Abstract
The desmosome is a highly organized plasma membrane domain that couples intermediate filaments to the plasma membrane at regions of cell–cell adhesion. Desmosomes contain two classes of cadherins, desmogleins, and desmocollins, that bind to the cytoplasmic protein plakoglobin. Desmoplakin is a desmosomal component that plays a critical role in linking intermediate filament networks to the desmosomal plaque, and the amino-terminal domain of desmoplakin targets desmoplakin to the desmosome. However, the desmosomal protein(s) that bind the amino-terminal domain of desmoplakin have not been identified. To determine if the desmosomal cadherins and plakoglobin interact with the amino-terminal domain of desmoplakin, these proteins were co-expressed in L-cell fibroblasts, cells that do not normally express desmosomal components. When expressed in L-cells, the desmosomal cadherins and plakoglobin exhibited a diffuse distribution. However, in the presence of an amino-terminal desmoplakin polypeptide (DP-NTP), the desmosomal cadherins and plakoglobin were observed in punctate clusters that also contained DP-NTP. In addition, plakoglobin and DP-NTP were recruited to cell–cell interfaces in L-cells co-expressing a chimeric cadherin with the E-cadherin extracellular domain and the desmoglein-1 cytoplasmic domain, and these cells formed structures that were ultrastructurally similar to the outer plaque of the desmosome. In transient expression experiments in COS cells, the recruitment of DP-NTP to cell borders by the chimera required co-expression of plakoglobin. Plakoglobin and DP-NTP co-immunoprecipitated when extracted from L-cells, and yeast two hybrid analysis indicated that DP-NTP binds directly to plakoglobin but not Dsg1. These results identify a role for desmoplakin in organizing the desmosomal cadherin–plakoglobin complex and provide new insights into the hierarchy of protein interactions that occur in the desmosomal plaque.
Desmosomes are highly organized adhesive intercellular junctions that couple intermediate filaments to the cell surface at sites of cell–cell adhesion (Farquhar and Palade, 1963; Staehelin, 1974; Schwarz et al., 1990; Garrod, 1993; Collins and Garrod, 1994; Cowin and Burke, 1996; Kowalczyk and Green, 1996). Desmosomes are prominent in tissues that experience mechanical stress, such as heart and epidermis, and the disruption of desmosomes or the intermediate filament system in these organs has devastating effects on tissue integrity (Steinert and Bale, 1993; Coulombe and Fuchs, 1994; Fuchs, 1994; McLean and Lane, 1995; Stanley, 1995; Bierkamp et al., 1996; Ruiz et al., 1996). Desmosomes are highly insoluble structures that can withstand harsh denaturing conditions (Skerrow and Matoltsy, 1974; Gorbsky and Steinberg, 1981; Jones et al., 1988; Schwarz et al., 1990). This property of desmosomes facilitated early identification of desmosomal components but has impaired subsequent biochemical analysis of the protein complexes that form between desmosomal components. Ultrastructurally, desmosomes contain a core region that includes the plasma membranes of adjacent cells and a cytoplasmic plaque that anchors intermediate filaments to the plasma membrane. The plaque can be further divided into an outer dense plaque subjacent to the plasma membrane and an inner dense plaque through which intermediate filaments appear to loop.
Molecular genetic analysis has revealed that the desmosomal glycoproteins, the desmogleins and desmocollins, are members of the cadherin family of cell–cell adhesion molecules (for review see Buxton et al., 1993, 1994; Cowin and Mechanic, 1994; Kowalczyk et al., 1996). The classical cadherins, such as E-cadherin, mediate calcium-dependent, homophilic cell–cell adhesion (Nagafuchi et al., 1987). The mechanism by which the desmosomal cadherins mediate cell–cell adhesion remains elusive (Amagai et al., 1994; Chidgey et al., 1996; Kowalczyk et al., 1996), although heterophilic interactions have recently been detected between desmogleins and desmocollins (Chitaev and Troyanovsky, 1997). Both classes of the desmosomal cadherins associate with the cytoplasmic plaque protein plakoglobin (Kowalczyk et al., 1994; Mathur et al., 1994; Roh and Stanley, 1995b ; Troyanovsky et al., 1994), which is part of a growing family of proteins that share a repeated motif first identified in the Drosophila protein Armadillo (Peifer and Wieschaus, 1990). This multigene family also includes the desmosomal proteins band 6/plakophilin 1, plakophilin 2a and 2b, and p0071, which are now considered to comprise a subclass of the armadillo family of proteins (Hatzfeld et al., 1994; Heid et al., 1994; Schmidt et al., 1994; Hatzfeld and Nachtsheim, 1996; Mertens et al., 1996).
The most abundant desmosomal plaque protein is desmoplakin, which is predicted to be a homodimer containing two globular end domains joined by a central α-helical coiled-coil rod domain (O'Keefe et al., 1989; Green et al., 1990; Virata et al., 1992). Previous studies have demonstrated that the carboxyl-terminal domain of desmoplakin interacts with intermediate filaments (Stappenbeck and Green, 1992; Stappenbeck et al., 1993; Kouklis et al., 1994; Meng et al., 1997), and the amino-terminal domain of desmoplakin is required for desmoplakin localization to the desmosomal plaque (Stappenbeck et al., 1993). Direct evidence supporting a role for desmoplakin in intermediate filament attachment to desmosomes was provided recently when expression of an amino-terminal polypeptide of desmoplakin was found to displace endogenous desmoplakin from cell borders and disrupt intermediate filament attachment to the cell surface in A431 epithelial cell lines (Bornslaeger et al., 1996).
The classical cadherins, such as E-cadherin, bind directly to both β-catenin and plakoglobin (Aberle et al., 1994; Jou et al., 1995; for review see Cowin and Burke, 1996). β-Catenin is also an armadillo family member (McCrea et al., 1991; Peifer et al., 1992), and both plakoglobin and β-catenin bind directly to α-catenin (Aberle et al., 1994, 1996; Jou et al., 1995; Sacco et al., 1995; Obama and Ozawa, 1997). α-Catenin is a vinculin homologue (Nagafuchi et al., 1991) and associates with both α-actinin and actin (Knudson et al., 1995; Rimm et al., 1995; Nieset et al., 1997). Through interactions with β- and α-catenin, E-cadherin is coupled indirectly to the actin cytoskeleton, and this linkage is required for the adhesive activity of E-cadherin (Ozawa et al., 1990; Shimoyama et al., 1992). In addition, E-cadherin association with plakoglobin appears to be required for assembly of desmosomes (Lewis et al., 1997), underscoring the importance of E-cadherin in the overall program of intercellular junction assembly. However, the hierarchy of molecular interactions that couple the desmosomal cadherins to the intermediate filament cytoskeleton is largely unknown, although the desmocollin cytoplasmic domain appears to play an important role in recruiting components of the desmosomal plaque (Troyanovsky et al., 1993, 1994). Since desmosomal cadherins form complexes with plakoglobin and because the amino-terminal domain of desmoplakin is required for desmoplakin localization at desmosomes, we hypothesized that the amino-terminal domain of desmoplakin interacts with the desmosomal cadherin– plakoglobin complex.
In previous studies, we used L-cell fibroblasts to characterize plakoglobin interactions with the cytoplasmic domains of the desmosomal cadherins and found that the desmosomal cadherins regulate plakoglobin metabolic stability (Kowalczyk et al., 1994) but do not mediate homophilic adhesion (Kowalczyk et al., 1996). To test the ability of the desmoplakin amino-terminal domain to interact with the desmosomal cadherin–plakoglobin complex, we established a series of L-cell lines expressing the desmosomal cadherins in the presence or absence of a desmoplakin amino-terminal polypeptide (DP-NTP).1 The results indicate that one important function of the desmoplakin amino-terminal domain is to cluster desmosomal cadherin–plakoglobin complexes. In addition, DP-NTP and plakoglobin were found to form complexes that could be co-immunoprecipitated from L-cell lysates. Using the yeast two hybrid system, DP-NTP was found to bind directly to plakoglobin but not Dsg1. These data suggest that plakoglobin couples the amino-terminal domain of desmoplakin to the desmosomal cadherins and that desmoplakin plays an important role in organizing the desmosomal cadherin–plakoglobin complex into discrete plasma membrane domains.
Materials and Methods
Cell Culture
L-cells were cultured in Dulbecco's minimal essential medium (DMEM), 10% fetal bovine serum, and penicillin/streptomycin. To establish cell lines, cells were transfected with various cDNA constructs by calcium phosphate precipitation as described previously (Kowalczyk et al., 1994, 1996). The cDNA construct encoding plakoglobin also included a neomycin resistance gene. The antibiotic G418 (GIBCO BRL, Grand Island, NY) was used at 400 μg/ml (active concentration) to select transfected cells. For cells that were not cotransfected with plakoglobin, a plasmid (pSV2neo) that encoded the neomycin resistance gene, was used as the selection marker. Colonies resistant to G418 were isolated and protein expression was assessed by immunoblot and immunofluorescence. In each case, at least three independently isolated clones expressing the proteins of interest were used for futher characterization. For transient transfections, COS cells were cultured in DMEM, 10% fetal bovine serum, and penicillin/streptomycin and transfected using the calcium phosphate precipitation method as described previously (Stappenbeck and Green, 1992; Kowalczyk et al., 1995). Cells were assayed by immunofluorescence 48 h after transfection. All media and serum were obtained from GIBCO BRL, and tissue culture plasticware was purchased from Becton Dickinson (Lincoln Park, NJ).
cDNA Constructs
Cadherins and Chimeric Cadherins.
cDNA constructs encoding full length human desmoglein-1 (Dsg1) and desmocollin-2a (Dsc2a) were generated as described previously (Kowalczyk et al., 1994, 1996). A chimeric cadherin comprising the extracellular domain of Dsg1 and the intracellular domain of mouse E-cadherin (Dsg1Ecad) was constructed as described previously (Kowalczyk et al., 1994; see Fig. 1 for structure). A second chimera consisting of the extracellular domain of E-cadherin and the cytoplasmic domain of Dsg1 was also generated (EcadDsg1). This chimera comprised the cDNA sequences encoding the extracellular domain of mouse E-cadherin (nucleotides 68–2,190; Nagafuchi et al., 1987) and the cytoplasmic domain of human Dsg1 (nucleotides 1,585–3,072; Nilles et al., 1991). The fusion between E-cadherin and Dsg1 sequences occurs within the transmembrane spanning domain and an 11-amino acid epitope tag from c-myc was added to the carboxyl terminus of the chimera. The E-cadherin portion of the chimera was generated by PCR using the 5′ E-cadherin primer LN37 (5′-TGATTTAAAGTCGACTCCACCATGGGAGCCCGGTGC-3′ to generate a SalI site at the 5′ end of the E-cadherin cDNA and the 3′ E-cadherin primer LN45 (5′-TATAGGTACCGCCAATCCTGCTGCCAC-3′), which generates a 3′ KpnI site within sequences encoding the central region of the membrane spanning domain. The Dsg1 cytoplasmic domain was also generated using PCR. The 5′ Dsg1 primer (LN46: 5′-ATGCGGTACCTCTCATCATGGGATTC-3′) was used to generate a 5′ Kpn I site within the sequences encoding the Dsg1 transmembrane spanning domain in frame with the Kpn I site from the 3′ E-cadherin primer (LN45). A 3′ Dsg1 primer (LN36 GCCAAGCTTCTACAAGTCCTCTTCAGAAATGAGCTTTTGCTCCACCTTGC T A T A T- TGCAC) was used to produce a 3′ HindIII restriction site and the carboxyl-terminal c-myc epitope tag. The PCR products were subcloned into pBluescript (Strategene, La Jolla, CA) and PCR-generated regions were replaced with cloned DNA or sequenced to verify that errors were not introduced into the coding sequence. The KpnI sites were used to ligate the cDNA encoding the extracellular domain of E-cadherin to the cytoplasmic domain of Dsg1. cDNAs encoding Dsg1, Dsc2a, and the Dsg1Ecad chimeras were each subcloned into the RC4B expression vector (Evans and Scarpulla, 1988) for transfection into eukaryotic cells. The EcadDsg1 chimera was subcloned into the mammalian expression vector LK-444, which uses the β-actin promoter as described previously (Kowalczyk et al., 1994).
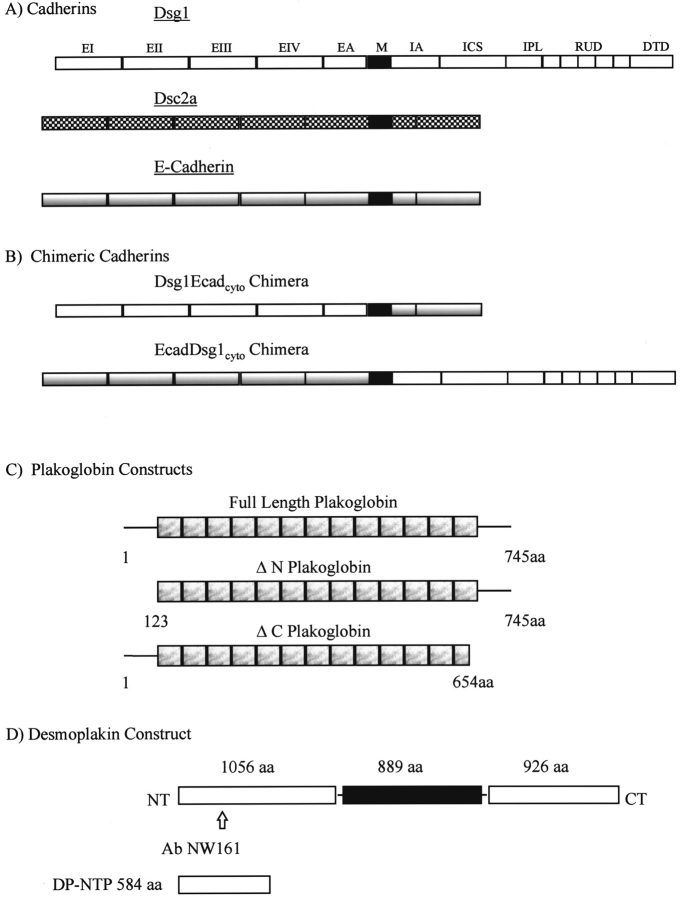
Figure 1.
Schematic representation of cDNA constructs. (A) Full length cDNAs encoding human Dsg1 (Nilles et al., 1991) or Dsc2a (Parker et al., 1991; Kowalczyk et al., 1994) were expressed. Nomenclature for domain structure of the cadherins is adopted from Koch et al. (1990). (B) Two chimeric cadherins were also constructed. The first chimera encodes the extracellular domain of Dsg1 and the cytoplasmic domain of mouse E-cadherin (Dsg1Ecad). The inverse chimera comprises the mouse E-cadherin extracellular domain and the Dsg1 cytoplasmic domain (EcadDsg1). (C) Plakoglobin cDNAs encoding full length human plakoglobin or deletion mutants lacking the amino- or carboxyl-terminal domains were constructed. The hatched boxes represent the central repeated unit domains homologous to the armadillo repeats (Peifer et al., 1994). (D) A cDNA encoding the first 584 amino acids of the desmoplakin amino-terminal domain (DP-NTP) was constructed as previously described (Bornslaeger et al., 1996).
Plakoglobin and Plakoglobin Deletion Mutants.
A full length plakoglobin cDNA was isolated and subcloned into the mammalian expression vector LK-444. A deletion of plakoglobin lacking the amino-terminal domain (PgΔN) was generated by PCR using primers LN123 (5′-ACGCGTCGACCCACGATGCTCAAGTCGGCCATTG-3′) and LN52 (5′-CTGCAGACCACATGCTGGCCGTGGAGCAAAAGCTCATTTCTGAAGAGG- ACTTGTAGAAGCTTGGCC-3′). The resuling PCR product of PgΔN (nucleotides 486–2,354; Franke et al., 1989) contains a SalI restriction site and Kozak consensus sequence at the 5′ end and an 11-amino acid c-myc epitope tag and a HindIII restriction site at the 3′ end. Using the engineered restriction sites, the PgΔN PCR product was inserted into the SalI/ HindIII site in pBluescript and subcloned into the SalI/HindIII sites of the mammalian expression vector LK-444.
A deletion of plakoglobin lacking the carboxyl-terminal domain and a portion of armadillo repeat number 13 (PgΔC) was generated by PCR using primers LN51 (5′-ACGTGTCGACCCACGATGGAGGTGATGAACCTGA-3′) and LN120 (5′-TGTTCCGCATCTCCGAGGTGGAGCAAAAGCTCATTTCTGAAGAGGACTTGTAGAAGCTTGCGCA-3′). The resulting PCR product of PgΔC (nucleotides 120–2,081) contains a SalI restriction site and Kozak consensus sequence at the 5′ end and an 11-amino acid c-myc epitope tag and a HindIII restriction site at the 3′ end. Using the engineered restriction sites, c-myc–tagged PgΔC was subcloned into the SalI/HindIII site in pBluescript and then into the mammalian expression vector LK-444. For each of the plakoglobin deletion constructs, PCR-generated regions were replaced with original cDNA sequences and sequenced to verify that no errors were introduced.
Desmoplakin Amino-terminal Construct.
A cDNA construct encoding a portion of the desmoplakin amino-terminal domain, termed DP-NTP, was constructed and subcloned into a mammalian expression vector using the CMV promoter (Bornslaeger et al., 1996). As previously reported (Bornslaeger et al., 1996), the protein product of the 2.1-kb cDNA is a 70-kD polypeptide, ∼24 kD smaller than predicted from the cDNA. DNA sequence analysis indicated the presence of a mutation that arose spontaneously in a precursor plasmid during bacterial replication, which terminates translation at amino acid number 584. Translation read through allows the accumulation of trace amounts of a larger polypeptide (∼94 kD), which is present at levels too low to be detected by the NW161 antibody (Bornslaeger et al., 1996).
Immunofluorescence and Electron Microscopy
The distribution of desmosomal proteins in L-cell lines and transiently transfected COS cells was analyzed by immunofluorescence. For most experiments, cells were rinsed in PBS and fixed in −20°C methanol. In some experiments, cells were washed in PBS containing calcium and magnesium (PBS+), incubated in 0.01% saponin for 5 min, rinsed gently in PBS+, and then fixed in −20°C methanol. The following antibodies were used to detect each protein: DP-NTP was detected using polyclonal antibody NW161 directed against an amino-terminal desmoplakin polypeptide (Bornslaeger et al., 1996); Dsg1 was monitored using a human pemphigus foliaceus antibody # 982 (Kowalczyk et al., 1995); Dsc2a was detected using monoclonal antibody 7G6 (Kowalczyk et al., 1994); plakoglobin was monitored using monoclonal antibody 11E4 or a polyclonal antibody (a gift from Dr. J. Papkoff, Megabios Corp. [Burlingame, CA]) directed against a carboxyl-terminal plakoglobin peptide (Hinck et al., 1994; Kowalczyk et al., 1994). The E-cadherin extracellular domain of the EcadDsg1 chimera was detected using rat monoclonal antibodies directed against the E-cadherin extracellular domain (anti-uvomorulin antibody DECMA-1 from Sigma Chemical Co. [St. Louis, MO]) or mAb ECCD-2, a gift from Dr. M. Takeichi, Kyoto University [Kyoto, Japan]). For electron microscopy, cells were grown to confluence on tissue culture grade plasticware or on glass coverslips and processed for conventional electron microscopy as described previously (Green et al., 1991).
Immunoprecipitation and Immunoblot Analysis
Immunoprecipitation was carried out as described (Kowalczyk et al., 1994) with the following changes. L-cells were scraped into Tris-buffered saline containing 0.5% Triton X-100, vortexed, and subjected to centrifugation at 14,000 g. Antibodies directed against Dsg1 (pemphigus serum # 982), the desmoplakin amino terminus (NW161), or preimmune serum from antibody NW161 were incubated with the cell lysate for 2 h at 4°C. Gamma bind plus sepharose beads (Pharmacia Fine Chemicals, Piscataway, NJ) were then added for 2 h at 4°C, immune complexes were captured by centrifugation, and the beads were washed 5 times in Tris-buffered saline containing 0.5% Triton X-100 for 10 min under gentle rotation at 4°C. Immune complexes were released by incubation in reducing SDS-PAGE sample buffer and analyzed by immunoblot using Enhanced Chemiluminescence (Amersham Intl, Arlington Heights, IL). Plakoglobin was detected using mAb 11E4, which is directed against the amino-terminal domain of plakoglobin, or a rabbit polyclonal antibody directed against the carboxyl-terminal domain (Hinck et al., 1994; Kowalczyk et al., 1994).
Yeast Two Hybrid Constructs and Assays
Yeast two hybrid vectors encoding the GAL4 DNA binding (pAS-CYH2; Harper et al., 1993) or transcription activation domain (pACTII; Bai and Elledge, 1995) were generously provided by Dr. Stephen Elledge (Baylor College of Medicine, Houston, TX). The amino-terminal deletion construct of human plakogobin (PgΔN) described above was subcloned from pBluescript into the activation domain construct pACTII. The cDNA in pBluescript was restriction digested with SalI and treated with Klenow fragment of DNA polymerase I to create a blunt 5′ end. The DNA was subsequently digested with EcoRI to remove the PgΔN insert and subcloned into the SmaI and EcoRI sites of pACTII to generate a PgΔN cDNA, which was inserted in frame with upstream sequences encoding the GAL4 transcription activation domain (p515). A construct comprising the cDNA sequences encoding the entire cytoplasmic tail of human Dsg1 in the pAS1-CYH2 DNA binding domain vector was a generous gift from Dr. Michael Klymkowsky (University of Colorado, Boulder, CO). The cDNA sequences encoding Dsg1 were removed from pAS1-CYH2 by digestion with EcoRI and BamHI and subcloned into the same sites of the Path 1 vector (p47), which contains a SalI restriction site 3′ of the BAM HI site. The Dsg1 sequences were then removed with EcoRI and SalI and subcloned into the EcoRI and XhoI sites of pACTII to generate a construct encoding nucleotides 1,633–3,072 of human Dsg1 in frame with upstream sequences encoding the GAL4 activation domain (p504). The cDNA encoding DP-NTP in pBluescript (p552) was restriction digested with BamHI and SalI and subcloned into the BamHI and XhoI sites of pACTII (p597). This construct was subsequently digested with NcoI and SmaI to subclone the DP-NTP sequences into the same restriction sites of the pAS1-CYH2 vector (p601). The first 584 amino acids of human desmoplakin are encoded in both the pACTII and pAS1-CYH2 constructs, similar to the DP-NTP expression vectors described above.
To assay interactions between proteins, yeast (strain HF7c) were transformed with the plasmids of interest and grown on synthetic defined media lacking either tryptophan (SD−trp) for the pAS1-CYH2 vector or leucine (SD−leu) for the pACTII vector to select for transformed clones. Transformations and β-galactosidase assays were performed according to methods published in the Matchmaker™ Two Hybrid product protocol (Clontech Laboratories Inc., Palo Alto, CA). Briefly, isolated colonies were transformed with 5 to 10 μg plasmid DNA using a standard lithium acetate/ polyethylene glycol method. Interactions between proteins were tested by streaking transformed colonies onto Whatman filter membranes and assaying β-galactosidase activity using the substrate X-Gal (5-Bromo- 4-chloro-3-indolyl β-d-galactopyranoside; Sigma Chemical Co.). As a second reporter for interactions, colonies were also tested for growth on SD-leu-trp-his in the presence of 20 mM 3-aminotriazole (Sigma Chemical Co.). Materials for base media and agar were obtained from DIFCO Laboratories (Detroit, MI), and materials for synthetic defined media were purchased from Clontech Laboratories Inc.
Results
The Desmoplakin Amino-terminal Domain Clusters Desmosomal Cadherin–Plakoglobin Complexes
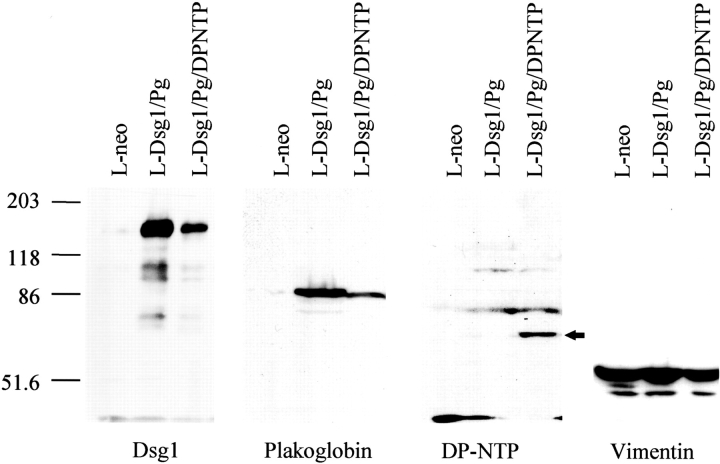
The amino-terminal domain of desmoplakin was previously demonstrated to target desmoplakin to the desmosomal plaque (Stappenbeck et al., 1993; Bornslaeger et al., 1996). We hypothesized that the amino-terminal region of desmoplakin may interact with the desmosomal cadherin– plakoglobin complex, thereby coupling the desmosomal cadherins to the intermediate filament cytoskeleton. To address this question, a series of stable L-cell fibroblast cell lines expressing various desmosomal molecules was established to reconstitute complexes of specific desmosomal components. A schematic diagram depicting the constructs used in this study is shown in Fig. 1. L-cell lines expressing Dsg1 and plakoglobin in the presence or absence of DP-NTP were generated, and the expression of each protein was monitored by immunoblot (Fig. 2). An immunoblot for vimentin is also shown to demonstrate that similar levels of protein were loaded in each lane. Parental L-cells (not shown) and neomycin-resistant L-cells do not express detectable levels of desmosomal components (Fig. 2), with the exception of plakoglobin, which is expressed at extremely low levels (Kowalczyk et al., 1994, 1996).
Figure 2.
Immunoblot analysis of Dsg1, plakoglobin, and DP-NTP expression in stable L-cell lines. L-cell lines expressing Dsg1 and plakoglobin (L-Dsg1/Pg), Dsg1, plakoglobin and DP-NTP (L-Dsg1/Pg/DP-NTP), or the neomycin resistance marker (L-neo) were analyzed by immunoblot using antibodies directed against Dsg1 (pemphigus foliaceus sera # 982; Kowalczyk et al., 1994, 1995), plakoglobin (monoclonal antibody 11E4; Kowalczyk et al., 1994), or the amino-terminal domain of human desmoplakin (NW161; Bornslaeger et al., 1996). The arrow in the DP-NTP blot identifies the 70-kD DP-NTP polypeptide; two higher molecular weight bands are detected nonspecifically. A rabbit polyclonal antibody directed against vimentin (ICN Pharmaceuticals, Inc. [Costa Mesa, CA]) was used to demonstrate that similar amounts of total protein were loaded in each lane. Note that plakoglobin in the L-Dsg1/Pg line contains a carboxyl-terminal c-myc epitope tag and migrates more slowly than untagged plakoglobin in the L-Dsg1/Pg/DP-NTP cell line.
The distribution of exogenously expressed desmosomal proteins in the L-cell lines was monitored by dual label indirect immunofluorescence (Fig. 3). In L-cells expressing Dsg1 and plakoglobin, the proteins colocalized and staining was largely uniform along the plasma membrane (Fig. 3, A and B; see also Kowalczyk et al., 1994). No endogenous desmoplakin staining was detected in these cells (Fig. 3 D). In contrast, in L-cell lines co-expressing DP-NTP, both Dsg1 (Fig. 3 E) and plakoglobin (Fig. 3 F) were present in punctate clusters. In addition, DP-NTP (Fig. 3 G) colocalized with plakoglobin (Fig. 3 H), suggesting that all three components were present in a single complex. It is important to note that the clustering of desmosomal cadherins and plakoglobin was observed only in the presence of DP-NTP. Out of a total of 84 L-cell lines expressing Dsg1 and plakoglobin that were generated, clustering was not observed in any of these cell lines (see also Kowalczyk et al., 1996). In contrast, 16 independent clones of L-cell lines expressing the desmoplakin polypeptide were generated, and clustering was observed in all 16 clones. Furthermore, in the representative Dsg1/Pg/DP-NTP cell line shown here, punctate Dsg1 staining was observed in 71 out of 111 cells. This is in comparison to 0 out of 133 cells for the Dsg1/Pg line, indicating that the clustering was due to the co-expression of the desmoplakin polypeptide.
Figure 3.
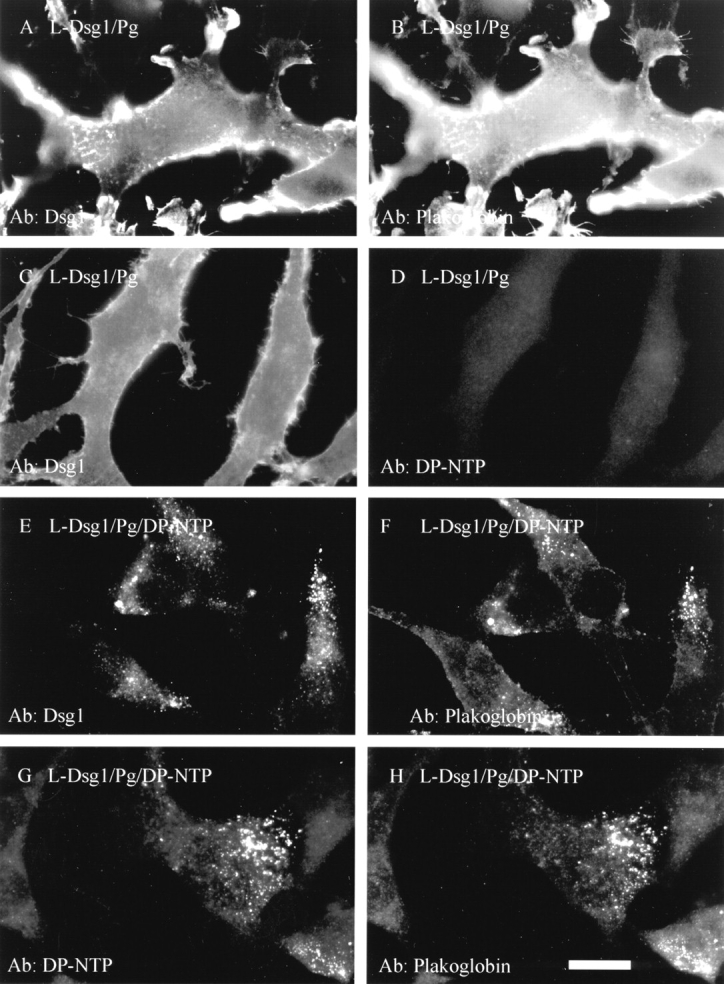
DP-NTP clusters Dsg1 and plakoglobin in L-cell lines. L-cells expressing Dsg1 and plakoglobin (A–D) or Dsg1, plakoglobin, and DP-NTP (E–H) were processed for immunofluorescence. Staining was carried out using antibodies directed against Dsg1 (A and C), plakoglobin (B, F, and H), or the desmoplakin amino-terminal domain (D and G). In the absence of DP-NTP, Dsg1 and plakoglobin are in a diffuse distribution, although some irregularities in the plasma membrane lead to variations in the staining pattern of Dsg1 and plakoglobin (A–C). However, the distribution of both Dsg1 and plakoglobin is punctate in cells co-expressing DP-NTP (E–H). Bar, 10 μm.
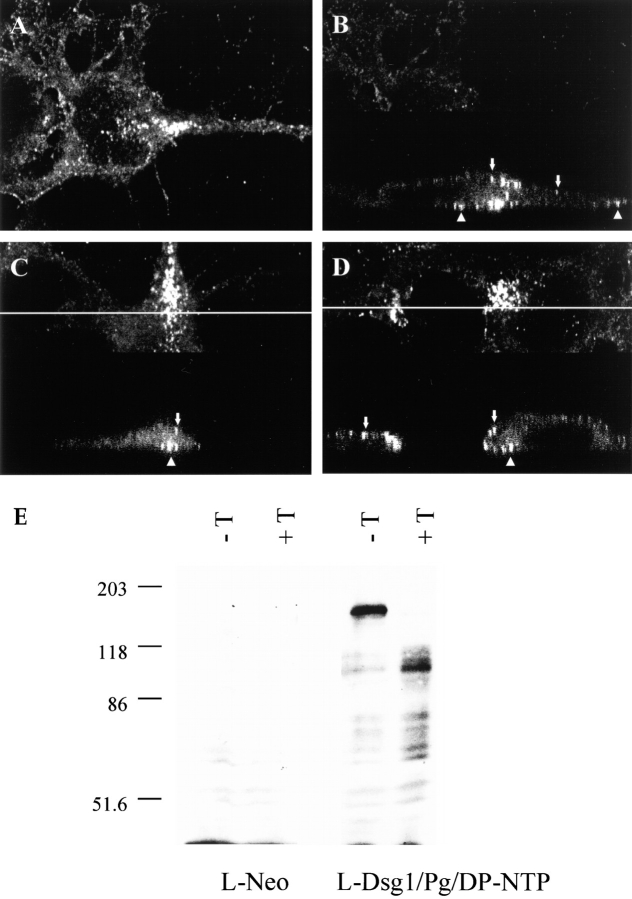
Laser scanning confocal microscopy was used to further characterize the distribution of the punctate clusters in cell lines co-expressing Dsg1, plakoglobin, and DP-NTP. In many instances, punctate clusters of Dsg1 were visualized along the ventral cell surface, as shown in Fig. 4 A, where the focal plane was set at the cell–substrate interface. These clusters colocalized with plakoglobin (not shown), consistent with the results shown in Fig. 3. Vertical sections of the cell in Fig. 4 A reveals the presence of clusters on both the ventral and dorsal cell surfaces (Fig. 4 B). This distribution was observed consistently, as demonstrated by the additional vertical sections shown in Fig. 4, C and D. In addition, treatment of cell lines expressing Dsg1, plakoglobin, and DP-NTP with trypsin demonstrated that the Dsg1 was sensitive to trypsin degradation (Fig. 4 E). The antibody used to detect Dsg1 (Ab NW1) is directed against the Dsg1 cytoplasmic domain (Kowalczyk, A.P., unpublished observations), indicating that the generation of a Dsg1 fragment of ∼100 kD in cells treated with trypsin is due to degradation of a portion of the Dsg1 extracellular domain. Together with the observations that the punctate clusters of Dsg1 were observed along the plasma membrane by confocal microscopy, the trypsinization results indicate that the vast majority of Dsg1 was present on the cell surface. This is consistent with a previous report demonstrating that both Dsg1 and Dsc2a are present on the cell surface when expressed in L-cells (Kowalczyk et al., 1995).
Figure 4.
Localization of Dsg1 to the cell surface by confocal microscopy and trypsinization. L-cells expressing Dsg1, plakoglobin, and DP-NTP were processed for immunofluorescence microscopy using an antibody (PF sera #982) directed against Dsg1 (A–D) and examined using an LSM10 confocal microscope (Carl Zeiss, Thornwood, NY). In A, a confocal image is shown in which the focal plane was set at the cell–substrate interface. In B–D, vertical sections (Φ z scans) were taken of the cells at the top of each panel. Arrows identify punctate Dsg1 staining at the apical membrane, and arrowheads identify staining along the membrane at the cell–substrate interface. To further verify that the Dsg1 was present on the cell surface, L-cells expressing Dsg1, plakoglobin, and DP-NTP were incubated in the presence or absence of 0.01% trypsin for 10 min at 37°C in PBS containing 1 mM EDTA. The cells were rinsed in serum to inactivate the trypsin, lysed in SDS-PAGE sample buffer, and immunoblot analysis performed. Note that trypsin treatment resulted in the appearance of a 100-kD fragment of Dsg1 that is recognized by the Dsg1 antibody (NW1) which is directed against the cytoplasmic domain. This demonstrates that the trypsin degraded the Dsg1 extracellular domain but did not gain access to the cytoplasmic compartment of the cells. In addition, no degradation of plakoglobin was observed (not shown), futher confirming that the trypsinization was limited to the exterior of the cells.
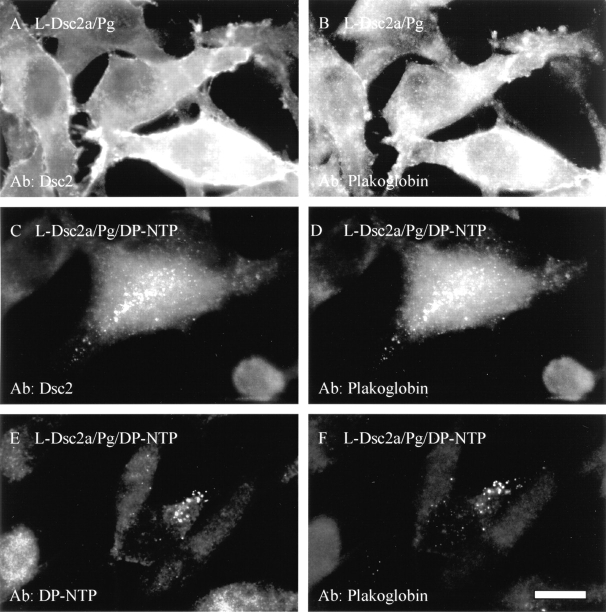
DP-NTP Also Clusters Dsc2a–Plakoglobin Complexes
In contrast to adherens junctions, desmosomes contain two distinct subclasses of cadherins, desmogleins and desmocollins. Therefore, the ability of DP-NTP to cause clustering of Dsc2a–plakoglobin complexes was also analyzed (Fig. 5). When expressed alone, Dsc2a (Fig. 5 A) and plakoglobin (Fig. 5 B) were distributed in a diffuse, largely uniform staining pattern along the membrane of the L-cells. However, in the presence of DP-NTP, both Dsc2a (Fig. 5 C) and plakoglobin (Fig. 5 D) were colocalized in distinct punctate clusters. In addition, DP-NTP (Fig. 5 E) and plakoglobin (Fig. 5 F) colocalized in these clusters, indicating that DP-NTP causes clustering of both classes of desmosomal cadherins.
Figure 5.
DP-NTP clusters Dsc2a and plakoglobin in L-cell lines. L-cells expressing Dsc2a and plakoglobin (A and B) or Dsc2a, plakoglobin, and DP-NTP (C–F) were processed for immunofluorescence. Staining was carried out using antibodies directed against Dsc2a (A and C), plakoglobin (B, D, and F), or the desmoplakin amino-terminal domain (E). Similar to results observed for Dsg1 and plakoglobin, Dsc2 and plakoglobin are distributed in a punctate staining pattern in cells expressing DP-NTP. Note that some cells within the population do not express desmosomal proteins due to heterogeneity in this cell line. Bar, 10 μm.
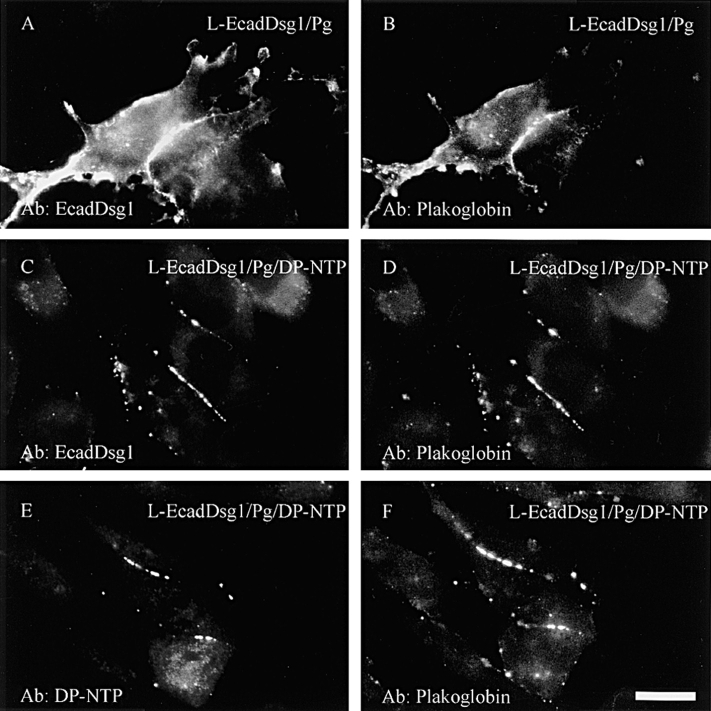
A Chimeric Cadherin with the E-cadherin Extracellular Domain and the Dsg1 Cytoplasmic Domain (EcadDsg1) Recruits Plakoglobin and DP-NTP to Cell–Cell Borders
A number of previous studies have demonstrated that the desmosomal cadherins do not mediate homophilic adhesion when expressed in L-cells (Amagai et al., 1994; Chidgey et al., 1996; Kowalczyk et al., 1996). A previous study demonstrated that a chimeric cadherin with the E-cadherin extracellular domain and the Dsg3 cytoplasmic domain mediates homophilic adhesion in L-cells (Roh and Stanley, 1995a ). Therefore, a chimera containing the E-cadherin extracellular domain and Dsg1 cytoplasmic domain (EcadDsg1) was co-expressed with plakoglobin and DP-NTP to determine if the chimera could recruit plakoglobin and DP-NTP to regions of cell–cell contact. In the absence of DP-NTP, the EcadDsg1 chimera (Fig. 6 A) and plakoglobin (Fig. 6 B) were largely diffuse on the plasma membrane, although some areas of cell–cell border staining were observed. However, when this chimera was co- expressed with plakoglobin and DP-NTP, the EcadDsg1 chimera (Fig. 6 C) and plakoglobin (Fig. 6 D) were codistributed in a punctate pattern. In addition, the EcadDsg1 chimera and plakoglobin were often localized at regions of cell–cell contact. Furthermore, DP-NTP (Fig. 6 E) was recruited to cell–cell contacts and colocalized with plakoglobin (Fig. 6 F). The punctate distribution of DP-NTP at cell–cell interfaces was observed only in L-cells expressing the EcadDsg1 chimera. The observation that the EcadDsg1 chimera, plakoglobin, and DP-NTP were all present at cell– cell borders suggests that all three proteins were present in the same complex and that these complexes were recruited to areas of intercellular contact by the homophilic interactions of the E-cadherin extracellular domain.
Figure 6.
A chimeric cadherin with the E-cadherin extracellular domain and the Dsg1 cytoplasmic domain (EcadDsg1) recruits plakoglobin and D-NTP to cell–cell borders. L-cell lines co- expressing the EcadDsg1 chimera and plakoglobin in the absence (A and B) or presence of DP-NTP (C–F) were generated, and the localization of the proteins was determined by immunofluorescence. In the absence of DP-NTP, the EcadDsg1 chimera (A) and plakoglobin (B) colocalized at cell–cell interfaces. In the presence of DP-NTP, the EcadDsg1 chimera (C) and plakoglobin (D) were present in a punctate staining pattern at cell–cell borders. In addition, DP-NTP was also present at cell interfaces (E) and colocalized with plakoglobin (F) in punctate clusters, indicating that DP-NTP was recruited to cell–cell interfaces by the cadherin–plakoglobin complex. Bar, 10 μm.
Cell lines expressing the EcadDsg1 chimera and plakoglobin in the presence or absence of DP-NTP were also examined by electron microscopy. In L-cell lines co-expressing DP-NTP, electron-dense regions subjacent to the plasma membrane were observed at regions of cell–cell contact (Fig. 7 B). Interestingly, these structures are similar to the junctions lacking association with intermediate filaments that were previously reported in A431 cell lines expressing DP-NTP (Fig. 7 C; Bornslaeger et al., 1996). Although the EcadDsg1 chimera and plakoglobin did colocalize at cell– cell borders in the absence of DP-NTP, the extensive electron-dense plaque-like structures such as those present in cells co-expressing DP-NTP (Fig. 7 B) were never observed in the absence of DP-NTP (Fig. 7 A). These data suggest that clustering of the cadherin–plakoglobin complex by DP-NTP leads to the formation of a submembrane plaque that can be identified ultrastructurally and which closely resembles the outer dense plaque of the desmosome.
Figure 7.
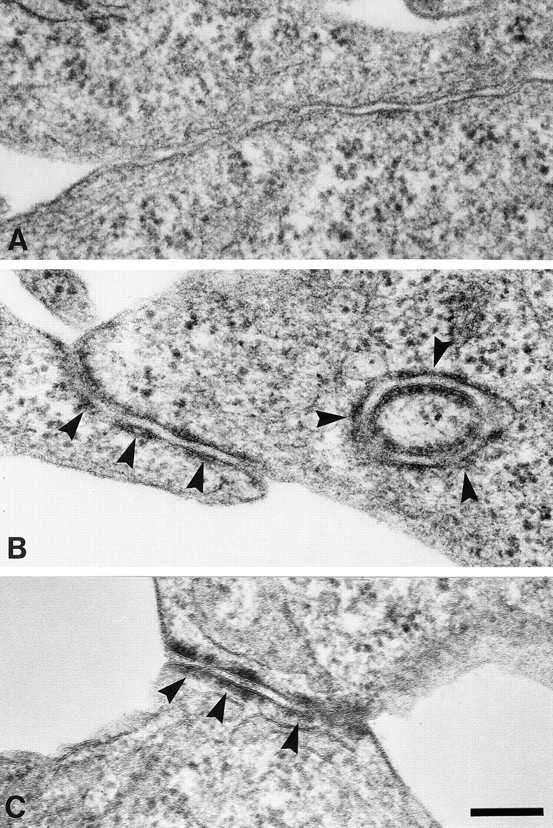
Electron microscopy of junctional structures assembled by L-cells co-expressing the EcadDsg1 chimera with plakoglobin and DP-NTP. L-cell lines expressing the EcadDsg1 chimera and plakoglobin in the absence (A) or presence (B) of DP-NTP were processed for conventional electron microscopy. Junctional structures with an electron-dense region subjacent to the plasma membrane were observed only in L-cell lines co-expressing DP-NTP (arrows). Note that the circular profile in B may represent a cross section of junctional plaques arising from interactions with a neighboring cell projection. These junctional structures were similar in organization to the junctions assembled in A431 cell lines expressing DP-NTP (C, arrows; Bornslaeger et al., 1996). Bar, 0.2 μm.
Plakoglobin Bound to the Desmoglein Cytoplasmic Domain Does Not Associate with α-Catenin
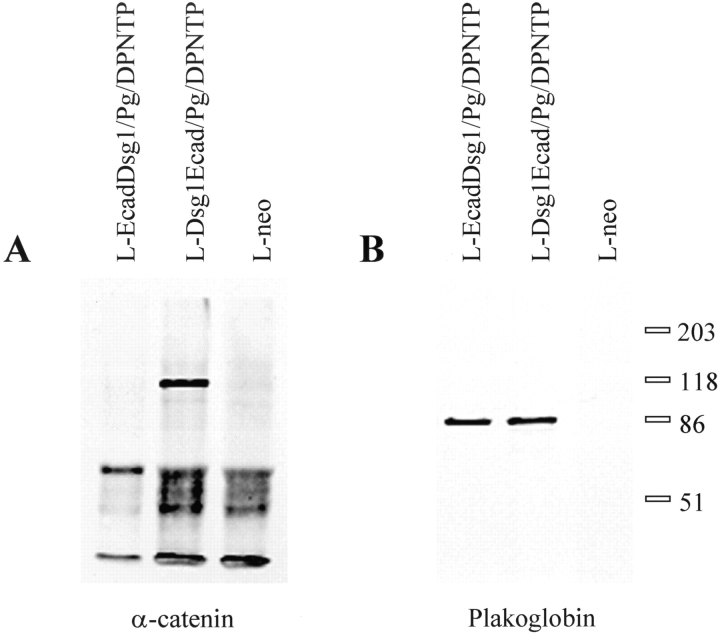
In contrast to the desmosomal cadherins, E-cadherin is coupled to the actin cytoskeleton through interactions with α-catenin, which is able to bind directly to both β-catenin and plakoglobin. The ability of plakoglobin to bind to α-catenin and to associate with both desmosomal cadherins and classical cadherins raises the question as to how these two classes of cadherins are coupled to distinct cytoskeletal filament networks. Therefore, we used the EcadDsg1 chimera along with a Dsg1Ecad chimeric cadherin to determine if α-catenin could associate with plakoglobin that is bound to the Dsg1 cytoplasmic domain. The chimeric cadherins were immunoprecipitated from cell lysates using antibodies directed against the extracellular domain of either chimera. Immunoblot analysis of the immunoprecipitated proteins revealed that α-catenin co-immunoprecipitated only with the Dsg1Ecad chimera and did not co-immunoprecipitate with the EcadDsg1 chimera (Fig. 8 A). However, plakoglobin co-immunoprecipitated with both the Dsg1Ecad and the EcadDsg1 chimeras (Fig. 8 B). These data indicate that plakoglobin that is associated with the Dsg1 cytoplasmic domain does not bind to α-catenin.
Figure 8.
Plakoglobin associates with both the Dsg1Ecad and the EcadDsg1 chimeras, but α-catenin is not associated with the EcadDsg1 chimera. The Dsg1Ecad or the EcadDsg1 chimeras were immunoprecipitated from L-cell lysates under mild detergent conditions as described in Materials and Methods. L-cells expressing only neomycin (L-neo) resistance were used as a control. The immunoprecipitates were analyzed by immunoblot for the presence of either plakoglobin (A) or α-catenin (B).
Plakoglobin Is Required for DP-NTP to Associate with the Dsg1 Cytoplasmic Domain
In adherens junctions, β-catenin is thought to link E-cadherin to α-catenin by binding directly to both proteins. Due to the fact that plakoglobin and β-catenin are closely related, we reasoned that plakoglobin may couple desmoplakin to the desmosomal cadherin cytoplasmic domains. To test this hypothesis, the distribution of Dsg1 was monitored in L-cell lines expressing DP-NTP in the presence of plakoglobin deletion mutants or in the absence of exogenously expressed plakoglobin (Fig. 9). Similar to control cells expressing full length plakoglobin (see Fig. 3), Dsg1 was distributed in punctate clusters in L-cells co-expressing DP-NTP in the presence of plakoglobin deletion mutants lacking either the amino- (Fig. 9 A) or carboxyl-terminal (Fig. 9 B) domains (refer to Fig. 1 for deletion map). In the absence of exogenously expressed plakoglobin, Dsg1 (Fig. 9, C and E) was distributed in a diffuse pattern in cells co-expressing Dsg1 and DP-NTP. Very low levels of endogenous plakoglobin are expressed in L-cells (Fig. 9 D), and only small amounts of endogenous plakoglobin can be detected in L-cells expressing Dsg1 (Kowalczyk et al., 1994). Although DP-NTP was occasionally observed in a punctate staining pattern, DP-NTP was more diffuse (Fig. 9 F) in the absence of exogenous plakoglobin than in cells co-expressing plakoglobin (see also Figs. 3, 5, and 6).
Figure 9.
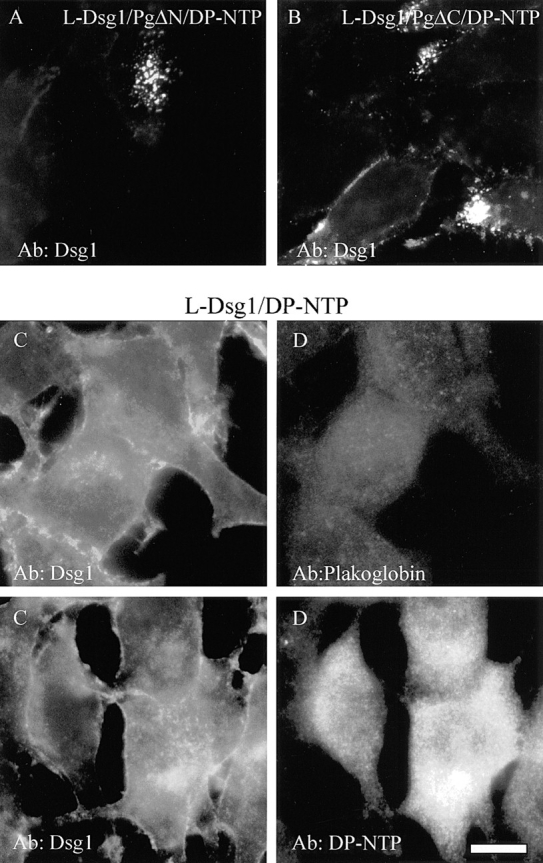
Plakoglobin is required for DP-NTP to cluster Dsg1. The distribution of Dsg1 was monitored by immunofluorescence in L-cell lines co-expressing DP-NTP in the presence of plakoglobin deletions lacking the amino (L-Dsg1/PgΔN/DP-NTP; A) or carboxyl (L-Dsg1/PgΔC/DP-NTP; B) domain. In both cell lines, Dsg1 was distributed in a punctate staining pattern. In contrast, in L-cells expressing Dsg1 and DP-NTP in the absence of exogenously expressed plakoglobin (C–F), Dsg1 was diffusely distributed (C and E). Endogenous mouse plakoglobin is expressed at very low levels in L-cells (Kowalczyk et al., 1994) and was not detectable by immunofluorescence (D). DP-NTP was occasionally found in aggregates, but the staining was more diffuse (F) than in L-cell lines co-expressing plakoglobin and the cadherins. Dsg1 was monitored with PF serum #982, DP-NTP was detected with NW161, and plakoglobin was monitored using a rabbit polyclonal antibody (Hinck et al., 1994) that recognizes mouse plakoglobin. Bar, 10 μm.
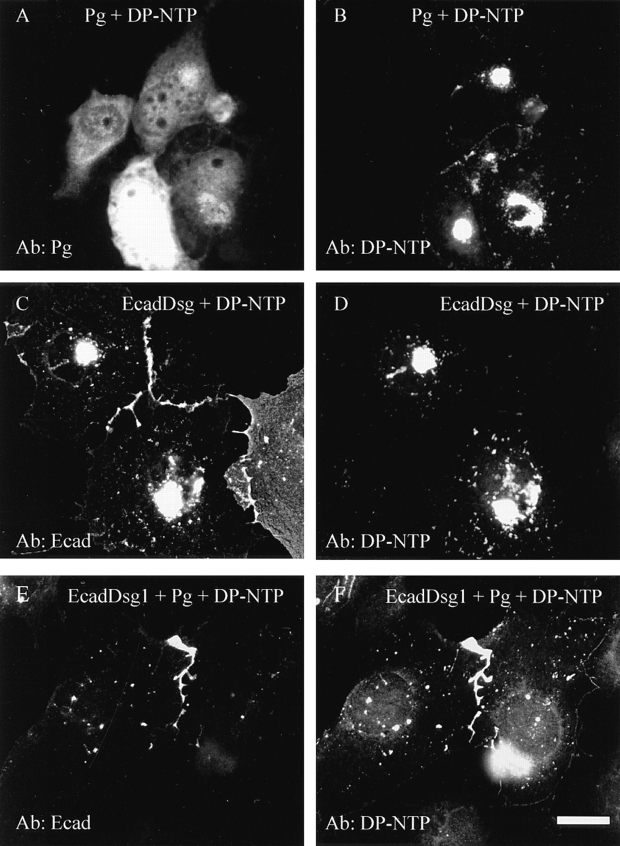
To further define the role of plakoglobin in the association of DP-NTP with the desmosomal cadherin cytoplasmic domains, the EcadDsg1 chimera, plakoglobin, and DP-NTP were transiently expressed in COS cells (Fig. 10). When co-expressed in the absence of the chimera, plakoglobin (Fig. 10 A) was distributed diffusely in the cytoplasm and DP-NTP (Fig. 10 B) was distributed in perinuclear aggregates. COS cells assemble some desmosomes, and low levels of both plakoglobin and desmoplakin staining at cell borders could be detected in transiently transfected cells, presumably reflecting incorporation of the transiently expressed proteins into existing desmosomes (Stappenbeck et al., 1993). However, the vast majority of transiently expressed DP-NTP was not recruited to cell borders by the EcadDsg1 chimera in the absence of plakoglobin (Fig. 10, A and B). In COS cells cotransfected with the EcadDsg1 chimera and DP-NTP, the EcadDsg1 chimera accumulated at cell–cell borders (Fig. 10 C), whereas DP-NTP remained in perinuclear aggregates (Fig. 10 D). In contrast, in cells transiently expressing plakoglobin, DP-NTP, and the EcadDsg1 chimera, the EcadDsg1 chimera (Fig. 10 E) colocalized at borders with both plakoglobin (not shown) and DP-NTP (Fig. 10 F). These findings are consistent with the results observed in L-cell lines and demonstrate that plakoglobin is required for the EcadDsg1 chimera to recruit DP-NTP to areas of cell–cell contact.
Figure 10.
Plakoglobin is required for the EcadDsg1 chimera to recruit DP-NTP to cell–cell borders in transiently transfected COS cells. When co-expressed transiently in COS cells, plakoglobin (A) was largely diffuse in the cytoplasm and DP-NTP (B) was present in aggregates that often surrounded the nucleus. When plakoglobin was co-expressed along with the EcadDsg1 chimera and DP-NTP, the chimera (C), DP-NTP (D), and plakoglobin (not shown) accumulated at cell–cell interfaces. In contrast, when the EcadDsg1 chimera was co-expressed with DP-NTP, the chimera accumulated at cell–cell interfaces (E), but DP-NTP remained in perinuclear aggregates (F). Bar, 10 μm.
Plakoglobin Forms Complexes with DP-NTP in L-cells and Binds Directly to DP-NTP
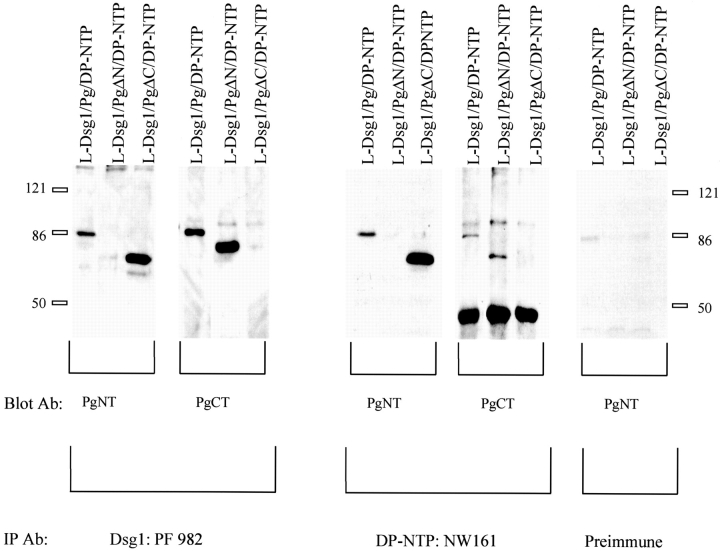
Co-immunoprecipitation experiments were conducted to determine if protein complexes could be identified in L-cells that exhibited clustered cadherin–plakoglobin complexes. L-cells co-expressing Dsg1, DP-NTP, and plakoglobin or deletion mutants of plakoglobin were solubilized in 0.5% Triton X-100, and immunoprecipitations were carried out using an antibody directed against either Dsg1 or DP-NTP (Fig. 11). Plakoglobin or plakoglobin deletion mutants were then detected using antibodies directed against the amino- (PgNT) or carboxyl-terminal domain (PgCT) of plakoglobin. The results indicate that plakoglobin and the plakoglobin deletion mutants co-immunoprecipitate with both Dsg1 and DP-NTP. Preimmune sera from antibody NW161 was used as a control to verify that plakoglobin was co- immunoprecipitated specifically with DP-NTP. These data demonstrate that plakoglobin is in a complex with DP-NTP in the L-cell lines. In addition, the fact that plakoglobin deletion mutants lacking either the amino- or carboxyl-terminal end domains of plakoglobin also co-immunoprecipitated with DP-NTP suggests that the central armadillo repeats of plakoglobin are important for association with both Dsg1 and DP-NTP.
Figure 11.
Plakoglobin co-immunoprecipitates with DP-NTP. Dsg1 or DP-NTP were immunoprecipitated from L-cell lines co-expressing Dsg1, DP-NTP, and full length plakoglobin or plakoglobin deletion mutants. The immunoprecipitates were then analyzed by immunoblot for the presence of plakoglobin using antibodies specific for the amino-terminal (PgNT) or carboxyl-terminal (PgCT) domain of plakoglobin. Full length plakoglobin, PgΔN, and PgΔC co-immunoprecipitated with Dsg1 and with DP-NTP. Dsg1 was immunoprecipitated with PF sera #982, and DP-NTP was immunoprecipitated with NW161. Note that no plakoglobin is detected when the preimmune serum from NW161 was used as the immunoprecipitating antibody.
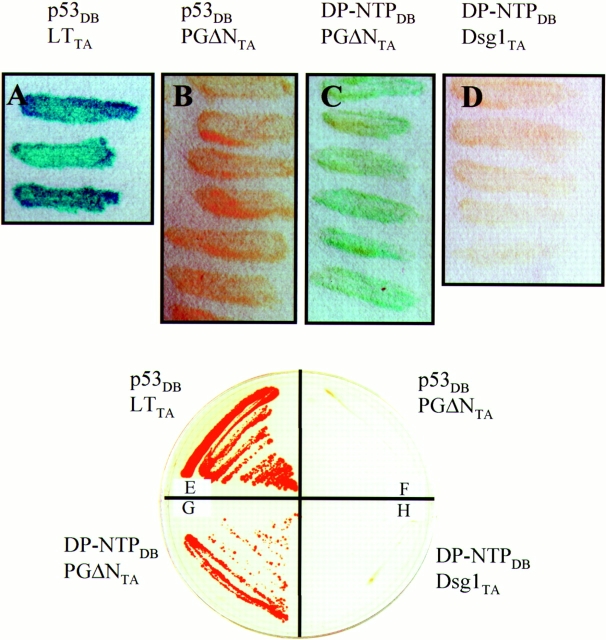
To determine if plakoglobin and DP-NTP interact directly, these proteins were expressed in the yeast two hybrid system and interactions were analyzed by monitoring β-galactosidase activity (Fig. 12, A–D). Cotransformation of plasmids containing the interacting domains from p53 and large T antigen (p53DB and LTTA) were used as positive controls for transcription activation (Fig. 12 A). Control experiments were also conducted to verify that the DP-NTP (DP-NTPDB) and plakoglobin constructs (PGΔNTA) did not spontaneously activate transcription. No β-galactosidase activity was observed when PGΔNTAwas tested for interaction with p53DB (Fig. 12 B). However, β-galactosidase activity was consistently observed when yeast were cotransformed with PGΔNTA and DP-NTPDB (Fig. 12 C), indicating that these proteins interact directly. Although DP-NTPDB was found to interact with PGΔNTA (Fig. 12 C), an interaction between DP-NTPDB and the Dsg1 cytoplasmic tail (Dsg1TA) was not detected (Fig. 12 D). Similar to previous studies (Witcher et al., 1996), the Dsg1 cytoplasmic tail exhibited strong interactions with a PGΔN-DNA– binding domain fusion, indicating that the Dsg1 construct was functional (not shown). Although DP-NTP did not activate transcription in the DNA-binding domain vector, PGΔN in the DNA-binding domain construct did exhibit low levels of transcription activation, precluding our ability to test PGΔN interaction with DP-NTP in the reverse orientation. However, these results were confirmed using growth in the absence of histidine as a second, independent reporter for the activation of transcription. Yeast cotransformed with DP-NTPDB and PGΔNTA exhibited growth on agar plates lacking histidine in the presence of 20 mM 3-aminotriazole (Fig. 12 G), whereas no growth was detected in parallel experiments in which the Dsg1 tail was tested for interactions with DP-NTP (Fig. 12 H). These data indicate that DP-NTP binds directly to plakoglobin but not Dsg1.
Figure 12.
Plakoglobin interacts directly with DP-NTP in the yeast two hybrid system. Colonies of yeast strain HF7c were transformed with the indicated constructs, and colonies were assayed for β-galactosidase activity (A–D) or the ability to grow in the absence of histidine (E–H). Positive control vectors encoding interacting domains of p53 in the DNA-binding domain vector (DB) and large T antigen in the transcription activation vector (TA) were cotransformed as controls for transcription activation. Yeast transformed with these vectors exhibited high levels of β-galactosidase activity (A) and large amounts of growth on histidine drop out medium (E). In contrast, cotransformation with p53DB and a plakoglobin deletion construct PGΔNTA did not exhibit β-galactosidase activity or growth in the absence of histidine. However, yeast cotransformed with PGΔNTA and DP-NTPDB exhibited both β-galactosidase activity (C) and growth on histidine drop out plates (G), indicating that these two proteins interact directly. In contrast, no activation of transcription was detected when DP-NTP was tested for interaction with the Dsg1 cytoplasmic domain (D and H).
Discussion
The molecular interactions that couple the desmosomal cadherins to the intermediate filament cytoskeleton have not been well characterized. Previous studies had shown that the amino-terminal domain of desmoplakin is important for localizing desmoplakin to the cytoplasmic plaque of the desmosome (Stappenbeck et al., 1993; Bornslaeger et al., 1996). In the current study, we sought to determine if desmosomal cadherin–plakoglobin complexes interact with the amino-terminal domain of desmoplakin. The results indicate that the amino-terminal domain of desmoplakin clusters desmosomal cadherin–plakoglobin complexes and binds directly to plakoglobin, but not desmoglein. These results provide new insights into the hierarchy of molecular interactions that occur in the cytoplasmic plaque of the desmosome.
In L-cell lines co-expressing DP-NTP, both Dsg1- (Fig. 3) and Dsc2a–plakoglobin (Fig. 5) complexes are clustered into punctate regions of staining, which appear to be at the plasma membrane. It is interesting to note that the desmosomal cadherins are clustered in L-cells co-expressing DP-NTP, even though these cells do not aggregate in suspension (not shown). The fact that the desmosomal cadherins are clustered but are not engaged in adhesion in L-cells indicates that clustering can occur independent of adhesion. These observations are consistent with the finding that desmosomal complexes resembling half desmosomes are assembled in HaCaT keratinocytes grown in low calcium medium (Demlehner et al., 1995). Clustering of desmosomal cadherins and plakoglobin represents only one aspect of desmosome assembly, and it will be interesting to determine if other desmosomal components such as pinin (Ouyang and Sugrue, 1992, 1996), envoplakin (Ruhrberg et al., 1996), and the plakophilins require desmosomal cadherin-mediated adhesion for assembly into the plaque.
Co-immunoprecipitation experiments demonstrated that plakoglobin and DP-NTP are present in complexes that can be extracted from L-cell lysates (Fig. 11). Although we could not detect the desmosomal cadherins in these complexes, it is likely that the complexes that form between DP-NTP and plakoglobin do contain the cadherins. This is supported by the fact that the desmosomal cadherins are clustered by DP-NTP along with plakoglobin. Furthermore, in cell lines expressing the EcadDsg1 chimera, plakoglobin and DP-NTP are both redistributed to cell–cell interfaces and colocalize with the chimera. These observations strongly suggest that the desmosomal cadherins, plakoglobin, and DP-NTP exist in the same complex.
In addition to the observation that plakoglobin and DP-NTP co-immunoprecipitate, yeast two hybrid analysis demonstrated that DP-NTP binds directly to plakoglobin, but not Dsg1 (Fig. 12). The lack of DP-NTP binding to Dsg1 in the two hybrid system is consistent with the observation that Dsg1 clustering did not occur in L-cell lines in the absence of plakoglobin and with the observation that DP-NTP was not recruited to COS cell–cell borders with the EcadDsg1 chimera in the absence of plakoglobin (Fig. 10). These results lead us to propose that plakoglobin couples desmoplakin to the desmosomal cadherins, thereby linking these cadherins to the intermediate filament cytoskeleton. Previous studies by Troyanovsky et al. (1993, 1994) demonstrated that a connexin-Dsc2a chimera recruited endogenous desmoplakin to the plasma membrane, even when the plakoglobin-binding region of the Dsc2a tail was deleted. The data presented here do not rule out the possibility that desmoplakin binds to desmocollin directly, or that other proteins in addition to plakoglobin can facilitate desmoplakin–desmosomal cadherin interactions. However, our data indicate that plakoglobin plays an important role in linking desmoplakin to the desmoglein cytoplasmic domain. In the desmosome, therefore, plakoglobin may play a role analogous to the adherens junction protein β-catenin, which couples the classical cadherins to α-catenin (Aberle et al., 1994; Jou et al., 1995; Cowin and Burke, 1996).
Recently, transgenic mice with a null mutation in the plakoglobin gene were reported to have defects in desmosome assembly in the intercalated discs of the heart (Bierkamp et al., 1996; Ruiz et al., 1996). In these mice, Dsg2 no longer clustered into distinct junctional structures but exhibited a diffuse distribution on the cell surface. In addition, normal desmosomes were no longer detected in the intercalated discs of the plakoglobin null mice, and adherens junctions and desmosomal components appeared to be mixed into the same junctional structures. Interestingly, desmoplakin localized in these structures, suggesting that this molecule can associate with other junctional proteins in addition to plakoglobin. A family of proteins related to plakoglobin, termed plakophilins, has been identified, and the desmosomal component originally termed band 6 is now known to be a plakophilin family member (Hatzfeld et al., 1994; Heid et al., 1994; Hatzfeld and Nachtsheim, 1996; Mertens et al., 1996). It is possible that plakophilin family members or other desmosomal components also play a role in linking the desmosomal cadherins to desmoplakin.
An interesting property of plakoglobin is that it binds to both classical and desmosomal cadherins. The observation that DP-NTP binds to plakoglobin and clusters desmosomal cadherin–plakoglobin complexes implies that desmoplakin binds to plakoglobin that is associated with the desmosomal cadherin cytoplasmic domain. In contrast, plakoglobin that is associated with the desmosomal cadherin cytoplasmic domain is unable to bind to α-catenin (Fig. 8; Plott et al., 1994; Roh and Stanley, 1995a ). Recent studies have demonstrated that the α-catenin and Dsg1 binding sites on plakoglobin overlap in the amino-terminal armadillo repeats of plakoglobin, suggesting that Dsg1 and α-catenin cannot bind to the same plakoglobin molecule simultaneously (Sacco et al., 1995; Aberle et al., 1996; Chitaev et al., 1996; Troyanovsky et al., 1996; Witcher et al., 1996). However, the classical cadherins bind to the central armadillo repeats of plakoglobin, leaving the amino-terminal armadillo repeats of plakoglobin available to bind α-catenin. In the present study, deletion mutants of plakoglobin lacking either the amino- or carboxyl-terminal domain co-immunoprecipitated with DP-NTP and were clustered in L-cells co-expressing DP-NTP. These results, along with the ability of the amino-terminal plakoglobin deletion to interact with DP-NTP in the two hybrid system, suggest that desmoplakin may bind to the central armadillo repeats of plakoglobin. In A431 cells, the expression of plakoglobin deletion mutants lacking the amino- and carboxyl-terminal end domains promoted the assembly of elongated and fused desmosomes, suggesting that the central armadillo repeats of plakoglobin can promote interactions between desmosomal proteins, which may include desmoplakin (Palka and Green, 1997). Future studies will be directed at mapping precisely where desmoplakin binds to plakoglobin in relation to the sites on plakoglobin that bind to the desmosomal cadherins. Such studies should provide insight into the mechanisms by which the cadherin cytoplasmic tails govern the domains on plakoglobin that are available for interactions with either α-catenin or desmoplakin.
Acknowledgments
The authors would like to thank Drs. M. Wheelock, K. Johnson, J. Papkoff, M. Udey, M. Takeichi, K. Trevor, and J. Stanley for sharing antibody –and cDNA reagents that made this work possible. Thanks also to Drs. S. Elledge and M. Klymkowsky for generously providing yeast two hybrid reagents and expertise. We also would like to thank S. Norvell for assistance with the confocal microscopy, D. Kopp for assistance with the COS transfections, S. Guy for cDNA sequence analysis, and K. Myung and M. Moody for assistance preparing samples for electron microscopy.
This work was supported by the National Institutes of Health (RO1AR43380 and RO1AR41836) and a March of Dimes Birth Defects Foundation grant (1-FY96-0146) to K.J. Green. A.P. Kowalczyk was supported by a postdoctoral fellowship from the Dermatology Foundation, and K.J. Green is a Faculty Research Award recipient of the American Cancer Society.
Footnotes
Address all correspondence to Kathleen Green, Department of Pathology, Northwestern University Medical School, 303 East Chicago Avenue, Chicago, IL 60611. Tel.: (312) 503-5300. Fax: (312) 503-8240.
1. Abbreviation used in this paper: DP-NTP, desmoplakin amino-terminal polypeptide.
References
- Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- Aberle H, Schwartz H, Hoschuetzky H, Kemler R. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to α-catenin. J Biol Chem. 1996;271:1520–1526. doi: 10.1074/jbc.271.3.1520. [DOI] [PubMed] [Google Scholar]
- Amagai M, Karpati S, Klaus-Kovtun V, Udey MC, Stanley JR. The extracellular domain of pemphigus vulgaris antigen (Desmoglein 3) mediates weak homophilic adhesion. J Investig Dermatol. 1994;102:402–408. doi: 10.1111/1523-1747.ep12372164. [DOI] [PubMed] [Google Scholar]
- Bai C, Elledge SJ. Cloning using the two-hybrid system. Methods Enzymol. 1995;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Mclaughlin KJ, Schwarz H, Huber O, Kemler R. Embryonic heart and skin defects in mice lacking plakoglobin. Dev Biol. 1996;180:780–785. doi: 10.1006/dbio.1996.0346. [DOI] [PubMed] [Google Scholar]
- Bornslaeger EA, Corcoran CM, Stappenbeck TS, Green KJ. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell–cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J Cell Biol. 1996;134:985–1001. doi: 10.1083/jcb.134.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RS, Cowin P, Franke WW, Garrod DR, Green KJ, King IA, Koch PJ, Magee AI, Rees DA, Stanley JR, Steinberg MS. Nomenclature of the desmosomal cadherins. J Cell Biol. 1993;121:481–483. doi: 10.1083/jcb.121.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey MAJ, Clarke JP, Garrod DR. Expression of full-length desmosomal glycoproteins (desmocollins) is not sufficient to confer strong adhesion on transfected L929 cells. J Investig Dermatol. 1996;106:689–695. doi: 10.1111/1523-1747.ep12345525. [DOI] [PubMed] [Google Scholar]
- Chitaev NA, Troyanovsky SM. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein, and desmocollin contributes to cell–cell adhesion. J Cell Biol. 1997;138:193–201. doi: 10.1083/jcb.138.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaev NA, Leube RE, Troyanovsky RB, Eshkind LG, Franke WW, Troyanovsky SM. The binding of plakoglobin to desmosomal cadherins: patterns of binding sites and topogenic potential. J Cell Biol. 1996;133:359–369. doi: 10.1083/jcb.133.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J.E., and D.R. Garrod. 1994. Molecular biology of desmosomes and hemidesmosomes. R.G. Landes Co., Austin, Texas. [DOI] [PubMed]
- Coulombe, P.A., and E. Fuchs. 1994. Molecular mechanisms of keratin gene disorders and other bullous diseases of the skin. In Molecular Mechanisms of Epithelial Cell Junctions: From Development to Disease. S. Citi, editor. R.G. Landes Co., Austin, Texas. 259–285.
- Cowin, P., and S. Mechanic. 1994. Desmosomal cadherins and their cytoplasmic interactions. In Molecular Mechanisms of Epithelial Cell Junctions: From Development to Disease. S. Citi, editor. R.G. Landes Co., Austin, Texas. 141–155.
- Cowin P, Burke B. Cytoskeleton-membrane interactions. Curr Opin Cell Biol. 1996;8:56–65. doi: 10.1016/s0955-0674(96)80049-4. [DOI] [PubMed] [Google Scholar]
- Demlehner MP, Schafer S, Grund C, Franke WW. Continual assembly of half-desmosomal structures in the absence of cell contacts and their frustrated endocytosis: a coordinated sisyphus cycle. J Cell Biol. 1995;131:745–760. doi: 10.1083/jcb.131.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. Both upstream and intron elements are required for elevated expression of rat somatic cytochrome c gene expression in COS-1 cells. Mol Cell Biol. 1988;8:35–41. doi: 10.1128/mcb.8.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Goldschmidt MD, Zimbelmann R, Mueller HM, Schiller DL, Cowin P. Molecular cloning and amino acid sequence of human plakoglobin, the common junctional plaque protein. Proc Natl Acad Sci USA. 1989;86:4027–4031. doi: 10.1073/pnas.86.11.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Intermediate filaments and disease: mutations that cripple cell strength. J Cell Biol. 1994;125:511–516. doi: 10.1083/jcb.125.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D. Desmosomes and hemidesmosomes. Curr Opin Cell Biol. 1993;5:30–40. doi: 10.1016/s0955-0674(05)80005-5. [DOI] [PubMed] [Google Scholar]
- Gorbsky G, Steinberg MS. Isolation of the intercellular glycoproteins of desmosomes. J Cell Biol. 1981;90:243–248. doi: 10.1083/jcb.90.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Parry DAD, Steinert PM, Virata MLA, Wagner RM, Angst BD, Nilles LA. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J Biol Chem. 1990;265:2603–2612. [PubMed] [Google Scholar]
- Green KJ, Stappenbeck TS, Noguchi S, Oyasu R, Nilles LA. Desmoplakin expression and distribution in cultured rat bladder epithelial cells of varying tumorogenic potential. Exp Cell Res. 1991;193:134–143. doi: 10.1016/0014-4827(91)90547-8. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami G, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M, Nachtsheim C. Cloning and characterization of a new armadillofamily member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M, Gunnar IK, Plessmann U, Weber K. Band 6 protein, a major constituent of desmosomes from stratified epithelia, is a novel member of the armadillo multigene family. J Cell Sci. 1994;107:2259–2270. doi: 10.1242/jcs.107.8.2259. [DOI] [PubMed] [Google Scholar]
- Heid HW, Schmidt A, Zimbelmann R, Schafer S, Winter-Simanowski S, Stumpp S, Keith M, Figge U, Schnolzer M, Franke WW. Cell type-specific desmosomal plaque proteins of the plakoglobin family: plakophilin 1 (band 6 protein) Differentiation. 1994;58:113–131. doi: 10.1046/j.1432-0436.1995.5820113.x. [DOI] [PubMed] [Google Scholar]
- Hinck L, Nelson WJ, Papkoff J. Wnt-1 modulates cell–cell adhesion in mammalian cells by stabilizing β-catenin binding to the cell adhesion protein cadherin. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Jones JCR, Goldman RD. Comparison of the polypeptide composition of desmosomes prepared from two bovine epithelial tissues. J Cell Biochem. 1988;36:223–236. doi: 10.1002/jcb.240360304. [DOI] [PubMed] [Google Scholar]
- Jou T-S, Stewart DB, Stappert J, Nelson WJ, Marrs JA. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell–cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch PJ, Walsh MJ, Schmelz M, Goldschmidt MD, Zimbelmann R, Franke WW. Identification of desmoglein, a constitutive desmosomal glycoprotein, as a member of the cadherin family of cell adhesion receptors. Eur J Cell Biol. 1990;53:1–12. [PubMed] [Google Scholar]
- Kouklis PD, Hutton E, Fuchs E. Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J Cell Biol. 1994;127:1049–1060. doi: 10.1083/jcb.127.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk AP, Palka HL, Luu HH, Nilles LA, Anderson JE, Wheelock MJ, Green KJ. Posttranslational regulation of plakoglobin expression: influence of the desmosomal cadherins on plakoglobin metabolic stability. J Biol Chem. 1994;269:31214–31223. [PubMed] [Google Scholar]
- Kowalczyk AP, Anderson JE, Borgwardt JE, Hashimoto T, Stanley JR, Green KJ. Pemphigus sera recognize conformationally sensitive epitopes in the amino-terminal region of desmoglein-1. J Investig Dermatol. 1995;105:147–152. doi: 10.1111/1523-1747.ep12316680. [DOI] [PubMed] [Google Scholar]
- Kowalczyk AP, Borgwardt JE, Green KJ. Analysis of desmosomal cadherin adhesive function and stoichiometry of the desmosomal cadherin/plakoglobin complex. J Investig Dermatol. 1996;107:293–300. doi: 10.1111/1523-1747.ep12363000. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, A.P., and K.J. Green. 1996. The desmosome: a component system for adhesion and intermediate filament attachment. In Current Topics in Membranes. Vol. 43. W.J. Nelson, editor. Academic Press, Inc., San Diego. 187–209.
- Lewis, J.E., III, J.K.W., K.M. Sass, P.J. Jensen, K.R. Johnson, and M.J. Wheelock. 1997. Cross-talk betwen adherens junctions and desmosomes depends on plakoglobin. J. Cell Biol. 136:919–934. [DOI] [PMC free article] [PubMed]
- Mathur M, Goodwin L, Cowin P. Interactions of the cytoplasmic domain of the desmosomal cadherin Dsg1 with plakoglobin. J Biol Chem. 1994;269:14075–14080. [PubMed] [Google Scholar]
- McCrea PD, Turck CW, Gumbiner B. A homologue of the armadillo protein in Drosophila(plakoglobin) associates with E-cadherin. Science (Wash DC) 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- McLean WHI, Lane EB. Intermediate filaments in disease. Curr Opin Cell Biol. 1995;7:118–125. doi: 10.1016/0955-0674(95)80053-0. [DOI] [PubMed] [Google Scholar]
- Meng J-J, Bornslaeger EA, Green KJ, Steinert PM, Ip W. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J Biol Chem. 1997;272:21495–21503. doi: 10.1074/jbc.272.34.21495. [DOI] [PubMed] [Google Scholar]
- Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi AY, Shirayoshi K, Okazaki K, Yasuda K, Takeichi M. Transformation of cell adhesion properties by exogenously introduced E-cadherin cDNA. Nature (Lond) 1987;329:341–343. doi: 10.1038/329341a0. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A, Takeichi M, Tsukita S. The 102kd cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell. 1991;65:849–857. doi: 10.1016/0092-8674(91)90392-c. [DOI] [PubMed] [Google Scholar]
- Nieset JE, Redfield AR, Jin F, Knudsen KA, Johnson KR, Wheelock MJ. Characterization of the interactions of α-catenin with α-actinin and β-catenin/plakoglobin. J Cell Science. 1997;110:1013–1022. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- Nilles LA, Parry DAD, Powers EE, Angst BD, Wagner RM, Green KJ. Structural analysis and expression of human desmoglein: cadherin-like component of the desmosome. J Cell Sci. 1991;99:809–821. doi: 10.1242/jcs.99.4.809. [DOI] [PubMed] [Google Scholar]
- Obama H, Ozawa M. Identification of the domain of α-catenin involved in its association with β-catenin and plakoglobin (γ-catenin) J Biol Chem. 1997;272:11017–11020. doi: 10.1074/jbc.272.17.11017. [DOI] [PubMed] [Google Scholar]
- O'Keefe EJ, Erickson HP, Bennett V. Desmoplakin I and desmoplakin II. Purification and characterization. J Biol Chem. 1989;264:8310–8318. [PubMed] [Google Scholar]
- Ouyang P, Sugrue SP. Identification of an epithelial protein related to the desmosome and intermediate filament network. J Cell Biol. 1992;118:1477–1488. doi: 10.1083/jcb.118.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang P, Sugrue S. Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J Cell Biol. 1996;135:1027–1042. doi: 10.1083/jcb.135.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Ringwald M, Kemler R. Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc Natl Acad Sci USA. 1990;87:4246–4250. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palka HL, Green KJ. Roles of plakoglobin end domains in desmosome assembly. J Cell Science. 1997;110:2359–2371. doi: 10.1242/jcs.110.19.2359. [DOI] [PubMed] [Google Scholar]
- Parker AE, Wheeler GN, Arnemann J, Pidsley SC, Ataliotis P, Thomas CL, Rees DA, Magee AI, Buxton RS. Desmosomal glycoproteins II and III. Cadherin-like junctional molecules generated by alternative splicing. J Biol Chem. 1991;266:10438–10445. [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophilahomolog of human plakoglobin. Cell. 1990;63:1167–1178. doi: 10.1016/0092-8674(90)90413-9. [DOI] [PubMed] [Google Scholar]
- Peifer M, McCrea P, Green KJ, Wieschaus E, Gumbiner B. The vertebrate adhesive junction proteins β-catenin and plakoglobin and the drosophila segment polarity gene armadillo form a multigene family with similar properties. J Cell Biol. 1992;118:681–691. doi: 10.1083/jcb.118.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Plott RT, Amagai M, Udey MC, Stanley JR. Pemphigus vulgaris antigen lacks biochemical properties characteristic of classical cadherins. J Investig Dermatol. 1994;103:168–172. doi: 10.1111/1523-1747.ep12392642. [DOI] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci USA. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh J-Y, Stanley JR. Intracellular domain of desmoglein 3 (pemphigus vulgaris antigen) confers adhesive function on the extracellular domain of E-cadherin without binding catenins. J Cell Biol. 1995a;128:939–947. doi: 10.1083/jcb.128.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh J-Y, Stanley JR. Plakoglobin binding by human Dsg3 (pemphigus vulgaris antigen) in keratinocytes requires the cadherin-like intracytoplasmic segment. J Investig Dermatol. 1995b;104:720–724. doi: 10.1111/1523-1747.ep12606963. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C, Hajibagheri MAN, Simon M, Dooley TP, Watt FM. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J Cell Biol. 1996;134:715–729. doi: 10.1083/jcb.134.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P, Brinkmann V, Ledermann B, Behrend M, Grund C, Thalhammer C, Vogel F, Birchmeier C, Gunthert U, Franke WW, et al. Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J Cell Biol. 1996;135:215–225. doi: 10.1083/jcb.135.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco PA, McGranahan TM, Wheelock MJ, Johnson KR. Identification of plakoglobin domains required for association with N-cadherin and α-catenin. J Biol Chem. 1995;270:20201–20206. doi: 10.1074/jbc.270.34.20201. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Heid HW, Schafer S, Nuber UA, Zimbelmann R, Franke WW. Desmosomes and cytoskeletal architecture in epithelial differentiation: cell type-specific plaque components and intermediate filament anchorage. Eur J Cell Biol. 1994;65:229–245. [PubMed] [Google Scholar]
- Schwarz MA, Owaribe K, Kartenbeck J, Franke WW. Desmosomes and hemidesmosomes: constitutive molecular components. Annu Rev Cell Biol. 1990;6:461–491. doi: 10.1146/annurev.cb.06.110190.002333. [DOI] [PubMed] [Google Scholar]
- Shimoyama Y, Nagafuch A, Fujita S, Gotoh M, Takeichi M, Tsukita S, Hirohashi S. Cadherin dysfunction in a human cancer cell line: possible involvement of loss of α-catenin expression in reduced cell-cell adhesiveness. Cancer Res. 1992;52:5770–5774. [PubMed] [Google Scholar]
- Skerrow CJ, Matoltsy AG. Chemical characterization of isolated epidermal desmosomes. J Cell Biol. 1974;63:524–530. doi: 10.1083/jcb.63.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA. Structure and function of intercellular junctions. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- Stanley JR. Autoantibodies against adhesion molecules and structures in blistering skin diseases. J Exp Med. 1995;181:1–4. doi: 10.1084/jem.181.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Green KJ. The desmoplakin carboxyl terminus coaligns with and specifically disrupts intermediate filament networks when expressed in cultured cells. J Cell Biol. 1992;116:1197–1209. doi: 10.1083/jcb.116.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Bornslaeger EA, Corcoran CM, Luu HH, Virata MLA, Green KJ. Functional analysis of desmoplakin domains: specification of the interaction with keratin versus vimentin intermediate filament networks. J Cell Biol. 1993;123:691–705. doi: 10.1083/jcb.123.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Bale SJ. Genetic skin diseases caused by mutations in keratin intermediate filaments. Trends Genet. 1993;9:280–284. doi: 10.1016/0168-9525(93)90014-9. [DOI] [PubMed] [Google Scholar]
- Troyanovsky SM, Eshkind LG, Troyanovsky RB, Leube RE, Franke WW. Contributions of cytoplasmic domains of desmosomal cadherins to desmosome assembly and intermediate filament anchorage. Cell. 1993;72:561–574. doi: 10.1016/0092-8674(93)90075-2. [DOI] [PubMed] [Google Scholar]
- Troyanovsky SM, Troyanovsky RB, Eshkind LG, Leube RE, Franke WW. Identification of amino acid sequence motifs in desmocollin, a desmosomal glycoprotein, that are required for plakoglobin binding and plaque formation. Proc Natl Acad Sci USA. 1994;91:10790–10794. doi: 10.1073/pnas.91.23.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Chitaev NA, Troyanovsky SM. Cadherin binding sites of plakoglobin: localization, specificity and role in targeting to adhering junctions. J Cell Sci. 1996;109:3069–3078. doi: 10.1242/jcs.109.13.3069. [DOI] [PubMed] [Google Scholar]
- Virata MLA, Wagner RM, Parry DAD, Green KJ. Molecular structure of the human desmoplakin I and II amino terminus. Proc Natl Acad Sci USA. 1992;89:544–548. doi: 10.1073/pnas.89.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witcher LL, Collins R, Puttagunta S, Mechanic SE, Munson M, Gumbiner B, Cowin P. Desmosomal cadherin binding domains of plakoglobin. J Biol Chem. 1996;271:10904–10909. doi: 10.1074/jbc.271.18.10904. [DOI] [PubMed] [Google Scholar]