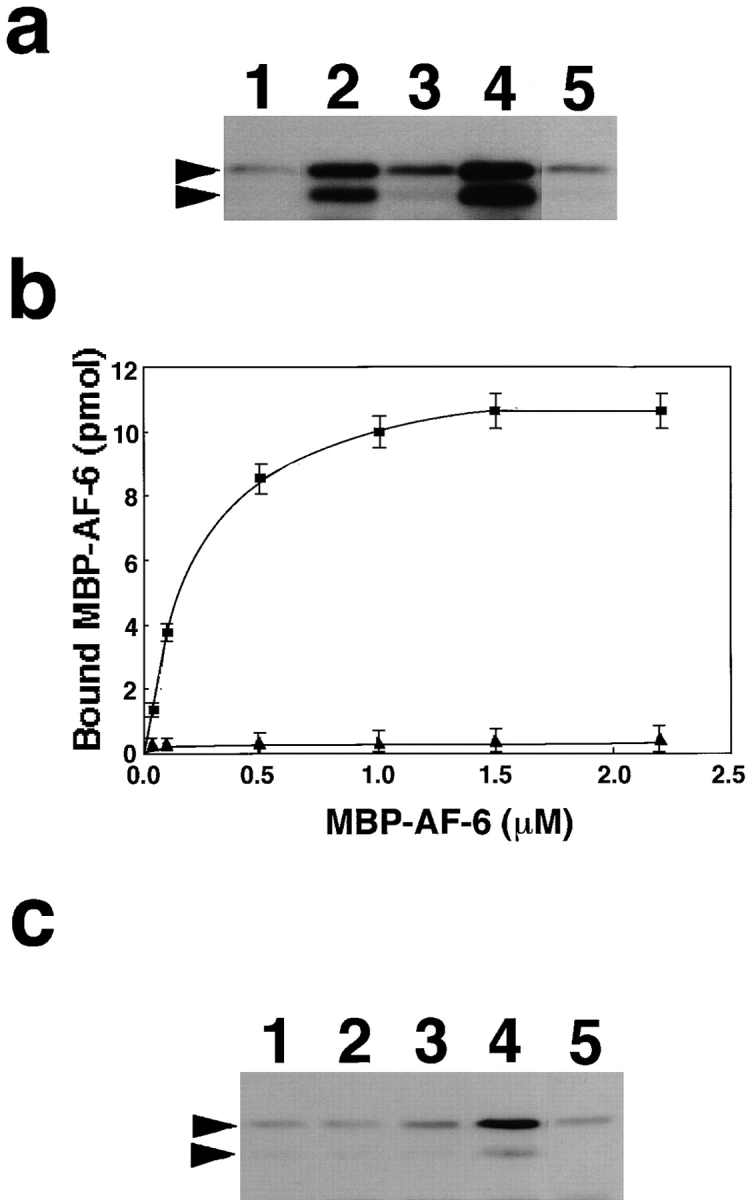
Dissociation of the Ras-interacting domain of AF-6 from ZO-1 by activated Ras. (a) Interaction of MBP-AF-6 (36– 206 amino acids) with ZO-1. Crude lysates of E. coli expressing MBP-AF-6 (36–206 amino acids) were loaded onto affinity columns immobilized with GST (lane 1), GST-ZO-1 (lane 2), GDP/ GST–Ha-Ras (lane 3), GTPγS/GST–Ha-Ras (lane 4), and GST-CD44 (lane 5). The interacting proteins were eluted with GST fusion proteins by the addition of glutathione. The eluates were subjected to SDS-PAGE and followed by immunoblot analysis with anti-MBP antibody. The arrowheads denote the positions of MBP-AF-6 (36–206 amino acids). (b) The kinetic study on the binding of MBP-AF-6 (36–206 amino acids) to GST-ZO-1. E. coli lysates containing various concentrations of MBP-AF-6 (36–206 amino acids) were loaded onto the GST-ZO-1 and GST-CD44 affinity columns (0.1 nmol). The proteins bound to GST-ZO-1 columns were eluted with GST-ZO-1 by the addition of glutathione. The eluates were subjected to SDS-PAGE and followed by immunoblot analysis with anti-MBP antibody. The immunodetected MBP-AF-6 were visualized and estimated with a densitograph. The values shown are means ± SEM of triplicate experiments. ▪, with GST-ZO-1; ▴, with GST-CD44. (c) Dissociation of MBP-AF-6 (36–206 amino acids) from ZO-1 by activated Ras. Crude lysates of E. coli expressing MBP-AF-6 (36–206 amino acids) were loaded onto affinity columns immobilized with GST-ZO-1. The proteins bound to the GST-ZO-1 columns were eluted by the addition of buffer containing GDP alone (lane 1), GTPγS alone (lane 2), GDP/Ras (lane 3), GTPγS/Ras (lane 4), and GTPγS/Rac (lane 5). The eluates were subjected to SDS-PAGE and followed by immunoblot analysis with anti-MBP antibody. The arrowhead denotes the position of MBP-AF-6 (36–206 amino acids). The results shown are representative of three independent experiments.
Localization of AF-6 in Activated Ras-expressing Rat1 Fibroblast Cells
As previously described, some ras-transformed fibroblast cells display anchorage-independent growth and reduced cell–cell contacts. We examined the localization of AF-6 in activated Ras-expressing Rat1 fibroblast cells. For this purpose, we used a Rat1 cell line that contains the activated ras (rasV12) gene under Lac repressor control, designated as Rat1 RasVal A1 cells. The addition of IPTG efficiently induced the expression of RasV12 (Fig. 11 A). The distributions of AF-6 and ZO-1 were examined as described above. In the Rat1 RasVal A1 cells in the absence of IPTG, AF-6 accumulated at cell–cell contact sites and was colocalized with ZO-1, as seen in the wild-type strain of Rat1 cells (Fig. 11, B, c and d). When Rat1 RasVal A1 cells were treated with IPTG, their morphology changed dynamically and the cell–cell contacts were perturbed. In these cells, the colocalization of AF-6 and ZO-1 was decreased at the surface of the cells dissociated from neighbor cells (Fig. 11, B, e and f). These results suggest that AF-6 may contribute to the perturbation of cell–cell contacts by activated Ras, or that the altered cell–cell contacts by activated Ras may change the cellular distribution of AF-6.
Figure 11.
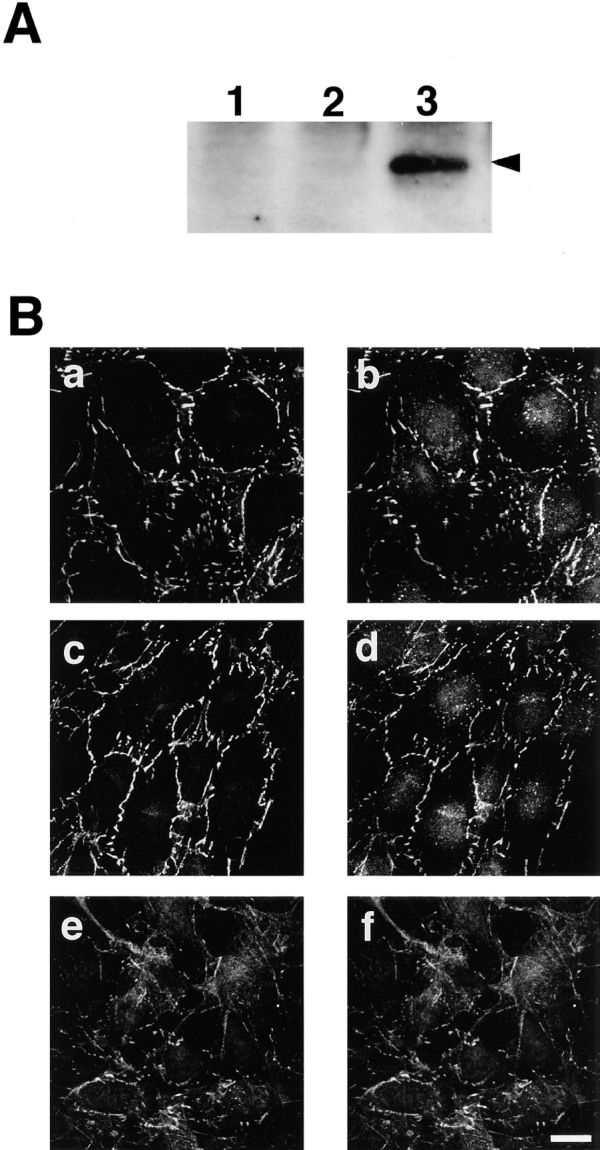
Localization of AF-6 in Ras-transformed Rat1 cells. (A) Immunoblot analysis of Rat1 RasVal A1 cells. The expression of RasV12 was induced in Rat1 RasVal A1 cells (lanes 3) by the addition of IPTG, and the cell lysates were subjected to SDS-PAGE, followed by immunoblot analysis with anti-Ras antibody (Rask4). Lane 1, Wild-type Rat1 cells; lane 2, Rat1 RasVal A1 cells in the absence of IPTG; lane 3, Rat1 RasVal A1 cells treated with IPTG for 24 h. (B) Localization of AF-6 in Ras-transformed Rat1 cells. Wild-type Rat1 cells (a and b), Rat1 RasVal A1 cells in the absence of IPTG (c and d), and Rat1 RasVal A1 cells treated with 5 mM IPTG for 24 h (e and f) were doubly stained with a rabbit polyclonal antibody against AF-6 (a, c, and e) and a mouse monoclonal antibody against ZO-1 (b, d, and f), followed by FITC-conjugated anti–rabbit IgG and Texas red-conjugated anti– mouse IgG antibodies. The results shown are representative of three independent experiments. Bar, 10 μm.
Discussion
AF-6 Is a Peripheral Component of Junctional Complex
Our immunofluorescent and immunoelectron microscopic analysis showed that AF-6 is concentrated at tight junctions in confluent MDCKII cells, and that its distribution closely overlaps with that of ZO-1. Native and recombinant AF-6 interacts with ZO-1 in vitro. ZO-1 is thought to directly interact with occludin and to serve as an essential peripheral component of tight junctions (Furuse et al., 1994). Thus, these results indicate that AF-6 is a peripheral component of tight junctions. Although the roles of AF-6 in tight junctions are not clear, it is possible that AF-6 modulates the functions of ZO-1 downstream of Ras. We have also found that AF-6 from bovine brain interacts with the GST-occludin affinity beads, whereas the purified AF-6 from insect cells, which lack ZO-1, only weakly interacts with the same beads. This raises the possibility that the binding of AF-6 from bovine brain may be mainly mediated by ZO-1.
A recent study showed that ZO-1 interacts with catenins during the early stages of the assembly of tight junctions (Rajasekaran et al., 1996). In cells lacking tight junctions such as fibroblasts and astrocytes, ZO-1 is localized at cell–cell contact sites with cadherin (Howarth et al., 1992; Itoh et al., 1993). We found here that AF-6 is localized with ZO-1 at cell–cell contact sites in Rat1 fibroblasts and PC12 rat pheochromocytoma cells lacking tight junctions. These results suggest that AF-6 is recruited with ZO-1 at cell–cell contact sites, where cadherin is localized, and serves as a peripheral component of cell–cell adhesions in cells lacking tight junctions.
ZO-1–binding Domain of AF-6
Since it has been reported that some PDZ domains can bind to other PDZ domains (Brenman et al., 1996), we first thought that the PDZ domain of AF-6 may interact with the PDZ domain of ZO-1. The present results, however, indicate that the NH2-terminal domain (36–206 amino acids) of AF-6 is mainly responsible for the interaction with ZO-1. A previous study showed that the NH2-terminal domain (36–206 amino acids) binds to activated Ras (Kuriyama et al., 1996). The present findings indicate that the Ras-binding domain and ZO-1–binding domain of AF-6 are very close or overlapped, though the ZO-1–binding domain of AF-6 is not necessarily coincident with the Ras-binding domain.
The GST-ZO-1 used in this study contains three PDZ domains, an SH3 domain, and a guanylate kinase domain. It is possible that the SH3 domain of ZO-1 interacts with the proline-rich domain of AF-6. Indeed, our in vitro binding assay showed that AF-6 (1,130–1,612 amino acids) interacts with ZO-1, but this interaction is weaker than that of AF-6 (36–494 amino acids). The AF-6–binding domain of ZO-1 remains to be identified. The interaction of recombinant AF-6 with occludin is very weak. It remains to be clarified whether AF-6 directly interacts with occludin in vivo.
Possible Roles of AF-6 in the Regulation of Cell–Cell Contacts
AF-6 is composed of several domains including Ras-binding, ZO-1–binding, myosin V-like, PDZ, and the COOH-terminal domains. Although the roles of AF-6 in the regulation of cell–cell contacts remain to be clarified, AF-6 may be a scaffolding protein as a peripheral component at the cell–cell contact sites. AF-6 shows the strong sequence homology with Drosophila Canoe and shares a common domain organization with Canoe (Kuriyama et al., 1996). Canoe is implicated in the formation of cone cells in the developing compound eye in Drosophila (Matsuo et al., 1997). The fates of cone cells are thought to be determined by cell–cell contacts. The phenotypic effect of canoe mutations on the cone cells depends on the state of Ras (Matsuo et al., 1997), and Canoe is shown to interact with the activated Ras in a cell-free system, indicating that Canoe serves as a target of Ras as described for AF-6 (Kuriyama et al., 1996). Genetic study of Canoe in Drosophila will provide us further information on the roles of Canoe/AF-6 in the regulation of cell–cell contacts.
We have shown here that AF-6 can interact with ZO-1 in vitro and is colocalized with ZO-1 at cell–cell contact sites including tight junctions. This suggests that AF-6 can modulate the function of ZO-1. ZO-1 is thought to be a linkage molecule between occludin and actin cytoskeleton and to play important roles in the rearrangement of cell– cell contacts and cytoskeletons (Furuse et al., 1994). It is possible that AF-6 modulates the function of ZO-1 and subsequently regulates the cell–cell contacts.
We have recently found that AF-6 interacts with filamentous actin by a cosedimentation assay (data not shown). Actin cytoskeleton is known to be linked to adhesion molecules through certain peripheral components and to support the cell–cell adhesions. Although the mode of interaction between AF-6 and filamentous actin is not known at present, the actin-binding activity of AF-6 may be necessary for the regulation of the cell–cell adhesions through AF-6. The COOH-terminal domain of AF-6 has the proline-rich region, which is thought to interact with certain proteins containing the SH3 or WW domain (Sudol, 1996). Searching for interacting proteins with the COOH-terminal domain is under investigation to understand the functions of AF-6.
Regulation of the Interaction of AF-6 and ZO-1 by Activated Ras
As described above, it is likely that the Ras-binding domain and ZO-1–binding domain of AF-6 are quite close. This led us to examine whether Ras inhibits the interaction of AF-6 with ZO-1. Indeed, activated Ras can dissociate MBP-AF-6 (36–206 amino acids) from ZO-1. This suggests that activated Ras inhibits the interaction of AF-6 and ZO-1 at cell–cell adhesions. Our preliminary experiments, however, showed that activated Ras dissociates full length recombinant AF-6 from ZO-1 less efficiently than MBP-AF-6 (36–206 amino acids) from ZO-1 (data not shown). The reason for the less efficient dissociation of full length AF-6 from ZO-1 is not known. Another domain of AF-6 in addition to the NH2-terminal domain containing the Ras-binding domain may strengthen the interaction of AF-6 and ZO-1. Alternatively, activated Ras may change the state of the AF-6–ZO-1 complex without dissociating AF-6 from ZO-1. In addition, since we used the recombinant Ras produced from bacteria, which was not modified with lipid at the COOH terminus, it is possible that lipid-modified, activated Ras can efficiently dissociate full length AF-6 from ZO-1.
Regulation of Cell–Cell Contacts by Ras
Ras is thought to regulate the dynamics of cell–cell contacts. Some ras-transformed epithelial cells display a fibroblastic morphology and reduced cell–cell contacts, as previously described (Basolo et al., 1991; Kinch et al., 1995). Several molecules interacting with activated Ras in addition to AF-6 have been identified as Ras targets in mammals. These include Raf (Daum et al., 1994), phosphatidylinositol-3-OH kinase (Rodriguez-Viciana et al., 1994), Ral guanine nucleotide dissociation stimulator (Kikuchi et al., 1994; Spaargaren and Bischoff, 1994), and Rin1 (Han and Colicelli, 1995). One of these Ras targets may account for the functions of Ras in the regulation of cell–cell contacts. We have shown here that AF-6 specifically accumulates at cell–cell contact sites. None of the above Ras targets except for AF-6 is reported to accumulate at cell–cell contact sites in an adhesion-dependent fashion. Expression of activated Ras perturbs cell–cell contacts in Rat1 fibroblast cells, followed by a decrease in the accumulation of AF-6 and ZO-1 at the cell surface. Moreover, we have shown here that AF-6 interacts with ZO-1, and this interaction is inhibited by activated Ras. Taken together, it is likely that the Ras-AF-6 pathway plays critical roles in the regulation of the cell–cell contacts through ZO-1. Alternatively, the altered cell–cell contacts by activated Ras may change the cellular distribution of AF-6 and ZO-1. The activated Ras-expressing cells change a plethora of characteristics. We do not know exactly what happens in these cells. Further studies are necessary for better understanding the mechanism of the regulation of cell–cell contacts by Ras.
Acknowledgments
We thank Drs. Masahiko Itoh, Mikio Furuse, and Shoichiro Tsukita (University of Kyoto, Kyoto, Japan) for kindly providing MDCKII cells, anti-ZO-1 antibody, anti–α-catenin antibody, and expression plasmids of GST-mouse ZO-1, GST-mouse occludin, GST-mouse E-cadherin, GST-mouse α-catenin, and GST-mouse β-catenin; Drs. Hiroshi Itoh and Yoshito Kaziro (Tokyo Institute of Technology, Yokohama, Japan) for kindly providing Rat1 and Rat1 RasVal A1 cells; and A. Takemura for secretarial assistance.
This study was supported by grants-in-aid for scientific research and for cancer research from the Ministry of Education, Science, and Culture of Japan (1997) and by grants from the Mitsubishi Foundation and Kirin Brewery Company Limited.
Abbreviations used in this paper
- GST
glutathione-S-transferase
- IPTG
isopropyl-β-d-thiogalactoside
- MAP
mitogen-activated protein
- MBP
maltose-binding protein
Footnotes
Address all corrrespondence to Kozo Kaibuchi, Division of Signal Transduction, Nara Institute of Science and Technology, Nara 630-01, Japan. Tel.: (81) 7437-2-5440. Fax: (81) 7437-2-5449. E-mail: kaibuchi@bs.aist-nara.ac.jp
References
- Anderson JM. Cell signaling: MAGUK magic. Curr Biol. 1996;6:382–384. doi: 10.1016/s0960-9822(02)00501-8. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Stevenson BR, Jesaitis LA, Goodenough DA, Mooseker MS. Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J Cell Biol. 1988;106:1141–1149. doi: 10.1083/jcb.106.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M. Ras genes. Ann Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Basolo F, Elliott J, Tait L, Chen XQ, Maloney T, Russo IH, Pauley R, Momiki S, Caamano J, Klein-Szanto AJ, et al. Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog. 1991;4:25–35. doi: 10.1002/mc.2940040106. [DOI] [PubMed] [Google Scholar]
- Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc Natl Acad Sci USA. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Burry RW, Vandre DD, Hayes DM. Silver enhancement of gold antibody probes in pre-embedding electron microscopic immunocytochemistry. J Histochem Cytochem. 1992;40:1849–1856. doi: 10.1177/40.12.1453003. [DOI] [PubMed] [Google Scholar]
- Cano E, Mahadevan LC. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- Catling AD, Reuter CWM, Cox ME, Parsons SJ, Weber MJ. Partial purification of a mitogen-activated protein kinase kinase activator from bovine brain. J Biol Chem. 1994;269:30014–30021. [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophiladiscs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Daum G, Eisenmann-Tappe I, Fries H-W, Troppmair J, Rapp UR. The ins and outs of Raf kinases. Trends Biochem Sci. 1994;19:474–480. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Diamond J. The epithelial junction: bridge, gate, and fence. Physiologist. 1977;20:10–18. [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Sa, Tsukita, Sh, Tsukita Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. The structure, biochemistry and assembly of epithelial tight junctions. Am J Physiol. 1987;253:C749–C758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Chavez de Ramirez B, Cereijido M. Tight junction formation in cultured epithelial cells (MDCK) J Membr Biol. 1985;86:113–125. doi: 10.1007/BF01870778. [DOI] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Han L, Colicelli J. A human protein selected for interference with ras function interacts directly with ras and competes with raf1. Mol Cell Biol. 1995;15:1318–1323. doi: 10.1128/mcb.15.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Howarth AG, Hughes MR, Stevenson BR. Detection of the tight junction-associated protein ZO-1 in astrocytes and other nonepithelial cell types. Am J Physiol. 1992;262:C461–C469. doi: 10.1152/ajpcell.1992.262.2.C461. [DOI] [PubMed] [Google Scholar]
- Hülsken J, Behrens J, Birchmeier W. Tumor- suppressor gene products in cell contacts: the cadherin- APC-armadillo connection. Curr Opin Cell Biol. 1994;6:711–716. doi: 10.1016/0955-0674(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Itoh M, Yonemura S, Nagafuchi A, Sa, Tsukita, Sh, Tsukita A 220-kD undercoat-constitutive protein: its specific localization at cadherin-based cell–cell adhesion sites. J Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Sa, Tsukita, Sh, Tsukita The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, R.K., S.A. Moodie, A. Wolfman, and G.E. Landreth. 1994. The mitogen-activated protein kinase cascade is activated by B-Raf in response to nerve growth factor through interaction with p21ras. Mol. Cell. Biol. 14:6944– 6953. [DOI] [PMC free article] [PubMed]
- Jongens TA, Ackerman LD, Swedlow JR, Jan LY, Jan YN. Germ cell-less encodes a cell type-specific nuclear pore-associated protein and functions early in the germ-cell specification pathway of Drosophila. . Genes Dev. 1994;8:2123–2136. doi: 10.1101/gad.8.18.2123. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature (Lond) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kinch MS, Clark GJ, Der CJ, Burridge K. Tyrosine phosphorylation regulates the adhesions of ras- transformed breast epithelia. J Cell Biol. 1995;130:461–471. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science (Wash DC) 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, Yamamoto D, Prasad R, Croce C, Canaani E, et al. Identification of AF-6 and canoe as putative targets for Ras. J Biol Chem. 1996;271:607–610. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995a;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Marshall MS. Ras target proteins in eukaryotic cells. FASEB J. 1995b;9:1311–1318. doi: 10.1096/fasebj.9.13.7557021. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Takahashi K, Kondo S, Kaibuchi K, Yamamoto D. Regulation of cone cell formation by Canoe and Ras in the developing Drosophilaeye. Development. 1997;124:2671–2680. doi: 10.1242/dev.124.14.2671. [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Possee RD, Overton HA, Bishop DH. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- McCormick F. Activators and effectors of ras p21 proteins. Curr Opin Genet Dev. 1994;4:71–76. doi: 10.1016/0959-437x(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Moodie SA, Paris MJ, Kolch W, Wolfman A. Association of MEK1 with p21ras·GMPPNP is dependent on B-Raf. Mol Cell Biol. 1994;14:7153–7162. doi: 10.1128/mcb.14.11.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale RP, Nowell PC, Kuriyama K, et al. Cloning of the ALL-1 fusion partner, the AF-6 gene, involved in acute myeloid leukemias with the t(6;11) chromosome translocation. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. 1996;132:451–463. doi: 10.1083/jcb.132.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol- 3-OH kinase as a direct target of Ras. Nature (Lond) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Satoh T, Nakafuku M, Kaziro Y. Function of Ras as a molecular switch in signal transduction. J Biol Chem. 1992;267:24149–24152. [PubMed] [Google Scholar]
- Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M. The WW module competes with the SH3 domain? . Trends Biochem Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Tsukita Sh, Itoh M, Nagafuchi A, Yonemura S, Tsukita Sa. Submembranous junctional plaque proteins include potential tumor suppressor molecules. J Cell Biol. 1993;123:1049–1053. doi: 10.1083/jcb.123.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junction bordering transmitter release zones. J Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. The tight junction protein ZO-1 is homologous to the Drosophiladiscs-large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophilaencodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Yamamori B, Kuroda S, Shimizu K, Fukui K, Ohtsuka T, Takai Y. Purification of a Ras-dependent mitogen-activated protein kinase kinase kinase from bovine brain cytosol and its identification as a complex of B-Raf and 14-3-3 proteins. J Biol Chem. 1995;270:11723–11726. doi: 10.1074/jbc.270.20.11723. [DOI] [PubMed] [Google Scholar]