Abstract
The mammalian nucleus is highly organized, and nuclear processes such as DNA replication occur in discrete nuclear foci, a phenomenon often termed “functional organization” of the nucleus. We describe the identification and characterization of a bipartite targeting sequence (amino acids 1–28 and 111–179) that is necessary and sufficient to direct DNA ligase I to nuclear replication foci during S phase. This targeting sequence is located within the regulatory, NH2-terminal domain of the protein and is dispensable for enzyme activity in vitro but is required in vivo. The targeting domain functions position independently at either the NH2 or the COOH termini of heterologous proteins.
We used the targeting sequence of DNA ligase I to visualize replication foci in vivo. Chimeric proteins with DNA ligase I and the green fluorescent protein localized at replication foci in living mammalian cells and thus show that these subnuclear functional domains, previously observed in fixed cells, exist in vivo. The characteristic redistribution of these chimeric proteins makes them unique markers for cell cycle studies to directly monitor entry into S phase in living cells.
The complexity of the mammalian genome and the multitude of different biochemical processes occurring in the nucleus call for an efficient mechanism of coordination and organization. At least in part this is achieved by subdivision into functional domains (for review see 23, 54). DNA replication occurs at discrete nuclear foci (14, 18, 36, 37, 40, 58) where replication proteins (proliferating cell nuclear antigen [PCNA],1 5; the 70-kD subunit of replication protein A [RPA 70], 7; DNA polymerase α, 17) are localized. In addition to these replication proteins, DNA methyltransferase (DNA MTase; 24) and cell cycle proteins (cyclin A and cdk2; 7, 52) were also found to redistribute to these foci during S phase. In view of the complexity of mammalian DNA replication, it is likely that many more proteins are localized at these nuclear foci.
One good candidate for localization at replication foci is DNA ligase I (34). DNA ligase activity is required for DNA replication, repair, and recombination. Yeast DNA ligase I mutants are lethal and show defects in DNA replication and repair (19, 38). The human cDNA of DNA ligase I was cloned by functional complementation of yeast conditional mutants and shows an open reading frame of 919 amino acids (2) and an active site lysine residue at position 568 (20). Mammalian DNA ligase I is essential for in vitro replication of simian virus 40 DNA and cannot be substituted by other DNA ligases (59). Inherited genetic defects of the human DNA ligase I gene demonstrate its requirement for Okazaki fragment ligation during lagging strand DNA synthesis and for repair of DNA damage (3, 44, 53, 56), which is a prerequisite for genome integrity. Recent studies with homozygous null mice showed that in less demanding selection conditions, DNA ligase I function can be substituted by another yet unknown ligase activity (4). The enzyme has a hydrophilic protease-sensitive NH2-terminal domain of 249 amino acids (57), which has a negative regulatory function that is relieved upon phosphorylation by casein kinase II (43). This NH2-terminal domain is dispensable for enzyme activity in vitro, as well as for complementation of yeast (2) and bacterial ligase mutants (20), though it is required in vivo in mammalian cells (42). Moreover, this domain has no counterpart in the recently identified human DNA ligases III and IV (8, 60), despite extensive homology throughout their catalytic domains.
In this study we show that DNA ligase I is localized at nuclear replication foci during S phase. We identified and characterized a bipartite protein sequence that is necessary and sufficient for this cell cycle-dependent redistribution. Moreover, this sequence works position independently, can target heterologous proteins to subnuclear sites of DNA replication and, therefore, meets the criteria for a targeting sequence. We propose that this targeting sequence contributes to the high efficiency of Okazaki fragment ligation during lagging strand DNA synthesis and plays an important role in the coordinate regulation of the different steps of DNA replication. We furthermore used fusions of DNA ligase I with GFP to directly visualize replication foci in vivo. The characteristic redistribution of these fusion proteins makes them unique S phase markers to monitor cell cycle progression in living cells.
Materials and Methods
Antibodies
Polyclonal antibodies against DNA ligase I were raised in rabbits using a peptide antigen spanning amino acids 1–23 from the mouse enzyme with the addition of N-chloracetyl-glycine at the NH2 terminus for coupling to the carrier protein KLH (30). The anti-peptide antibody was purified from serum by affinity chromatography as described by Sawin et al. (47) using the peptide antigen coupled to Affi-Gel 10 (Bio Rad, Hercules, CA).
In addition, the following primary antibodies were used: mouse monoclonal anti-PCNA antibody (clone PC 10; Dako, Carpinteria, CA; Zymed, San Francisco, CA), 12CA5 hybridoma supernatant recognizing the Flutag epitope, β-galactosidase (β-gal)-specific mouse monoclonal antibody (Promega, Madison, WI), rabbit polyclonal anti-MTase antiserum (24), and mouse monoclonal anti-5-bromo-2′-deoxyuridine (BrdU) antibody FITC conjugated (Boehringer Mannheim, Mannheim, Germany).
DNA Ligase I Fusion Constructs
Expression plasmids were derived from pJ3Ω (35) or pEVRF0 (33). The latter was used to add a heterologous NLS (see below). Oligonucleotides encoding a nine-amino acid epitope (YPYDVPDYA; 61) from the hemagglutinin of influenza virus (Flutag) were inserted at the Bsp EI restriction site (codon 307), at the Eag I restriction site (codon 773) or between both sites in the cDNA of human DNA ligase I (ATCC 65856; 2). The β-gal epitope was derived from the β-gal gene of Escherichia coli (amino acids 361-1,069) and was added at the COOH-terminus of all but two deletion constructs. A short nuclear localization signal (NLS) derived from SV40 large T antigen (PKKKRKV) was added at the NH2 terminus of some deletion constructs to compensate for the loss of their own NLS using a translational fusion vector, as previously described (24). The green fluorescent protein (GFP) fusions were derived from the pRSGFP-C1 and the pEGFPC1 vectors (Clontech, Palo Alto, CA), by inserting a fragment containing the full length or the first 250 amino acids of the human DNA ligase cDNA at the COOH terminus of the open reading frame of both GFP mutants. It is noteworthy that the GFP expressed in these constructs, with an additional 26 amino acids derived from the multiple cloning site added to its COOH terminus, is by itself nuclear and cytoplasmic. Plasmid DNA was purified using columns (Qiagen, Hilden, Germany) according to the instructions of the manufacturer.
Cell Culture and Transfection
Human HeLa cells, monkey Cos 7 cells, and mouse MEL and C3H10T1/2 cells were grown in a humidified incubator at 37°C and 5% CO2 in DME supplemented with 10% fetal calf serum. Mouse (C2C12) and rat (L6E9) myoblast cells were grown as above except that the media was supplemented with 20% fetal calf serum.
Cos 7 cells were transfected by the DEAE-dextran pretreatment method as described by Leonhardt et al. (24). 2 d later, cells were scraped, extracted, and analyzed as described below. Exponentially growing mouse fibroblasts were transfected by the calcium phosphate-DNA coprecipitation method (15) with glycerol shock treatment (41) ∼8–12 h later. 36–48 h after DNA addition, cells were fixed and stained.
For visualization of GFP-ligase in living cells, transfected fibroblasts plated onto glass-bottom petri dishes were changed to Hepes-buffered medium (10 mM Hepes, pH 7.0, 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM Glucose) and screened with a fluorescence microscope using an FITC filter.
Whole Cell Extracts and Western Blotting
Transfected Cos cells were scraped and extracted as described by Leonhardt et al. (24) in 0.2 M NaCl, 0.32 M sucrose, 0.3% Triton X-100, 20 mM Tris-HCl (pH 7.4), 3 mM MgCl2, 0.5 mM dithiothreitol, and 0.2 mM phenylmethylsulfonylfluoride. Cell extracts used to characterize the anti-DNA ligase I antibody by immunoblots were made by extraction of cells for 30 min on ice with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) containing the following protease inhibitors: 1 mM EDTA, 1 mM Pefabloc, 10 μM leupeptin, 10 μM pepstatin, and 10 μM aprotinin. After 5 min centrifugation in the cold, the soluble fraction was analyzed by immunoblot. The protein extracts were dissolved in Laemmli sample buffer, separated by SDS-PAGE under reducing conditions, and transferred to PVDF membranes. After incubation with epitope tag-specific antibodies or anti-DNA ligase I antibodies, followed by HRP-conjugated secondary antibodies, the blots were developed using the ECL detection procedure (Amersham, Buckinghamshire, UK).
Immunofluorescence
Transfected cells were washed in PBS and fixed for 10 min in 3.7% formaldehyde in PBS or for 5 min in cold methanol. All following incubations were performed at room temperature. Formaldehyde-fixed cells were permeabilized for 10 min in 0.25% Triton X-100. Fixed cells were blocked in 5% goat serum or 0.2% gelatin and incubated for 60 min with the respective primary antibodies as described before (7, 24). After extensive washes in 0.1% NP-40, cells were incubated for another 60 min in FITC-conjugated goat anti–mouse or anti–rabbit IgG and biotinylated goat anti–rabbit or anti–mouse IgG antibodies, followed by washing and 30 min incubation in streptavidin-Texas red, as described before (7). DNA was counterstained with Hoechst 33258 and cells were mounted in mowiol with 2.5% DABCO (6). Pulse labeling with BrdU and detection of incorporated BrdU were done as described before (7, 24).
Microscopy
Stained specimens or live cultures were examined and photographed in microscopes (Axiophot and Axiovert; Zeiss, Inc., Thornwood, NY) equipped with phase-contrast and epifluorescence optics, using 40, 63, and 100× oil immersion Plan-Neofluor and Planapochromat objectives. Single Hoechst, FITC, and Texas red filters were used as well as FITC-Texas red dual filters. Pictures were taken with Kodak Ektar and Royal Gold films. Micrographs were scanned, assembled, and annotated with Adobe Photoshop and Canvas software in a Power MacIntosh computer and printed with a Phaser 440 dye sublimation printer (Tektronix). Overlays were mostly generated by photographic double exposure with the two respective filters or directly with a dual filter. In two cases (Fig. 2 C, the upper left early S phase nucleus, and F), overlays were generated digitally using Adobe Photoshop layers.
Figure 2.
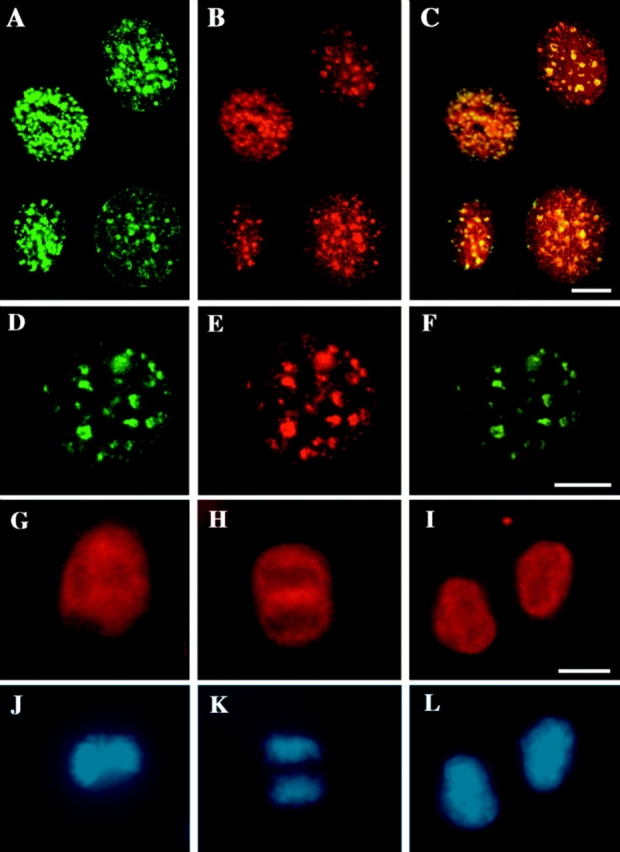
Localization of DNA ligase I throughout the cell cycle. Asynchronously growing mouse fibroblasts (C3H10T1/2 cells; A–C and G–L) and myoblasts (C2C12 cells; A–F) were pulse labeled with BrdU for 5 to 10 min (A–C) and formaldehyde (A–C, G, and J) or methanol fixed (D–F, H, I, K, and L). Cells were stained for DNA ligase I with the affinity-purified anti-DNA ligase I rabbit antibodies (see Fig. 1 red; B, E, and G–I), for sites of BrdU incorporation with anti-BrdU mouse monoclonal antibody (green; A), and for PCNA with anti-PCNA–specific mouse monoclonal antibody (green; D), and DNA was visualized by counterstaining with Hoechst 33258 dye (J–L). A–F show the distribution of DNA ligase I (B and E) in interphase cells relative to sites of ongoing DNA replication labeled with BrdU (A) and to PCNA (D), which has previously been shown to redistribute in S phase nuclei to replication centers (5, 7). A–C is a composite of cell nuclei at different stages of S phase. As can be better visualized in the overlay of the green and red images in C and F, DNA ligase I takes on a pattern of subnuclear foci that colocalize with sites of BrdU incorporation (C) and with PCNA (F). G–L show the distribution of DNA ligase I in mitotic cells. A cell in metaphase as evidenced by the absence of nuclear membrane and the alignment of the chromosomes in the metaphase plate in J, shows that DNA ligase I is excluded from the condensed chromosomes and upon nuclear envelope breakdown distributes in the cytoplasm (G). During chromatid separation and movement to the spindle poles at anaphase in K, DNA ligase I is still dispersed in the cytoplasm and excluded from the condensed chromosomes (H). At the end of telophase and cytokinesis, when the nuclear envelope reforms around the decondensing chromosomes (L), there is an immediate import of DNA ligase I into the nucleus as seen in I. Bars, 10 μm.
Results
Localization of DNA Ligase I throughout the Cell Cycle
To identify additional components of nuclear replication foci and to probe the dynamics of nuclear architecture, we analyzed the subcellular localization of DNA ligase I, which is involved in joining Okazaki fragments during lagging-strand DNA synthesis (for review see 29).
Polyclonal antibodies were raised against mammalian DNA ligase I. We used a peptide spanning the first 23 amino acid residues of mouse DNA ligase I, which differs from the human protein in two positions. The antisera were then affinity purified and tested for specificity and species reactivity by immunoblot. As shown in Fig. 1 A, both elution methods (0.2 M glycine-HCl, pH 2.0, and 6 M guanidine hydrochloride) yielded antibody fractions that specifically reacted with mouse DNA ligase I. The predicted size of mammalian DNA ligase I is 101–102 kD, but endogenous as well as recombinant proteins migrate anomalously slowly in SDS-PAGE, which may be due to its high proline content, to the presence of phosphoserine residues, and to the high hydrophilicity of the NH2-terminal domain (43, 57). The result obtained with our antibodies is in agreement with the previously observed migration of mammalian DNA ligase I. This signal is competed away by pre-incubating the antibodies with the peptide used as antigen but not with a nonspecific peptide, as shown in Fig. 1 B, further confirming its specificity. Finally, we tested for cross-reactivity with homologues from other mammalian species. Fig. 1 C shows that the affinity-purified antibodies specifically recognize proteins of the expected size in extracts from mouse, rat, monkey, and human cells. This result was expected because of the relatively high degree of conservation between human and mouse DNA ligase I cDNAs. Under the conditions used (50 μg of extract) some additional signals can be seen in the monkey cell extracts, even though at much lower intensity.
Figure 1.
Purification and characterization of antibodies against DNA ligase I. (a) Immunoblot analysis of MEL whole cell extract (20 μg/lane) with anti-DNA ligase I antibodies. Lane 1, Preimmune serum; lane 2, affinity-purified antibodies eluted with 0.2 M glycine-HCl, pH 2.0; lane 3, affinity-purified antibodies eluted with 6 M guanidine hydrochloride. A band with an apparent molecular weight of 120 to 130 kD reacts specifically with the affinity-purified antibodies. (b) Specifity of the antibody (lane 1) was further tested by pre-incubation with specific (S, lane 2) and unspecific (U, lane 3) peptides at a 100-fold molar excess. The DNA ligase I signal is competed out only with the NH2-terminal DNA ligase I peptide and not with the same amount of an unrelated peptide, confirming the specificity of the antibody. (c) Species reactivity of anti-DNA ligase I antibodies: lane 1, C2C12 (mouse) cell extract; lane 2, L6E9 (rat) cell extract; lane 3, Cos 7 (monkey) cell extract; lane 4, HeLa (human) cell extract. A total of 50 μg of whole cell extracts was loaded in each lane. The anti-DNA ligase I antibodies specifically detect a protein band of similar size in rat and mouse cell extracts and slightly bigger in monkey and human cell extracts, which correspond most likely to the DNA ligase I protein from these species.
We then used the affinity-purified antibodies to analyze the subcellular localization of DNA ligase I throughout the cell cycle. Asynchronously growing mouse fibroblasts and myoblasts were labeled with the thymidine analogue BrdU for 5 to 10 min and immediately fixed, to detect subnuclear sites of DNA replication. BrdU incorporation was visualized by staining with BrdU-specific mouse monoclonal antibodies, and samples were also stained for DNA ligase I with the affinity-purified rabbit antibodies. The cells that did not incorporate BrdU, and are therefore in G1 or G2 phase, exhibit a dispersed nucleoplasmic distribution of DNA ligase I with exclusion from nucleoli (data not shown). As depicted in Fig. 2, A–C, the nuclei with a punctate subnuclear BrdU pattern show colocalizing DNA ligase-labeled foci, as can be better seen in the overlay of the two images in C. This figure is a composite of nuclei, illustrating different patterns of subnuclear foci observed in both mouse fibroblasts and myoblasts. We further investigated the distribution of DNA ligase in interphase cells by costaining with anti-PCNA antibodies. One subnuclear pattern is presented in Fig. 2, D–F, and clearly shows the colocalization of both PCNA and DNA ligase I at sites of DNA replication. Nuclei with disperse PCNA nucleoplasmic signal, showed the same homogenous distribution of DNA ligase I (data not shown). The DNA ligase I distribution was identical using formaldehyde and methanol fixation.
In mitotic cells, at the metaphase stage shown by the alignment of the chromosomes in the metaphase plate in Fig. 2 J, DNA ligase I is excluded from the condensed chromosomes and distributes in the cytoplasm (Fig. 2 G). Later, during chromatid separation and movement to the spindle poles at anaphase in Fig. 2 K, DNA ligase I continues to be excluded from the condensed chromosomes (Fig. 2 H). At the end of telophase and cytokinesis, when the nuclear envelope reforms around the decondensing chromosomes (Fig. 2 L), DNA ligase I is immediately imported into the nucleus, as shown in Fig. 2 I.
In summary, DNA ligase I is excluded from condensed chromosomes during mitosis and immediately reenters the nucleus upon formation of the nuclear envelope. In interphase cells, it presents a homogenous nucleoplasmic pattern except during S phase, when it redistributes to subnuclear sites of DNA replication.
Mapping the DNA Ligase I Sequence Responsible for Targeting to Replication Foci
Since there are no dividing membranes in the nucleus, which could explain the concentration of proteins at these foci, we decided to further investigate the basis for this cell cycle-dependent redistribution. In the case of DNA MTase, a distinct targeting sequence had been identified that is necessary and sufficient for association with replication foci (24). The cDNA of human DNA ligase I has an open reading frame of 919 amino acids (2), and the protein resembles DNA MTase in that it also has a protease-sensitive NH2-terminal domain, in this case of 249 amino acids.
To test whether we could reproduce the endogenous DNA ligase patterns with recombinant proteins, the enzyme was tagged by adding an epitope from the hemagglutinin of influenza virus (Flutag; Fig. 3 A) and visualized in transfected cells by immunofluorescence microscopy. Double staining with antibodies against DNA MTase, which labels replication foci (24), showed co-localization of DNA ligase I at these sites (Fig. 3 B, a–c). Tagged DNA ligase I, like the endogenous protein, redistributed during the cell cycle and showed the same patterns as DNA MTase and PCNA (Fig. 3 B, and data not shown).
Figure 3.

Mapping of the human DNA ligase I targeting sequence. (A) Sixteen different epitope-tagged deletion mutations of DNA ligase I were constructed. Their structure is schematically outlined, and their respective capability to associate with nuclear replication foci is indicated with + (targeting proficient) and − (targeting deficient). Numbers on the left refer to the amino acids of human DNA ligase I remaining in the deletion constructs. The structure of DNA ligase I is outlined on the top, showing the location of the regulatory NH2-terminal domain, which is dispensable for enzyme activity in vitro (43), and the position of the active site lysine residue 568 (20). Notice that the lower part of the graph is an enlargement of the first 263 amino acids to better display the results of the fine mapping. Shaded boxes highlight the bipartite targeting sequence that is necessary and sufficient for association with replication foci, as defined by these deletion constructs. As indicated, the first four constructs are full length or deletion mutants of DNA ligase I tagged with the Flutag epitope inserted at codons 307, 773, or between both. A representative example of one S phase pattern of these Flu-tagged proteins is shown in part (B, a–c). The following 12 DNA ligase I deletion mutants are fused at their COOH terminus to β-gal (amino acids 361–1,069) as described in 24. The β-gal part of the fusion proteins is not depicted again in the lower half of this graph. The black diamond represents a short NLS derived from SV40 large T antigen and was added at the NH2 terminus of some deletion constructs to compensate for the potential loss of their own NLS using a translational fusion vector. The expression of all listed fusion proteins was monitored by Western blot analysis (data not shown). (B) The subnuclear patterns of the Flu epitope-tagged and β-gal fusion proteins with human DNA ligase I were determined by transiently expressing the fusion construct into mouse C3H10T1/2 cells and double staining the formaldehyde-fixed cells for: DNA ligase I (red; a, d, and g) using an epitope-specific monoclonal antibody; DNA MTase (which redistributes to replication sites during S phase) using rabbit polyclonal antiserum (green; b, e, and h); and overlay of DNA ligase and DNA MTase staining (c, f, and i). a–c depict a nucleus of a cell transfected with full length DNA ligase I tagged at codon 773 with the Flu epitope (corresponding to the first construct in A) in late S phase, and the double exposure in c shows the colocalization of DNA ligase I and DNA MTase at nuclear replication foci. Both proteins show similar redistribution within the nucleus during the cell cycle, which also parallels the one of PCNA, as shown in Fig. 2. d–f show one example of a targeting-proficient fusion construct with β-gal (d; containing amino acids 1–28 and 111–263 of DNA ligase I) that colocalizes with DNA MTase (e) at replication foci as can be seen in the double exposure (f). g–i show a targeting- deficient fusion construct with β-gal (g; containing amino acids 62–212 of DNA ligase I) that takes on a dispersed distribution and does not redistribute during S phase to replication foci as visualized with DNA MTase antibodies (h) and also in the double exposure (i). Bars, 10 μm.
To investigate whether or not DNA ligase I uses a mechanism similar to DNA MTase for association with replication foci, a series of deletion mutants was generated in search for a potential targeting sequence (Fig. 3 A). All fusion proteins were tested for stability in vivo as described before (24). In brief, Cos cells were transfected with the respective expression constructs; protein extracts were made and tested by Western blot analysis (data not shown). All constructs gave stable fusion proteins, which argues against possible artefacts due to instability problems. The apparent molecular weight of the fusion proteins differed in most cases from values deduced from the amino acid sequence. This discrepancy is consistent with the observation that DNA ligase I itself migrates anomalously slow (see Fig. 1). Two different epitope tags, Flutag and a β-gal–derived epitope, were inserted at different positions in the DNA ligase I cDNA, to rule out potential tagging artefacts. Two mutations deleting amino acids 308–772 and 264–919, immediately indicated that the entire catalytic domain was dispensable for association with replication foci. The latter were visualized as in Fig. 3 B, a–c, or by costaining with PCNA-specific antibodies (data not shown).
To fine map the region required for targeting, we constructed a series of COOH-terminal, NH2-terminal, and internal deletions scanning the entire NH2-terminal regulatory domain. The structure of these fusion proteins and their respective phenotype is shown in Fig. 3 A. Fig. 3 B, d–f, illustrates one example of a targeting proficient deletion mutant. On the other hand, several deletions within the regulatory domain abolished targeting, meaning the respective fusion proteins did not redistribute to replication foci in S phase nuclei and showed, instead, a dispersed nucleoplasmic distribution (Fig. 3 B, g–h). The fine mapping identified a bipartite-targeting sequence, which encompasses amino acids 1–28 and 111–179. This bipartite targeting sequence is necessary and sufficient to target heterologous proteins (like β-gal from Escherichia coli) to replication foci, since deletion of either part eliminates targeting (Fig. 3 A). Furthermore, this is the only targeting sequence within DNA ligase I, since deletion of just the first 28 amino acids in the context of the full length protein, abrogates targeting (Fig. 3 A). In other words, the catalytic domain (amino acids 250–919) by itself does not localize at replication foci, emphasizing the modular arrangement of targeting sequence and catalytic domain. While this work was in progress, a study geared towards the mapping of the NLS of DNA ligase I was published (34). In that report, one deletion construct lacking the first 115 amino acids was analyzed for subnuclear localization and found to be absent from replication foci, which is consistent with our mapping results.
The regulatory domain also contains the NLS. We found that the first 206 amino acids of DNA ligase I are sufficient for nuclear localization, while fusion proteins containing only the first 112 amino acids were clearly cytoplasmic (data not shown). This finding indicates that nuclear localization of DNA ligase I requires sequences spanning amino acids 112–206, which contains stretches of basic amino acids (e.g., amino acids 120–131 and/or 149–152) that fit the description of a potential NLS (12, 50). A recent report described the mapping of the DNA ligase I NLS to amino acids 119–131 (34), however, deletion of this region (removing amino acids 112–178 in the last ligase–β-gal fusion depicted in Fig. 3 A) does reduce but not abolish nuclear localization, i.e., the overproduced recombinant protein can be found in the nucleus and cytoplasm (data not shown). On the other hand, deletion of the first 28 amino acids also affected nuclear localization. These discrepancies with the previous study (34) can in part be explained because their experiments were based on fusions to a reporter protein that by itself can enter the nucleus.
Our results show that the classical NLSs (amino acids 120–131 and/or 149–152) are not absolutely required and also not sufficient for nuclear localization and that other regions (amino acids 1–28 and 178–206) contribute to the nuclear localization of DNA ligase I, which may be a combination of nuclear uptake and retention, as described for other proteins (48).
The identification of the targeting sequence in the human DNA ligase I raised the question whether or not this sequence was conserved in other proteins present at replication foci and in DNA ligases from other species. The comparison with the DNA MTase targeting sequence (24) shows no detectable similarity. On the contrary, both sequences are rather different; the DNA ligase targeting sequence is extremely hydrophilic, while the DNA MTase is rather hydrophobic (Fig. 4 A). This difference makes sense in view of their different biological function and may simply reflect their targeting to different parts of the replication factories. That is, these various targeting sequences most likely constitute protein–protein interaction surfaces in vivo, which are as diverse as their binding partners.
Figure 4.
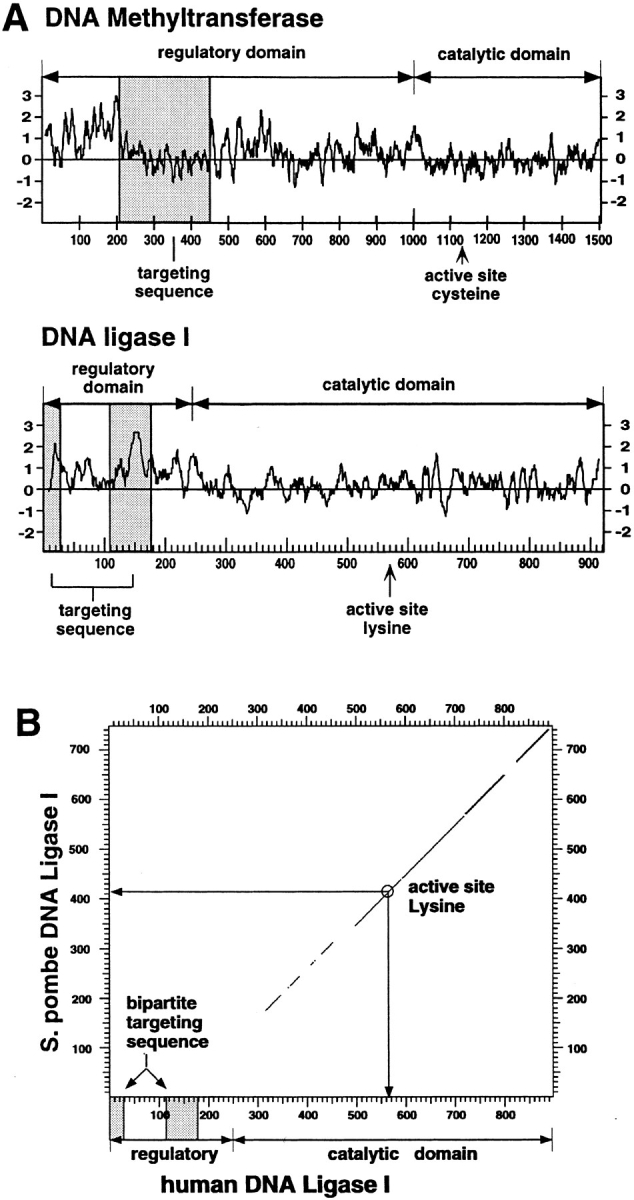
(A) Comparison between the targeting sequences of DNA ligase I and of DNA MTase. A hydrophilicity plot was prepared for both enzymes, and the respective targeting sequences were highlighted by shaded boxes. The overall structure of both enzymes was outlined by delineating the respective regulatory and catalytic domains and indicating the position of the active site residues. The targeting sequences do not share any sequence homology and are on the contrary very different; the bipartite targeting sequence of the DNA ligase I is extremely hydrophilic, while the DNA MTase sequence falls into a rather hydrophobic domain. In both cases the targeting sequence is located in the protease-sensitive, regulatory domain and is dispensable for enzyme activity in vitro. (B) Mammalian DNA ligase-targeting sequence is not conserved in lower eukaryotic homologues. The amino acid sequence of the human DNA ligase I was compared with the Schizosaccharomyces pombe homologue using the DNA Strider program version 1.2 (C. Marck) and the results are displayed in a dot plot format. The yeast and human enzymes show a high degree of homology throughout the catalytic domain, which is outlined in the graph; however, no homologous sequences for the NH2-terminal domain of the human enzyme (including the targeting sequence) could be detected in the fission yeast enzyme. Similar results are obtained comparing the human to the budding yeast protein.
A comparison with other ATP-dependent DNA ligases shows that the catalytic domain is highly conserved throughout evolution; however, no sequences with homology to the human targeting sequence were detected in lower eukaryotic or prokaryotic homologues (e.g., the comparison with DNA ligase I from Schizosaccharomyces pombe, Fig. 4 B). On the other hand, the targeting sequence is conserved between human and mouse (72% identity; 46), and the human targeting sequence also functions in mouse cells (Figs. 3 B and 5 A). There is also considerable conservation (data not shown) with the Xenopus laevis DNA ligase I (25). The fact that the targeting sequence is missing or is very different in lower eukaryotic and prokaryotic DNA ligases and is also not required for complementation may indicate that it only recently developed in evolution.
Figure 5.
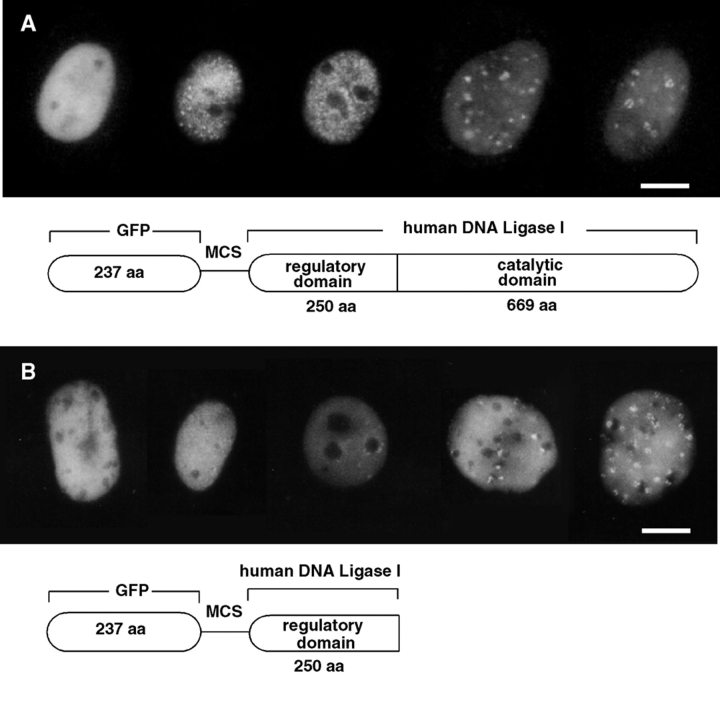
Visualization of DNA ligase I subnuclear localization in living cells. Asynchronous populations of mouse fibroblast and myoblast cells were transfected with plasmid DNA containing the full length (A) and the NH2-terminal 250 amino acids (B) of human DNA ligase I fused at the COOH terminus of the GFP. One day after DNA addition, cells were split onto glass bottom petri dishes, and the following days media was changed for a Hepes-buffered media. Cells expressing GFP-ligase fusion were screened under the microscope using an FITC filter and photographed. Below the micrographs are the respective schematic representations of the GFP fusion proteins with the full length human DNA ligase I (A) and with the NH2-terminal 250 amino acids of human DNA ligase I (B) containing the targeting sequence responsible for association with replication foci during S phase. The regulatory and catalytic domains of DNA ligase I are depicted, and MCS stands for multiple cloning site, which provides appropriate restriction sites for translational fusions. Bars, 10 μm.
Visualization of DNA Ligase I Subnuclear Redistribution in Living Cells
So far, the existence of subnuclear replication foci has only been shown by immunofluorescence. We therefore decided to directly probe these structures in living cells and fused the full length as well as the NH2 terminus alone containing the targeting sequence to the COOH-terminal end of the GFP, as schematically drawn in Fig. 5. The chimeric proteins were expressed in mouse cells and observed live under the microscope using an FITC filter. In addition to a nuclear dispersed distribution (first nucleus on the left hand side of Fig. 5, A and B), several discrete patterns of dots of different sizes and shapes were observed with the chimeric GFP-ligase proteins outlined in Fig. 5. Representative examples of the latter patterns are shown in the four nuclei at the right hand side of each panel. These patterns in live cells match the early, mid, and late S phase patterns reported in fixed and stained cells (14, 37, 40, 58). Based on that classification, the second nucleus from the left exhibits a fine punctate pattern of foci throughout the nucleoplasm characteristic of early S phase. The nucleus in the middle shows a concentration of foci at the nucleolar periphery corresponding to a later S phase stage. The two nuclei at the right hand side have larger foci with often irregular shape or loop-like structures, which are typical of late S phase. All these patterns were also observed by staining fixed cells with anti-DNA ligase I antibodies (Fig. 2) as well as with the other tagged constructs (Fig. 3, and data not shown). These same GFP-ligase–expressing cells were fixed and stained with anti-PCNA and anti-MTase antibody (Cardoso, M.C., R. Reusch, and H. Leonhardt, unpublished results), confirming the colocalization results presented in Figs. 2 and 3. The observation that the NH2-terminal domain of DNA ligase I (which contains the targeting sequence mapped in Fig. 3) fused at the COOH terminus of GFP (schematically shown in Fig. 5 B) shows the same patterns as the full length DNA ligase I chimera with GFP indicate that the targeting sequence works as a ‘module' and can function in the context of unrelated proteins and irrespective of its location in the fusion construct. Furthermore, they allow direct visualization of subnuclear replication foci in living mammalian cells. The very characteristic redistribution of the GFP-ligase fusion proteins during the cell cycle makes these constructs a unique S phase marker for studies in living cells.
Discussion
In this study, we describe the subcellular localization of DNA ligase I and analyze the basis for its cell cycle-dependent redistribution. We show that DNA ligase I is excluded from condensed chromosomes in mitotic cells and is rapidly imported into the nucleus upon formation of the nuclear envelope (Fig. 2). In interphase cells, DNA ligase I is homogenously distributed throughout the nucleoplasm except in S phase cells, when it is localized at subnuclear replication foci (Fig. 2). We mapped a bipartite targeting sequence, which is necessary and sufficient for this S phase-dependent redistribution (Fig. 3 A).
It should be mentioned that these results clearly differ from a previous report (34). That report claimed the identification of a targeting sequence solely based on a single deletion mutant not detected at replication foci, which, as a negative result, is by definition, inconclusive. Deletions often alter the overall folding and stability of proteins and can unspecifically affect functions that are far apart in the primary protein structure. For these reasons, a subcellular targeting sequence is usually defined as a protein sequence that (a) is necessary and (b) sufficient for subcellular localization; (c) works position independently; (d) is separated from the catalytic domain, and (e) can target heterologous proteins to the respective subcellular domain. In this study we mapped the targeting sequence of DNA ligase I and demonstrated that this sequence by itself is sufficient to recruit different heterologous proteins to replication foci in S phase cells. The targeting sequence functions as a module independently of its location in the fusion protein (see Figs. 3 and 5) and deletion of this sequence in the context of the entire DNA ligase I protein leaves the catalytic properties untouched (20) but abrogates its S phase-dependent association with replication foci. We show that the DNA ligase I targeting sequence is conserved between the human and mouse enzymes, and the human sequence also targets fusion proteins to replication foci in mouse cells (Fig. 3 B).
In lower eukaryotic homologues (e.g., yeast or Drosophila DNA ligases I) the NH2-terminal domain is also dispensable for enzyme activity in vitro (1, 45, 55). However, these domains are clearly shorter and exhibit no similarity among each other or to the higher eukaryotic counterparts (Fig. 4 B, and data not shown). Furthermore, a truncated human DNA ligase I protein, with the entire NH2-terminal domain deleted, is able to rescue yeast DNA ligase I mutant strains (2) but cannot rescue homozygous null DNA ligase I mutant mouse cells (42). The latter indicates that the NH2-terminal domain of human DNA ligase I has essential functions in mammalian cells that are not required in lower eukaryotes. The identification and mapping of the targeting sequence (Fig. 3) now assigns a function to this domain in vivo and provides a possible explanation for those presumably conflicting in vitro and in vivo results.
Interestingly, the NH2-terminal regulatory domain is also not conserved in the recently cloned mammalian DNA ligases III and IV, despite a high degree of homology throughout the catalytic domains (8, 60). These two enzymes are able to substitute for DNA ligase I in in vitro DNA repair assays (22, 49) but not in replication assays (31, 59). Altogether, these results suggest a function of the NH2-terminal domain of DNA ligase I in DNA replication rather than repair, which fits well with our mapping of the targeting to replication foci in this domain. A comparison with the previously identified targeting sequence of the DNA MTase showed no discernible similarities (Fig. 4 A), which suggests that these sequences interface with different components of the replication factory as illustrated in Fig. 6. In this context it is interesting that biochemical fractionation experiments have shown that DNA ligase I is present in megadalton, multiprotein complexes with multiple catalytic activities associated with DNA replication (27, 28, 32, 39, 62). The identification of the targeting sequence now renders possible a directed search for interacting factors and the elucidation of the molecular architecture of these replication factories.
Figure 6.

Model of a DNA replication and methylation factory. The DNA double strand (thick lines) is spooled through a multiprotein complex, often referred to as “replication factory,” which is attached to the nuclear matrix (17). The newly synthesized strands are represented by thin lines, and interruptions represent Okazaki fragments of the lagging strand. The numerous participating enzymes in these factories (only two are depicted) are organized in an assembly line-like fashion, which ensures that, upon passage through these factories, DNA is fully replicated, all Okazaki fragments are ligated, and all methyl groups (−CH3) are added to the new strand at hemimethylated sites. This organization is in part achieved by the tethering of DNA ligase I and DNA MTase to the respective sites of these factories via the targeting sequences mapped in these enzymes (see Fig. 3 and reference 24). Targeting sequences are depicted as separate domains since, in both cases (DNA ligase I and DNA MTase), they are protease-sensitive domains, dispensable for enzyme activity in vitro and they are necessary and sufficient for localization at replication foci.
Similar targeting principles seem to apply also to other functional domains of the mammalian nucleus (for review see 23). Thus, targeting sequences were identified in the Drosophila splicing regulators su(wa) and tra, which are concentrated at distinct nuclear foci called “speckled compartment” (26). Further, the nucleolar localization of the retroviral proteins HTLV-1 Rex (51), HIV-1 Tat (11, 13), and HIV-1 Rev (9, 21) was shown to be mediated by nucleolar targeting sequences. Moreover, nucleolar targeting of HIV-1 Rev was shown to be required for in vivo function (9, 21). Finally, the NH2-terminal 102 amino acids of p80-coilin, a protein that localizes to nuclear coiled bodies, are necessary and sufficient for targeting to this organelle (63). All of these different sequences have one thing in common: they are necessary and sufficient for targeting to subnuclear domains; however, they do not share similar protein motifs. On the contrary, they are as diverse as their function and the domain to which they target.
The vast majority of studies on nuclear organization have been done on fixed and stained cells, with the caveat of possible artefacts (for review see 10). Several efforts have been undertaken to use physiological buffers and unfixed cells to study subnuclear replication sites. Nevertheless, these studies were performed using permeabilized cells that underwent still extensive manipulation before these subnuclear compartments were visualized (16). Until now, localization of proteins at replication sites had not yet been demonstrated in live cells. The localization of DNA ligase I at replication foci (Fig. 2), the identification of a bipartite targeting sequence in its NH2-terminal regulatory domain, which mediates cell cycle-specific association (Fig. 3), and the availability of the GFP allowed us to address this issue directly. Chimeric proteins comprising GFP and DNA ligase I (full length or NH2-terminal domain) allow the visualization of these subnuclear structures in living cells and show patterns indistinguishable from fixed and stained cells (Fig. 5). These findings enable direct visualization of S phase in living cells, which should be very useful for cell cycle studies.
In summary, we describe the subcellular distribution of DNA ligase I during the cell cycle and map the sequence responsible for its association with replication foci in S phase cells. We showed that the targeting sequence works as an independent module and can direct unrelated proteins (like β-gal from Escherichia coli and GFP from Aequorea victoria) to sites of DNA replication irrespective of its position in the fusion protein. The targeting sequence works across species (human and mouse) as well as in different cell types.
Altogether, these results suggest that targeting might be a general principle of nuclear organization and might be a means to cope with the growing complexity in higher eukaryotes as evolution progressed. The targeting sequence, by bringing the catalytic domain to the right place at the right time, allows catalysis to occur at higher order kinetics, which meets the needs of a competitive and demanding environment such as the mammalian nucleus. We propose that the efficient coordination of complex processes such as DNA replication, from controlled initiation to ligation of Okazaki fragments and DNA methylation, might be accomplished in the mammalian nucleus by integration into assembly line-like protein factories (Fig. 6), which increase the effective enzyme concentration, specificity, and processivity. This dynamic nuclear architecture thus constitutes a higher order mechanism of regulating enzyme activity in vivo.
Acknowledgments
We are grateful to F.C. Luft and H. Haller for encouragement, support, and critical reading of the manuscript. We are indebted to M. Cochran, C. Clark, and L. Steinberg for their very special support of this work, and we would also like to acknowledge D. Hänlein for untiring help and advice with our computers. We thank G. Vargas for constructing a GFP–DNA ligase fusion.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (Le 721/2-1) and from the Max Delbrück Center for Molecular Medicine.
Abbreviations used in this paper
- β-gal
β-galactosidase
- GFP
green fluorescent protein
- MTase
methyltransferase
- NLS
nuclear localization sequence
- PCNA
proliferating cell nuclear antigen
Footnotes
References
- 1.Banks GR, Barker DG. DNA ligase-AMP adducts: identification of yeast DNA ligase polypeptides. Biochim Biophys Acta. 1985;826:180–185. doi: 10.1016/0167-4781(85)90004-1. [DOI] [PubMed] [Google Scholar]
- 2.Barnes DE, Johnston LH, Kodama K, Tomkinson AE, Lasko DD, Lindahl T. Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1990;87:6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 4.Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 5.Bravo R, MacDonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardoso, M.C., and H. Leonhardt. 1995. Immunofluorescence techniques in cell cycle studies. In Cell Cycle: Materials and Methods. M. Pagano, editor. Springer-Verlag, Heidelberg. 15–28.
- 7.Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Tomkinson AE, Ramos W, Mackey ZB, Danehower S, Walter CA, Schultz RA, Besterman JM, Husain I. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane AW, Perkins A, Rosen CA. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook PR. The nucleoskeleton and the topology of replication. Cell. 1991;66:627–635. doi: 10.1016/0092-8674(91)90109-c. [DOI] [PubMed] [Google Scholar]
- 11.Dang CV, Lee WM. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 12.Dingwall C, Laskey RA. Nuclear targeting sequences-a consensus? . Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 13.Endo S, Kubota S, Siomi H, Adachi A, Oroszlan S, Maki M, Hatanaka M. A region of basic amino-acid cluster in HIV-1 Tat protein is essential for transacting activity and nucleolar localization. Virus Genes. 1989;3:99–110. doi: 10.1007/BF00125123. [DOI] [PubMed] [Google Scholar]
- 14.Fox MH, Arndt-Jovin DJ, Jovin TM, Baumann PH, Robert-Nicoud M. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J Cell Sci. 1991;99:247–253. doi: 10.1242/jcs.99.2.247. [DOI] [PubMed] [Google Scholar]
- 15.Graham FL, Van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 16.Hassan AB, Cook PR. Visualization of replication sites in unfixed human cells. J Cell Sci. 1993;105:541–550. doi: 10.1242/jcs.105.2.541. [DOI] [PubMed] [Google Scholar]
- 17.Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 18.Huberman JA, Tsai A, Deich RA. DNA replication sites within nuclei of mammalian cells. Nature (Lond) 1973;241:32–36. doi: 10.1038/241032a0. [DOI] [PubMed] [Google Scholar]
- 19.Johnston LH, Nasmyth KA. Saccharomyces cerevisiaecell cycle mutant cdc9 is defective in DNA ligase. Nature (Lond) 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 20.Kodama K, Barnes DE, Lindahl T. In vitro mutagenesis and functional expression in Escherichia coliof a cDNA encoding the catalytic domain of human DNA ligase I. Nucleic Acids Res. 1991;19:6093–6099. doi: 10.1093/nar/19.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubota S, Siomi H, Satoh T, Endo S, Maki M, Hatanaka M. Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein. Biochem Biophys Res Commun. 1989;162:963–970. doi: 10.1016/0006-291x(89)90767-5. [DOI] [PubMed] [Google Scholar]
- 22.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO (Eur Mol Biol Organ) J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 23.Leonhardt H, Cardoso MC. Targeting and association of proteins with functional domains in the mammalian nucleus: the insoluble solution. Int Rev Cytol. 1995;162B:303–335. doi: 10.1016/s0074-7696(08)62620-0. [DOI] [PubMed] [Google Scholar]
- 24.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 25.Lepetit D, Thiebaud P, Aoufouchi S, Prigent C, Guesne R, Theze N. The cloning and characterization of a cDNA encoding Xenopus laevisDNA ligase I. Gene (Amst) 1996;172:273–277. doi: 10.1016/0378-1119(96)00175-8. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Cao LG, Wang YL, Baril EF. Further purification and characterization of a multienzyme complex for DNA synthesis in human cells. J Cell Biochem. 1993;53:405–419. doi: 10.1002/jcb.240530418. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Goodchild J, Baril EF. DNA ligase I is associated with the 21 S complex of enzymes for DNA synthesis in HeLa cells. Nucleic Acids Res. 1994;22:632–638. doi: 10.1093/nar/22.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindahl T, Barnes DE. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 30.Lindner W, Robey FA. Automated synthesis and use of N-chloroacetyl-modified peptides for the preparation of synthetic peptide polymers and peptide-protein immunogens. Int J Pept Protein Res. 1987;30:794–800. doi: 10.1111/j.1399-3011.1987.tb03388.x. [DOI] [PubMed] [Google Scholar]
- 31.Mackenney VJ, Barnes DE, Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J Biol Chem. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 32.Malkas LH, Hickey RJ, Li C, Pedersen N, Baril EF. A 21S enzyme complex from HeLa cells that functions in simian virus 40 DNA replication in vitro. Biochemistry. 1990;29:6362–6374. doi: 10.1021/bi00479a004. [DOI] [PubMed] [Google Scholar]
- 33.Matthias P, Müller MM, Schreiber E, Rusconi S, Schaffner W. Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res. 1989;17:6418. doi: 10.1093/nar/17.15.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, Villa A, Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO (Eur Mol Biol Organ) J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgenstern JP, Land H. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 37.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nasmyth KA. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. . Cell. 1977;12:1109–1120. doi: 10.1016/0092-8674(77)90173-8. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi H, Prem veer Reddy G, Pardee AB. Rapid incorporation of label from ribonucleoside disphosphates into DNA by a cell-free high molecular weight fraction from animal cell nuclei. Cell. 1983;32:443–451. doi: 10.1016/0092-8674(83)90464-6. [DOI] [PubMed] [Google Scholar]
- 40.O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker BA, Stark GR. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus and viral DNA. J Virol. 1979;31:360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrini JH, Xiao Y, Weaver DT. DNA ligase I mediates essential functions in mammalian cells. Mol Cell Biol. 1995;15:4303–4308. doi: 10.1128/mcb.15.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prigent C, Lasko DD, Kodama K, Woodgett JR, Lindahl T. Activation of mammalian DNA ligase I through phosphorylation by casein kinase II. EMBO (Eur Mol Biol Organ) J. 1992;11:2925–2933. doi: 10.1002/j.1460-2075.1992.tb05362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prigent C, Satoh MS, Daly G, Barnes DE, Lindahl T. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol Cell Biol. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabin BA, Hawley RS, Chase JW. DNA ligase from Drosophila melanogasterembryos. Purification and physical characterization. J Biol Chem. 1986;261:10637–10645. [PubMed] [Google Scholar]
- 46.Savini E, Biamonti G, Ciarrocchi G, Montecucco A. Cloning and sequence analysis of a cDNA coding for the murine DNA ligase I enzyme. Gene. 1994;144:253–257. doi: 10.1016/0378-1119(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 47.Sawin KE, Mitchison TJ, Wordeman LG. Evidence for kinesin-related proteins in the mitotic apparatus using peptide antibodies. J Cell Sci. 1992;101:303–313. doi: 10.1242/jcs.101.2.303. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt-Zachmann MS, Dargemont C, Kuhn LC, Nigg EA. Nuclear export of proteins: the role of nuclear retention. Cell. 1993;74:493–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- 49.Shivji MK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase ε in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 50.Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 51.Siomi H, Shida H, Nam SH, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 52.Sobczak-Thepot J, Harper F, Florentin Y, Zindy F, Brechot C, Puvion E. Localization of cyclin A at the sites of cellular DNA replication. Exp Cell Res. 1993;206:43–48. doi: 10.1006/excr.1993.1118. [DOI] [PubMed] [Google Scholar]
- 53.Somia NV, Jessop JK, Melton DW. Phenotypic correction of a human cell line (46BR) with aberrant DNA ligase I activity. Mutat Res. 1993;294:51–58. doi: 10.1016/0921-8777(93)90057-n. [DOI] [PubMed] [Google Scholar]
- 54.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi M, Senshu M. Two distinct DNA ligases from Drosophila melanogasterembryos. FEBS Lett. 1987;213:345–352. doi: 10.1016/0014-5793(87)81520-x. [DOI] [PubMed] [Google Scholar]
- 56.Teo IA, Broughton BC, Day RS, James MR, Karran P, Mayne LV, Lehmann AR. A biochemical defect in the repair of alkylated DNA in cells from an immunodeficient patient (46BR) Carcinogenesis. 1983;4:559–564. doi: 10.1093/carcin/4.5.559. [DOI] [PubMed] [Google Scholar]
- 57.Tomkinson AE, Lasko DD, Daly G, Lindahl T. Mammalian DNA ligases. Catalytic domain and size of DNA ligase I. J Biol Chem. 1990;265:12611–12617. [PubMed] [Google Scholar]
- 58.van Dierendonck JH, Keyzer R, van de Velde CJ, Cornelisse CJ. Subdivision of S-phase by analysis of nuclear 5-bromodeoxyuridine staining patterns. Cytometry. 1989;10:143–150. doi: 10.1002/cyto.990100205. [DOI] [PubMed] [Google Scholar]
- 59.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 60.Wei Y F, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, Shell BK, Nash RA, Schär P, et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 62.Wu Y, Hickey R, Lawlor K, Wills P, Yu F, Ozer H, Starr R, Quan JY, Lee M, Malkas L. A 17S multiprotein form of murine cell DNA polymerase mediates polyomavirus DNA replication in vitro. J Cell Biochem. 1994;54:32–46. doi: 10.1002/jcb.240540105. [DOI] [PubMed] [Google Scholar]
- 63.Wu ZA, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell. 1994;5:1119–1127. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]