Abstract
Abstract. Transforming growth factor β (TGF-β) signaling involves interactions of at least two different receptors, types I (TβRI) and II (TβRII), which form ligand-mediated heteromeric complexes. Although we have shown in the past that TβRII in the absence of ligand is a homodimer on the cell surface, TβRI has not been similarly investigated, and the site of complex formation is not known for either receptor. Several studies have indicated that homomeric interactions are involved in TGF-β signaling and regulation, emphasizing the importance of a detailed understanding of the homooligomerization of TβRI or TβRII. Here we have combined complementary approaches to study these homomeric interactions in both naturally expressing cell lines and cells cotransfected with various combinations of epitope-tagged type I or type II receptors. We used sedimentation velocity of metabolically labeled receptors on sucrose gradients to show that both TβRI and TβRII form homodimer-sized complexes in the endoplasmic reticulum, and we used coimmunoprecipitation studies to demonstrate the existence of type I homooligomers. Using a technique based on antibody-mediated immunofluorescence copatching of receptors carrying different epitope tags, we have demonstrated ligand-independent homodimers of TβRI on the surface of live cells. Soluble forms of both receptors are secreted as monomers, indicating that the ectodomains are not sufficient to mediate homodimerization, although TGF-β1 is able to promote dimerization of the type II receptor ectodomain. These findings may have important implications for the regulation of TGF-β signaling.
Transforming growth factor-β (TGF-β)1 is a multipotent cytokine involved in a wide range of biological functions including cell growth, apoptosis, production of extracellular matrix, wound healing, and differentiation (35, 37). Three high-affinity transmembrane receptors for TGF-β were identified, first through cross-linking of radiolabeled ligand and later by cDNA cloning: the type I (TβRI, 55 kD), type II (TβRII, 75 kD), and type III (280 kD) receptors (3, 9, 23, 25, 39). TβRI and TβRII appear to be the signaling receptors, while the type III receptor presents ligand to TβRII and I (21, 26, 29, 30, 44). TβRI and TβRII are serine-threonine kinases with cysteine-rich extracellular domains and 41% identity between their kinase domains (9, 22, 24, 33). In the absence of TβRI, the type II receptor can bind TGF-β but does not transduce signal (27, 43). On the other hand, TβRI can be cross-linked to radiolabeled TGF-β only in the presence of TβRII (9, 13, 19, 43). The TβRII kinase is constitutively active (23), and autophosphorylation on several serine residues regulates its activity and interactions with TβRI (28). The binding of TGF-β1 to TβRII mediates the formation of a heteromeric complex of TβRI and TβRII and the phosphorylation of specific serine residues in TβRI by TβRII (36, 43, 44; Wells, R., L. Gilboa, Y. Henis, and H. Lodish, manuscript in preparation). This phosphorylation activates TβRI kinase activity and promotes its interactions with downstream effector molecules, including members of the SMAD family (1, 2, 31, 32, 45).
Both the type II (6, 12) and the type III (12) TGF-β receptors form ligand-independent homooligomers (probably dimers) on the cell surface. That this is functionally important for the type II receptor is shown by studies demonstrating that homooligomerization of TβRII is involved in both positive and negative regulation of signal transduction via intermolecular autophosphorylation of specific serine residues (28). So far, there is no direct physical evidence for TβRI homomeric complexes. Two lines of evidence, however, suggest that the TGF-β receptor signaling complex contains at least two type I receptors: chimeric proteins with the extracellular domain of the erythropoietin (Epo) receptor and the cytoplasmic domain of a constitutively active TβRI mutant are not active unless dimerized by Epo (27), and functionally complementary TβRI mutants falling into two classes, termed kinase defective and activation defective, have been isolated (40).
We report here a detailed investigation of TβRI and TβRII homooligomer formation. We have studied cell lines that express native TGF-β receptors as well as cells cotransfected with various combinations of epitope-tagged receptors using several complementary approaches: sucrose gradient velocity centrifugation to determine the size of the receptor complexes, coimmunoprecipitation, and immunofluorescence copatching studies to detect receptor oligomerization on the surface of live cells. We show that both TβRI and TβRII form homodimers in the ER and that the extracellular region alone is insufficient for dimerization of either receptor. Our results further demonstrate that, similar to the type II receptor (12), TβRI forms ligand- and DTT-independent homooligomers on the surface of live COS7 cells. These results have important implications for our understanding of the events involved in TGF-β signaling.
Materials and Methods
Materials
TGF-β1 was supplied by Celltrix Laboratories (Palo Alto, CA) and R & D Systems, Inc. (Minneapolis, MN) and was radioiodinated for affinity labeling of tagged receptors as described (34, 39). For affinity labeling of soluble receptors, radioiodination was modified as described (41). 9E10 (α-myc) mouse ascites, which recognizes a specific c-myc sequence (8), was purchased from Harvard Monoclonals (Boston, MA). 12CA5 (α-HA) mouse ascites, which recognizes an epitope of the influenza hemagglutinin (HA) protein (42), was from BAbCO (Richmond, CA). IgG fractions were prepared from mouse ascites by ammonium sulfate precipitation followed by DEAE-cellulose chromatography (10). F (ab′)2 fragments were generated by pepsin digestion, following the protocol of Kurkela et al. (18). Fluorophore-labeled affinity-purified antibodies and Cy3-streptavidin were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). All the F (ab′)2 preparations were reduced by mercaptoethanol and alkylated with iodoacetamide to generate monovalent Fab′ fragments (11). To eliminate possible traces of IgG, the Fab′ preparations were treated with protein A–Sepharose (for 12CA5 or goat IgG) or protein G–Sepharose (for 9E10 IgG). The resulting Fab′ were free of contamination by F (ab′)2 or IgG, as judged by SDS-PAGE under nonreducing conditions.
Cell Lines
COS7 and L6 cells (CRL 1651 and CRL 1458, respectively; American Type Culture Collection, Rockville, MD) were grown in DME supplemented with 10% FCS (Biological Industries, Beit Haemek, Israel or GIBCO-BRL, Gaithersburg, MD), 100 U/ml penicillin, 100 μg/ml streptomycin, and 4 mM glutamine (Biological Industries or JRH Biosciences, Lenexa, KS).
Epitope Tagging of TβRI
Epitope tagging was performed according to Kolodziej and Young (16) using site-directed mutagenesis on uracil-containing single-stranded phagemids (17). The human TGF-βRI (ALK5) cDNA (9) was inserted into the SV-40 expression vector pcDNA-I (Invitrogen Corp., Carlsbad, CA) via HindIII and NotI sites on the polylinker. Using this vector, which also carries an M13 origin of replication, single-stranded phagemids were grown in a dut− ung− Escherichia coli strain (CJ236/P3; Invitrogen Corp.) with M13K07 (Bio-Rad Laboratories, Hercules, CA) as helper phage. Site- directed mutagenesis on this template was performed with mutagenic oligonucleotides encoding the epitope-tag sequence flanked by the sequences corresponding to nucleotides 66–81 (on the 5′ end) and 82–97 (3′ end) of the human TβRI (counting the A of the methionine codon of the cDNA as 1). The tag sequences used encoded either the HA epitope YPYDVPDYA or the human c-myc epitope EQKLISEEDL. Each tag was inserted in-frame, nine bases downstream of the putative signal peptide sequence. Double-stranded pcDNA-I preparations with inserts of wild-type TβRI or the epitope-tagged receptors were digested by XbaI, cutting in two places (at the 3′ of the polylinker and at position 706 in the TβRI coding sequence). The 1.6-kb piece from the wild-type, untagged receptor encoding most of the coding region of the cDNA was ligated with the 4.9-kb piece from the myc- or HA-tagged NH 2-terminal TβRI sequences and the cloning vector pcDNA-I. The resulting constructs contained as insert all of the TβRI coding region, most of which was derived from the wild-type, untagged receptor clone, and the NH 2-terminal region (including the tag) derived from the tagged receptor clone. The coding regions derived from the tagged cDNAs were verified by DNA sequencing.
Construction of HA-tagged Soluble Receptors
To construct an HA-tagged secreted form of the TβRII ectodomain (II-SF), a HindIII-PstI fragment of the full-length HA-tagged TβRII (TβRII-HA; 12) containing the 5′ region of the ectodomain was ligated into a PstI-HindIII fragment of the type II–secreted receptor (containing the 3′ region of the ectodomain) in pcDNA-I (24). For the experiments below, the construct was moved into the vector pcDNA-I/Amp (Invitrogen Corp.) via the HindIII-XbaI sites of the multiple cloning region.
A secreted form of TβRI was constructed by using PCR to insert a stop codon at the 5′ end of the transmembrane domain, just upstream of a PvuII site, replacing Leu126. The mutated cDNA was then cut with EcoRI (at the insertion site on the 5′ end) and PvuII and was inserted into the EcoRI and EcoRV sites of pcDNA-I/Amp (untagged I-SF). The HA-tagged full-length receptor was cut with HindIII (in the vector) and XhoI (cuts in the ectodomain sequences). The fragment containing the HA-tag was inserted into a backbone of the untagged I-SF cut with HindIII and XhoI (partial digest, with cut in the ectodomain), to yield the HA-tagged secreted form of TβRI (I-SF). The sequence in regions derived from PCR amplification was confirmed by sequencing.
COS7 Cell Transfections
COS7 cells were transiently transfected by the DEAE-dextran method (38) using pcDNA-I or pcDNA-I/Amp containing the TβRI and TβRII constructs (tagged, untagged, or the tagged soluble receptors). Cells were used for experiments 48 h after transfection.
Metabolic Labeling
Cells were grown to subconfluence in standard media on 10 cm plates. They were rinsed once with PBS, then starved in serum-free DME minus methionine and cysteine (ICN Biomedicals, Costa Mesa, CA) for 3–4 h at 37°C. The medium was replaced with fresh starvation medium supplemented with 0.2 mM oxidized glutathione (Boehringer Mannheim Corp., Indianapolis, IN) and 0.5 mCi/ml of [35S]methionine and [35S]cysteine (Express; New England Nuclear, Beverly, MA) and incubated at 37°C for 15 or 30 min. Plates were then rinsed three times with ice-cold PBS and incubated in 0.2 M iodoacetamide (1 M stock solution in 0.5 M Tris-HCl, pH 8.8, diluted to 0.2 M in PBS) for 10 min at 4°C. Cells were transferred into Eppendorf tubes (Madison, WI), lysed in 150 μl of MNT buffer (20 mM MES pH 6.0, 30 mM Tris, pH 7.4, 100 mM NaCl) supplemented with 2% n-octyl-polyoxyethylene (octyl-POE; Bachem Biosciences, Philadelphia, PA), and cleared with a 10–15-min spin (14,000 g) at 4°C. “+SDS” samples were lysed in MNT lysis buffer with 0.5% SDS. The HA-tagged secreted receptors (II-SF and I-SF) were metabolically labeled by rinsing transfected COS7 cells with PBS, followed by incubating the cells in 6 ml/ 10-cm dish of starvation medium with oxidized glutathione and 0.1 mCi/ml 35S-Express for 10.5 h. Media from two plates were then collected, filtered with a 0.22-μm filter, brought to 20 mM Hepes, pH 7.5, and 1 mM PMSF, concentrated in a Centricon 10 concentrator (Amicon, Inc., Berverly, MA) rinsed several times with MNT, brought to 150 μl with MNT (no detergent), and loaded on gradients as below.
Velocity Sedimentation on Sucrose Gradients
50 μl of size markers—carbonic anhydrase (29 kD), BSA (66 kD), alcohol dehydrogenase (150 kD), β-amylase (200 kD), apoferritin (443 kD), and thyroglobulin (669 kD), each 9 mg/ml except apoferritin, which was 2.2 mg/ml (Sigma Chemical Co., St. Louis, MO)—in MNT lysis buffer were added to the 150-μl lysates described above and layered over 4.0-ml gradients of 7.5–30% sucrose in MNT/1% octyl-POE. +SDS samples included an additional 0.5% SDS added to each gradient. Gradients were centrifuged in an SW60 rotor (Beckman Instruments, Fullerton, CA) at 60,000 rpm for 8 h at 4°C. 250-μl fractions were removed sequentially from the top of each gradient using a hand-held pipettor. To analyze size standards, 25-μl aliquots were removed from each fraction and were separated by 8–15% gradient SDS-PAGE, followed by Coomassie blue staining. The remainder of each fraction was brought to 0.5 ml (1 ml for +SDS samples) with IP buffer (0.5% deoxycholate, 1% Triton X-100, 10 mM EDTA, in PBS, pH 8.0) and immunoprecipitated with (a) 10 μl/ml of a polyclonal rabbit antiserum raised against the COOH-terminal 16 amino acids of TβRII (α-IIC; reference 36); (b) 15 μl/ml of a polyclonal rabbit antiserum raised against the juxtamembrane cytoplasmic domain of TβRI (9); or (c) 1 μl/ml α-HA ascites. After overnight incubation at 4°C, 50 μl of protein A–Sepharose CL-4B beads (1:1 in PBS; Sigma Chemical Co.) were added, and the lysates were incubated with rotation for an additional 30 min. Beads were rinsed twice with IP buffer (with 0.5% SDS for type II receptor immunoprecipitations) and once with PBS and eluted by boiling in SDS-PAGE sample buffer. Control immunoprecipitants (from lysates treated identically, but not run on gradients) were eluted into 0.5% SDS and one half of each was digested overnight at 37°C with endoglycosidase H (Endo H; Genzyme Corp., Boston, MA; 100 mIU/ml) in the presence of 100 mM sodium citrate, pH 6. Samples were analyzed by 7.5% SDS-PAGE; gels were fluorographed with 2,5 diphenyl-oxazole, dried, and placed on Kodak XAR film (Rochester, NY), which was quantitated with a LaCie Silverscanner II and MacBAS (Fuji Photo Film Co., Tokyo, Japan) software. The secreted receptors in the media collected from the cells (see former section) were treated similarly, except that the gradients were 5–15% sucrose in MNT without detergent, and markers were cytochrome C (12.4 kD), carbonic anhydrase (29 kD), and BSA (66 kD). Fractions from the sucrose gradients were brought to 750 μl with IP buffer, immunoprecipitated with α-HA, and analyzed on 15% polyacrylamide/1.2% bisacrylamide SDS gels.
Receptor Cross-Linking
Binding and cross-linking of 100 pM 125I–TGF-β1 to transfected COS7 cells grown on 10-cm dishes was as described (36). Cross-linked proteins were resolved by 7.5% SDS-PAGE under reducing conditions and exposed to Kodak XAR film at -70°C. For affinity labeling of II-SF, transfected COS7 cells were rinsed with PBS and incubated at 37°C for 10.5 h in 6 ml/10-cm dish of DME without serum. The media from two dishes were filtered and concentrated as for the metabolically labeled material, brought to 1.0 ml with KRH (50 mM Hepes, pH 7.5, 128 mM NaCl, 1.3 mM CaCl2, 5 mM MgSO4, 5 mM KCl), and incubated for 3.5 h at 4°C with 250 pM 125I–TGF-β1. Disuccinimidyl suberate (0.1 mg/ml) was added (4°C, 15 min). The resulting material was quick-spun to remove insoluble cross-linker, concentrated with several washes of MNT, and brought up to 150 μl with MNT (no detergent) before being loaded on a 5–15% sucrose gradient.
Receptor Coimmunoprecipitation
48 h after transfection, cells were washed and incubated with serum-free medium (30 min, 37°C) to eliminate residual ligand. 2 mM DTT was added for the last 10 min of the incubation to selected dishes. Cells were then washed twice in ice-cold HBSS/Hepes/BSA (Hanks' balanced salt solution with 20 mM Hepes, pH 7.4, supplemented with 1% fatty acid–free BSA; Sigma Chemical Co.) and incubated in the same medium with or without 250 pM TGF-β1 (4°C, 1.5 h). The cells were washed twice more in HBSS/Hepes and lysed in a lysis buffer (PBS containing 1% Triton X-100, 10 mM EDTA, 1 mM p-aminoethylbenzenesulfonyl fluoride). Extracts were precleared (4°C, 1 h) by incubation with protein A–Sepharose (Pharmacia Biotech, Piscataway, NJ) for immunoprecipitation with 12CA5 antibodies or with protein G-Sepharose (for immunoprecipitation with 9E10 antibodies). Immunoprecipitation was carried out (4°C, 2 h) with 20 μg/ml of the appropriate IgG together with protein A– or protein G–Sepharose. Immunoprecipitates were washed three times with the lysis buffer, dissolved in Laemmli loading buffer with or without mercaptoethanol, and run on 7% SDS-PAGE. For N-glycosidase F treatment, immunoprecipitates were dissolved in 20 μl 0.5% SDS. 20 μl of 2× enzyme buffer (100 mM sodium phosphate, pH 7.5, 2% NP-40), and 2 U of N-glycosidase F (Boehringer Mannheim Corp.) was added to the eluate (37°C, 4 h). The gel was blotted onto nitrocellulose and then blocked in TBST (50 mM Tris, 100 mM NaCl, 0.1% Tween 20, pH 7.4) with 2% BSA and 5% skim milk (4°C, 1 h). The blot was then sequentially incubated with 20 μg/ml α-myc, 1:2,000 biotinylated GαM (goat IgG anti–mouse IgG; Jackson ImmunoResearch Laboratories), and 1:1,000 streptavidin-horseradish peroxidase (GIBCO-BRL). After each incubation, the blot was washed three times in TBST (4°C, 10 min). Enhanced chemiluminescence was used to visualize the precipitated receptors by autoradiography on an x-ray film.
Immunofluorescence Copatching Experiments
The method used is based on Henis et al. (12), labeling the first epitope with monovalent Fab′ fragments (to avoid adsorption of the mouse antibodies against the second tag to secondary antibodies prebound to the first tag). The method was modified by the use of a biotinylated secondary Fab′ followed by Cy3-streptavidin to mediate patching of the first epitope as well.
COS7 cells were grown on coverslips and transfected with TβRI-HA and TβRI-myc constructs. 48 h after transfection, cells were washed twice with serum-free DME and incubated 30 min at 37°C to allow endocytosis of ligand-bound receptors. After washing twice with cold HBSS/Hepes/ BSA, the cells were incubated in the same buffer (4°C, 2 h) with normal goat IgG (200 μg/ml) to block nonspecific binding. This was followed by successive incubations (4°C, 1 h each, with three washes between incubations) with: (a) α-myc Fab′(50 μg/ml); (b) Fab′ fragments of biotinylated GαM (5 μg/ml); (c) Fab′ of unlabeled GαM (200 μg/ml), used to block all free binding epitopes on the α-myc antibody; and (d) α-HA IgG (20 μg/ml); (e) FITC-labeled GαM (20 μg/ml). The cells were then washed and fixed by two incubations (15 min, 4°C and then 15 min, 22°C) with 3.2% paraformaldehyde in PBS, pH 7.4, supplemented with 1.1% lysine and 0.24% NaIO4. The reaction was quenched by three incubations with 50 mM glycine in PBS, pH 7.4 (5 min, 22°C). Cells were then incubated with 0.5 μg/ml Cy3-streptavidin and mounted with mowiol (Hoechst AG, Frankfurt, Germany) containing 29 mM n-propyl gallate (Sigma Chemical Co.). Fluorescence digital images were acquired with a Zeiss Axioskop (63× oil objective; Thornwood, NY), coupled to a cooled CCD camera (model CC/CE 200; Photometrics, Tuscon, AZ). The computerized microscope system (15) and Priism software driving the focus, excitation, and emission filter wheels were described earlier (5, 14). Fluorescein and Cy3 images were taken using selective filter sets that essentially eliminate leakage. Superposition of the two images and contrast enhancement were performed after correction for shift and magnification using Priism interactive multicolor visualization (5). The images were exported in TIFF format to Photoshop (Adobe, San Jose, CA) and printed.
Results
TβRI Forms Homodimers in the ER
Two different experiments in the literature have provided functional evidence that TβRI forms homooligomers within the signaling complex and that this oligomerization is functionally relevant (27, 40). The oligomeric structure of TβRI either in the ER or the plasma membrane has not been thoroughly characterized, however, and there is no conclusive evidence for physical interactions between TβRI polypeptides in the absence of ligand and the type II receptor. We therefore used sucrose gradient velocity centrifugation of metabolically labeled receptors to determine relative complex size. This approach was chosen since chemical cross-linking of TβRI and TβRII has not been possible (data not shown). We used L6 rat myoblasts, which are TGF-β responsive and which express both of the signaling receptors but lack the type III TGF-β receptor that may form complexes with TβRI and TβRII and could therefore complicate size determinations. L6 cells were pulse-labeled for 30 min, conditions under which all labeled TβRI is Endo H sensitive (41; Fig. 1 B). Endo H sensitivity was used as a marker for localization in the ER since only proteins in the ER and early Golgi carry the high mannose sugars, which are susceptible to the enzyme. Labeled L6 cells were lysed in a buffer containing octyl-POE and analyzed by velocity sedimentation on sucrose gradients in the same detergent, with or without SDS (see Materials and Methods). Octyl-POE was chosen as the detergent for several reasons: (a) It is nonionic; (b) it has a density of 1; (c) it has a high CMC; and (d) it is freely miscible in aqueous solutions, enabling analysis of the migration of detergent-solubilized proteins rather than of micelles.
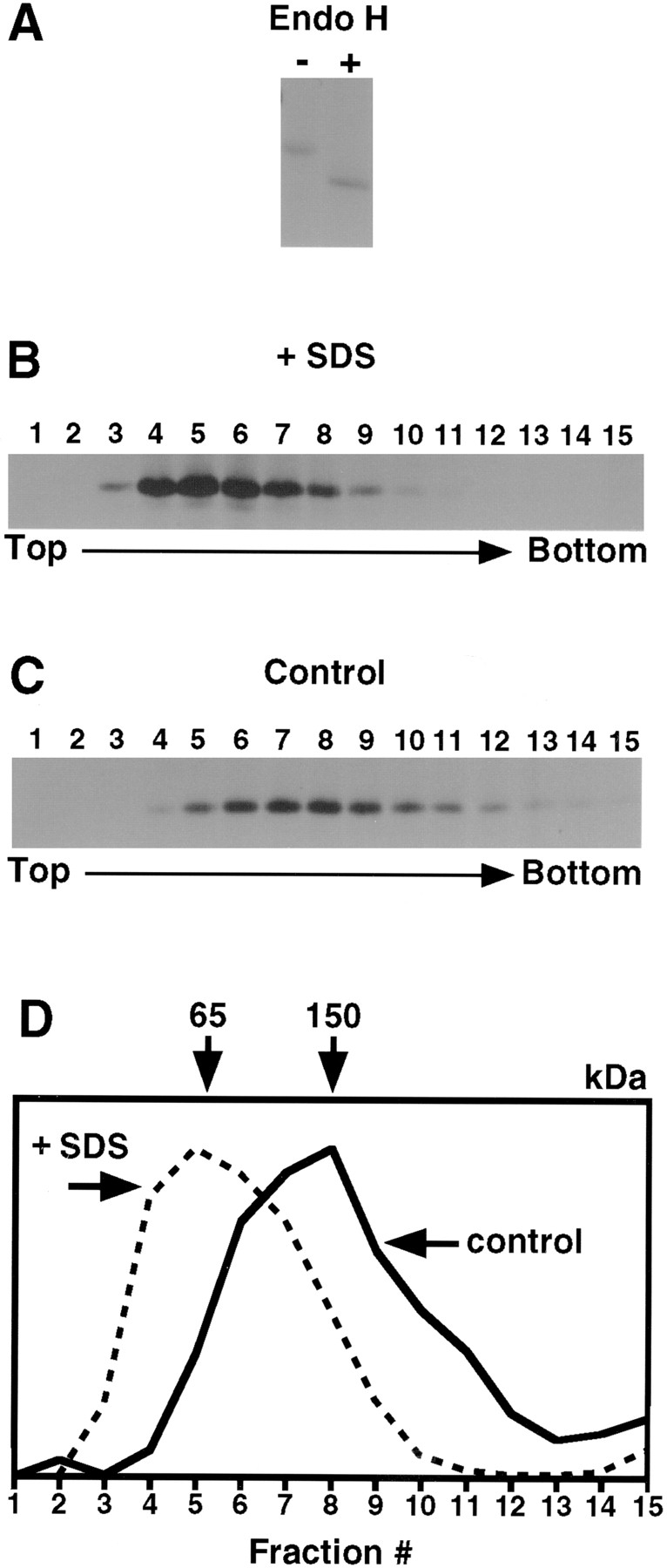
Figure 1.
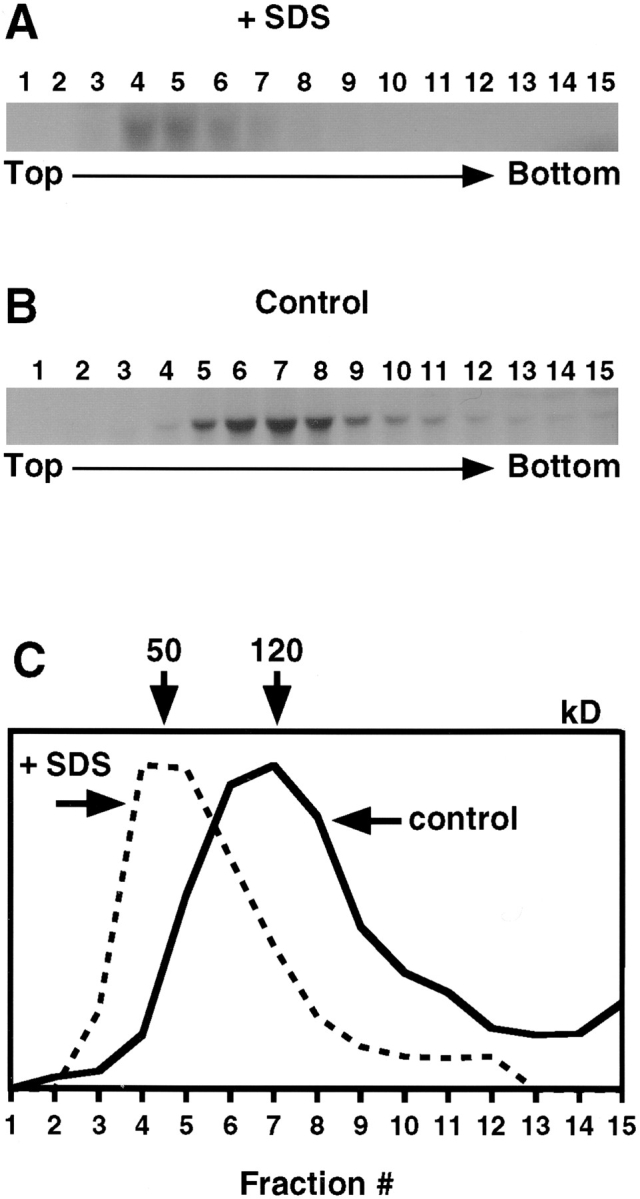
The type I receptor forms homodimers in the ER of L6 cells. L6 cells were pulse-labeled with 35S-Express for 30 min, cooled, and derivatized with iodoacetamide. After lysis in MNT buffer containing octyl-POE (with or without 0.5% SDS), samples were centrifuged on sucrose gradients, and the collected fractions (numbered 1 to 15, with 1 at the top of the gradient) were analyzed by immunoprecipitation with anti-TβRI antibodies followed by SDS-PAGE and autoradiography (see Materials and Methods). Endo H treatment (not shown) of control material confirms that all of the labeled material is Endo H sensitive, representing the ER form. (A) SDS (0.5%) was included in the lysis buffer and in the gradient. Migration of the receptor is consistent with monomers. (B) SDS was not present. Samples were treated with Endo H before SDS-PAGE. Migration of the receptor is consistent with dimers. (C) Graphical representation of the quantified bands. Values were normalized to the peak fraction. Numbers at the top represent the protein size predicted by markers to migrate at each peak.
Receptors from cells lysed and run in the presence of 0.5% SDS yielded a peak at fractions 4–5, corresponding to 50 kD, as determined by size marker analysis carried out for each individual gradient (Fig. 1, A and C). This is compatible with the molecular mass of the monomeric receptor, predicted to be 53 kD. Receptors from cells not treated with SDS migrated with a peak in fractions 6–8 (Fig. 1 , B and C), consistent with a size of 120 kD. Although absolute size determinations from a gradient can be inaccurate for detergent-solubilized membrane proteins, the size of this complex compared with the SDS-treated monomers is consistent with TβRI dimers. TβRI from cells labeled similarly but chased for 4 h (such that labeled TβRI carried mature N-linked oligosaccharides and was at the trans-Golgi or beyond; reference 41) migrated with similar velocity. We conclude that TβRI forms dimer-sized complexes in the ER. Similar results were obtained with Mv1Lu cells (not shown) and with COS7 cells transfected with tagged or untagged TβRI (Fig. 2 and data not shown), demonstrating the generality of TβRI dimerization. The similarity between COS7 cells and the naturally expressing cell lines regarding TβRI dimerization indicates that overexpression in COS cells does not affect dimerization. This allowed us to use COS7 cells for immunofluorescence copatching studies, which require receptor density at the cell surface high enough to be visualized by immunofluorescence.
Figure 2.
TβRI forms homodimers in the ER of COS7 cells. COS7 cells were transiently transfected with TβRI-HA and treated 48 h later as described in Fig. 1. Immunoprecipitation of fractions from the sucrose gradients was with α-HA antibodies. Numbers at the top represent protein size predicted by size markers included in the run. Similar results were obtained with untagged TβRI. (A) SDS (0.5%) was included. (B) No SDS.
Coimmunoprecipitation Studies Identify Ligand- and DTT-independent TβRI Homooligomers
The velocity sedimentation experiments described above demonstrate that TβRI forms complexes of a size expected for a dimer. However, it is also possible that TβRI associates with another protein of roughly the same size. To demonstrate that TβRI indeed forms homooligomers, we performed coimmunoprecipitation studies on cells cotransfected with two TβRI forms, each carrying a different epitope tag. HA or myc epitopes were inserted at the ectodomain NH2 terminus, three amino acids downstream of the putative signal sequence cleavage site. They were introduced at the NH2 terminus to enable immunofluorescence copatching studies (see next section). Fig. 3 demonstrates that the tagged receptors were expressed at the surface of COS7 cells and labeled by 125I–TGF-β1 similar to untagged TβRI. Very low levels of TβRI and TβRII were observed in control cells transfected with vector alone (Fig. 3, lane 5) or with TβRI cDNA in the same vector (lane 4). These low levels may be attributed to the native receptor population in COS7 cells. Cotransfection of TβRI (tagged or untagged) together with TβRII resulted in a significant increase in the labeling of TβRI (lanes 1–3). The insertion of the epitope tags did not eliminate receptor signaling, as tested by cotransfection with p3TP-Lux (a construct that carries the luciferase gene under a TGF-β response element) into L17 cells that lack endogenous TβRI activity (not shown). Furthermore, in the sedimentation velocity studies (Fig. 2), the tagged TβRI was able to form dimer-sized complexes, as observed for the untagged receptor.
Figure 3.
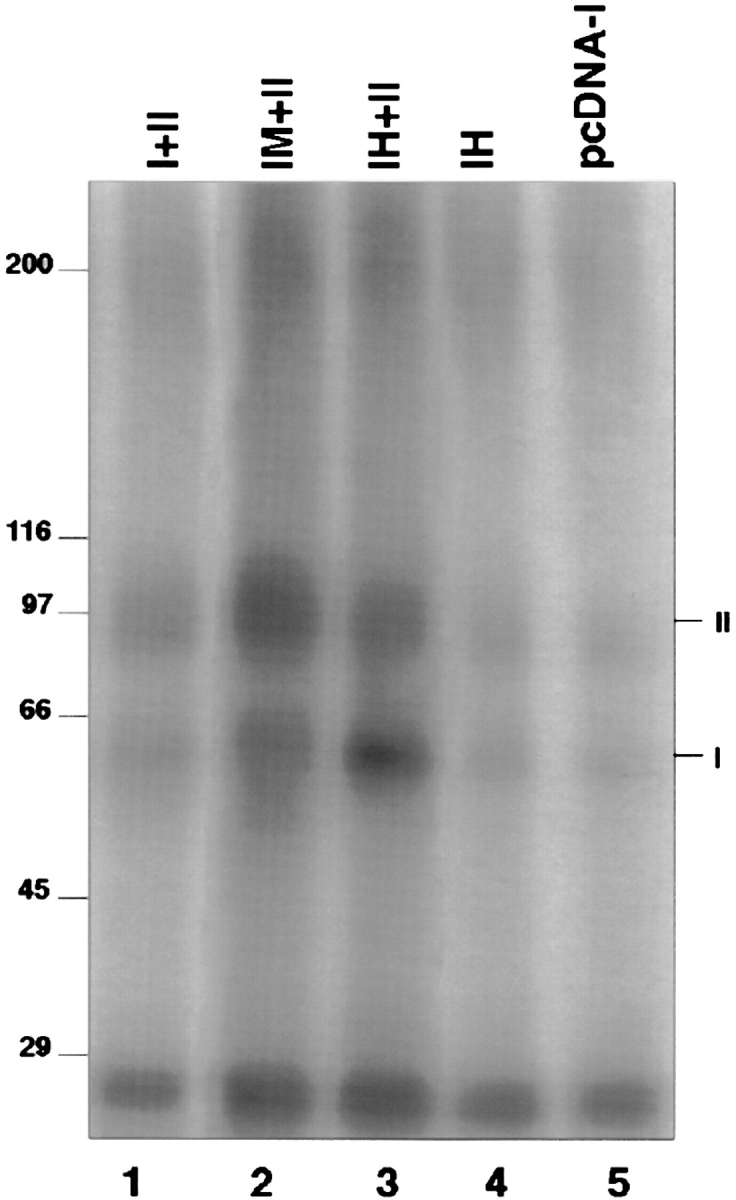
Affinity labeling of wild-type and epitope-tagged TβRI. COS7 cells were transiently transfected with human TβRII (lanes 1–3) together with wild-type TβRI (lane 1), TβRI-myc (lane 2), or TβRI-HA (lane 3). Alternatively, cells were singly transfected with TβRI-HA (lane 4) or with the vector pcDNA-I (lane 5). Cells were incubated with 100 pM of 125I–TGF-β1, and bound ligand was cross-linked to cell surface receptors. Detergent extracts of the transfected cells were analyzed by 7.5% SDS-PAGE and exposed to an x-ray film. Positions of molecular weight markers and of TβRI and TβRII are indicated. Cells that were singly transfected with wild-type TβRI or TβRI-myc did not show labeling above the background of mock-transfected cells, as shown for TβRI-HA (lane 4). Note that under the transient expression conditions, many cells also express singly either TβRI or II, and the relative labeling intensities of the two receptor types vary between experiments.
We used the tagged type I receptors in coimmunoprecipitation experiments designed to confirm that the dimer-sized TβRI complexes observed in the sedimentation velocity studies did in fact represent dimers and to determine whether exposure to ligand or DTT influence TβRI homooligomers. The latter experiment was motivated by the observation that cross-linking of iodinated TGF-β1 to TβRI in the presence of TβRII is eliminated by pretreatment of cells with DTT (4), an effect that may involve disruption of the TβRI/TβRII heterooligomeric structure (Rodriguez, C., R. Lin, P.E. Scherer, and H.F. Lodish, manuscript in preparation).
COS7 cells were cotransfected with TβRI-HA and TβRI-myc and lysates were immunoprecipitated with α-HA antibodies. The immunoprecipitates were subjected to SDS-PAGE and Western blotting, and the blots were analyzed with α-myc antibodies followed by biotinylated GαM and streptavidin–horseradish peroxidase. The results show that TβRI-myc coprecipitates with TβRI-HA (Fig. 4 A, lane 5, and B, lane 2). Preincubation of cells with TGF-β1 (Fig 4 A, lane 6, and B, lane 3), DTT (Fig 4 A, lane 7), or both (lane 8) did not change the quantity of TβRI-myc coprecipitated with TβRI-HA. Blots from control cells, singly transfected with TβRI-HA, were not stained by α-myc, showing the specificity of the blotting antibody (Fig. 4 A, lane 4). Transfection with TβRI-myc alone was used as control for the immunoprecipitating antibody, α-HA (Fig. 4 A, lane 3). A positive control of cells singly expressing TβRI-myc, where α-myc was used both for immunoprecipitation and blotting, is depicted in lanes 1 and 2 (Fig. 4 A). The type I receptors appear in two bands (∼59 and 53 kD; Fig. 4 A, lane 1). The upper band is N-glycosylated TβRI since treatment of the precipitated receptors with N-glycosidase F led to disappearance of the 59-kD band, leaving only the lower band (Fig. 4 B). The 53-kD band in Fig. 4 A gets somewhat distorted and swept downward in the coprecipitation lanes because of the presence of high amounts of the α-HA antibody used for precipitation (seen in the lowest band). This antibody, unlike α-myc, runs very close to the lower TβRI band. When the gels are run under nonreducing conditions (providing better separation from the antibody band), this distortion disappears (Fig 4 B). These results demonstrate that TβRI forms homooligomers that are unaffected by TGF-β1 or DTT and are stable in the presence of detergents.
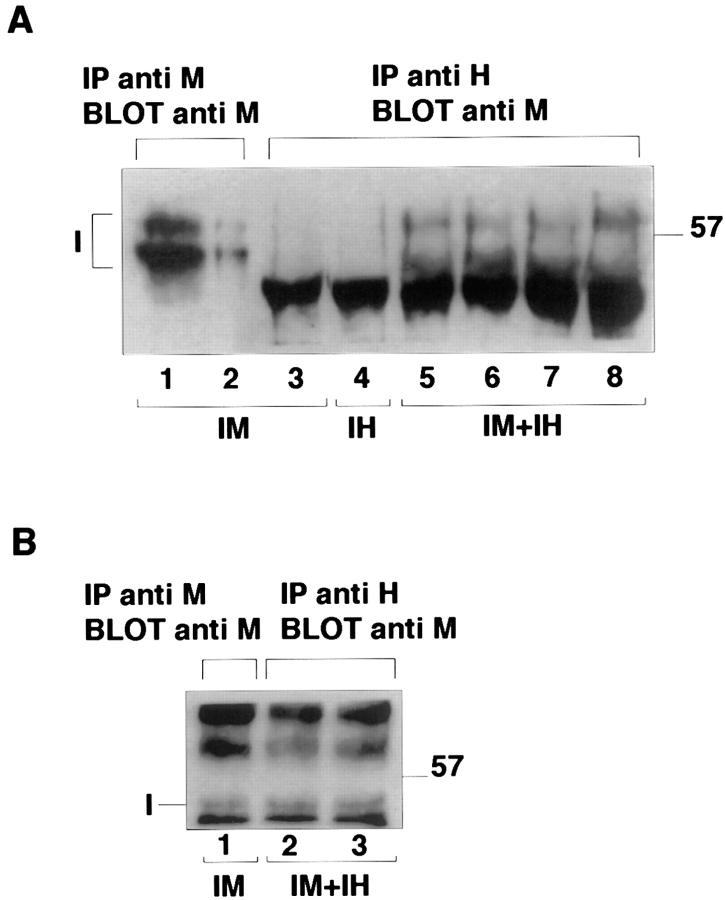
Figure 4.
Coimmunoprecipitation of TβRI-myc with TβRI-HA. COS7 cells were transiently transfected with TβRI-myc (IM), TβRI-HA (IH), or both (IM+IH), as indicated at the bottom of each lane. Immunoprecipitation was either with α-myc or α-HA, as indicated at the top of the gel. Blotting was always with α-myc, followed by biotinylated GαM, streptavidin–horseradish peroxidase, and ECL. All lanes were loaded with immunoprecipitates derived from the same amounts of cell lysates (three 10-cm dishes), except where both immunoprecipitation and blotting were with α-myc (1/3 of the above lysate in A, lane 1; 1/18 in A, lane 2 and in B, lane 1). (A) Effects of ligand and/or DTT treatment. The gels were run under reducing conditions. In lanes 6 and 8, the cells were preincubated with 250 pM TGF-β1 (2 h, 4°C). In lanes 7 and 8, the cells were pre-treated with DTT (see Materials and Methods). TβRI appears in two bands, around 59 and 53 kD. The nonspecific band at the bottom of lanes 3–8 is a component of the α-HA antibodies used for the immunoprecipitation in these lanes (see text). (B) Effect of N-glycosidase F. The 59-kD band collapses to 53 kD, indicating that it represents N-glycosylated receptors. The immunoprecipitates were treated with N-glycosidase F as described under Materials and Methods. The gel was run under nonreducing conditions. In lane 3, the cells were preincubated with 250 pM TGF-β1. The band seen at 62 kD under these conditions is a nonspecific band. (The specific 59-kD band disappears).
Immunofluorescence Copatching Experiments Show Ligand-independent TβRI Homodimers on the Surface of Live COS7 Cells
The experiments described in the former sections suggest that TβRI forms dimer-sized complexes and that at least some of them are caused by homooligomer formation. To measure the TβRI oligomeric structure at the native cell membrane, we performed immunofluorescence copatching studies (for method description see Methods and reference 12). These studies have the advantage that they are performed on live cells (without any possible interference by detergents), measure the bulk of cell surface receptors on coexpressing cells only, and supply a semiquantitative measure for oligomer size. In this method, a tagged receptor at the surface of live, unfixed cells is forced into patches by a double layer of bivalent IgGs. The second receptor, which carries a different tag, is labeled by antibodies coupled to another fluorophore. The samples are then examined to explore whether the second receptor is swept into the same micropatches or whether the two receptors congregate in separate patches. The experiment is performed in the cold to avoid internalization. If the two fluorophores used are Cy3 (red emission) and fluorescein (green emission), then mutual patches appear yellow, while separate patches are either red or green. It is therefore possible to count the percentage of mutual and of separate patches, thus obtaining a semiquantitative determination of the level of complex formation and of complex size (12). It should be noted that the tags (HA and myc) and antibodies used in the current studies do not mediate receptor complex formation or nonspecific sweeping into mutual patches since similar experiments using the same tags and antibodies did not show copatching between TβRII and TβRIII (12).
COS7 cells cotransfected with TβRI-HA and TβRI-myc were subjected to successive incubations with different antibodies to mediate patching and fluorescent labeling of the tagged receptors, as specified under Materials and Methods. Typical results are shown in Fig. 5. The labeling procedure is fully specific, as can be seen in the inset of Fig. 5 A, showing control cells singly transfected with TβRI-myc and subjected to the same labeling procedure; the cells exhibit Cy3 labeling and almost no fluorescein labeling, which would be associated with TβRI-HA. In all cells expressing both TβRI-myc and TβRI-HA, ∼50% of the patches were yellow. This indicates that the majority of TβRI at the cell surface resides in dimers. Statistically, this is the percentage expected to reside in mutual complexes in the case of a dimer since only half of the dimers would carry two different tags (see Discussion). Preincubation with ligand (Fig. 5, B and D) or DTT (Fig. 5 , C and D) did not alter the percentage of mutual (yellow) patches formed by the antibodies. It should be noted that 50% copatching is not a pattern that is commonly observed in such experiments; thus, no copatching was detected for coexpressed TβRII and III (12), and only a low percentage of copatching is observed for TβRI/TβRII in the absence of ligand (Wells, R.G., L. Gilboa, Y.I. Henis, and H.F. Lodish, manuscript in preparation). The results presented in Fig. 5 suggest that all (or most of) the type I receptors appear as ligand-independent dimers on the surface of live cells and that this structure is not altered by preincubation with DTT.
Figure 5.
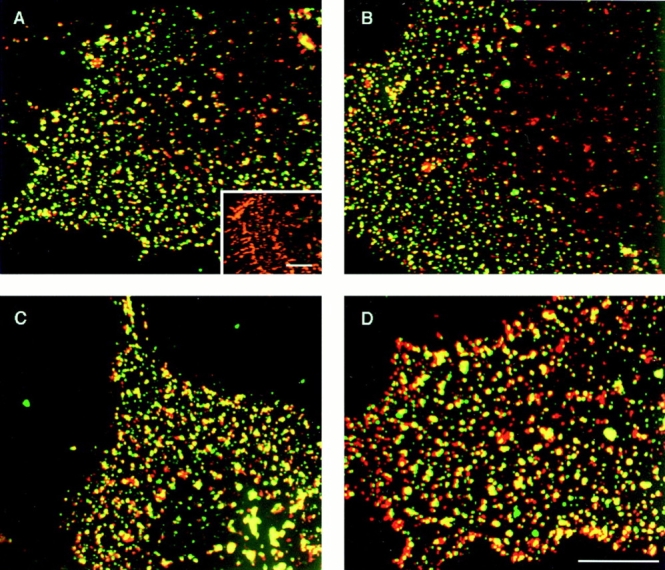
Copatching of epitope-tagged TβRI on the surface of live cells. COS7 cells were transiently cotransfected with TβRI-myc and TβRI-HA (A–D) or with TβRI-myc alone (A, inset). They were labeled in the cold consecutively by a series of antibodies to mediate patching and fluorescent labeling, as described under Materials and Methods. After this labeling procedure, TβRI-myc is labeled by Cy3 (red), TβRI-HA by FITC (green), and patches containing both tags are yellow. The labeling specificity is demonstrated by the exclusive labeling with Cy3 (red) of cells expressing only TβRI-myc (A, inset) and by the existence of separately labeled patches (only red or only green) in the coexpressing cells. For each experiment, the patches on 20–30 cells were counted on the computer screen to determine the percentage of double-labeled (yellow) patches. Flat cell regions were selected for counting, avoiding the region of the nucleus that is out of focus and contains more nonspecific staining. The percentage of copatching was 46 ± 5% in all cases except for the control. (A) No DTT treatment, no preincubation with ligand. (Inset) Transfection with TβRI-myc only. (B) Incubation with 250 pM TGF-β1; the ligand was added together with the normal goat IgG preincubation and kept in during all successive incubations. (C) DTT-treated cells, no ligand. Pretreatment with DTT was done as described under Materials and Methods, before incubations with the antibodies. (D) DTT-treated cells, incubation with 250 pM TGF-β1. Bars, 20 μm.
TβRII Forms Homodimers in the ER
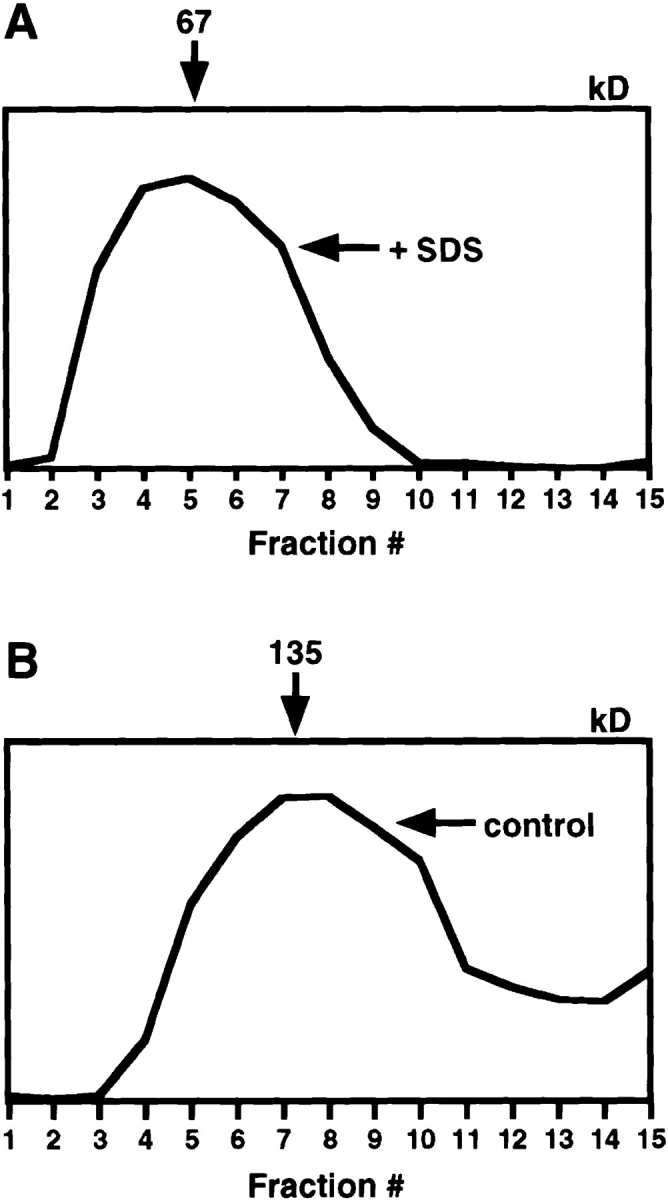
Homooligomerization of TβRII might have an important role in TGF-β signal transduction. We have previously used immunofluorescence copatching to show that TβRII forms small homooligomers on the surface of live COS7 cells (12), similar to TβRI (Fig. 5). To determine whether TβRI homooligomerization occurs in the ER, we performed velocity centrifugation experiments with metabolically labeled, Endo H–sensitive type II receptors. The experiments used L6 cells as described above for TβRI, except that a shorter labeling pulse (15 min) was required to give a TβRII population entirely sensitive to Endo H (Fig. 6 A; reference 41). Velocity centrifugation of TβRII from cells lysed and run in the presence of 0.5% SDS yielded a peak at fractions 4–6 (Fig. 6, B and D), consistent with a monomeric TβRII. Receptors from lysates lacking SDS migrated with a peak at 150 kD (Fig. 6, C and D), consistent with the ER form being a dimer. Metabolically labeled Endo H–resistant receptor, measured after a 2 h chase, migrated at the same point (data not shown), consistent with our immunofluorescence copatching experiments in COS7 cells (12). Similar results were obtained with COS7 cells transiently expressing TβRII (tagged or untagged) as well as with Mv1Lu (not shown), confirming that dimerization of TβRII is not cell type specific. These findings show that the type II receptors form homodimers in the ER, which are then transported, presumably in this form, to the cell surface.
Figure 6.
The type II receptor forms homodimers in the ER. L6 cells were pulse-labeled with 35S-Express for 15 min, chilled, lysed with (B) or without (C) 0.5% SDS, and loaded on sucrose gradients with (B) or without (C) 0.5% SDS, as in Fig. 1. D is a graphic representation of the data as in Fig. 1. Endo H digestion of control material (A), labeled similarly, confirms that all labeled receptor is in the ER form (Endo H sensitive).
The Ectodomains of TβRII and TβRI Are Monomeric, but TGF-β1 Can Induce Dimerization of II-SF
To determine whether the ectodomain of either signaling receptor is responsible for the homodimerization we have observed, we performed sucrose gradient velocity centrifugation studies on the secreted ectodomains of both receptors. The soluble exoplasmic domain of TβRII is efficiently secreted and has binding characteristics equivalent to the full-length receptor (24). I-SF is also efficiently secreted, suggesting that it is correctly folded; however, it cannot be cross-linked to ligand in the presence or absence of II-SF and likely requires transmembrane and cytoplasmic domains for full interaction with the type II receptor and for ligand binding (Wells, R.G., L. Gilboa, Y.I. Henis, and H.F. Lodish, manuscript in preparation). COS7 cells were transfected with a plasmid carrying an epitope-tagged form of the soluble type II receptor (II-SF), and the media from metabolically labeled cells were separated on detergent-free sucrose gradients. In the absence of ligand, II-SF peaked at fractions 3–4 (Fig. 7, A and C), predicted by size markers to be 20 kD, consistent with a monomer. The truncated receptor has complex and heterogeneous N-linked glycosylation, accounting for the multiple labeled bands. The same receptor, when collected from the media of unlabeled cells and cross-linked to 125I–TGF-β1, migrated as a 60–65-kD protein (Fig. 7, B and C), consistent with a dimeric receptor bound to the ligand. This dimerization did not occur if detergent (1% Triton X-100) was added (not shown).
Figure 7.
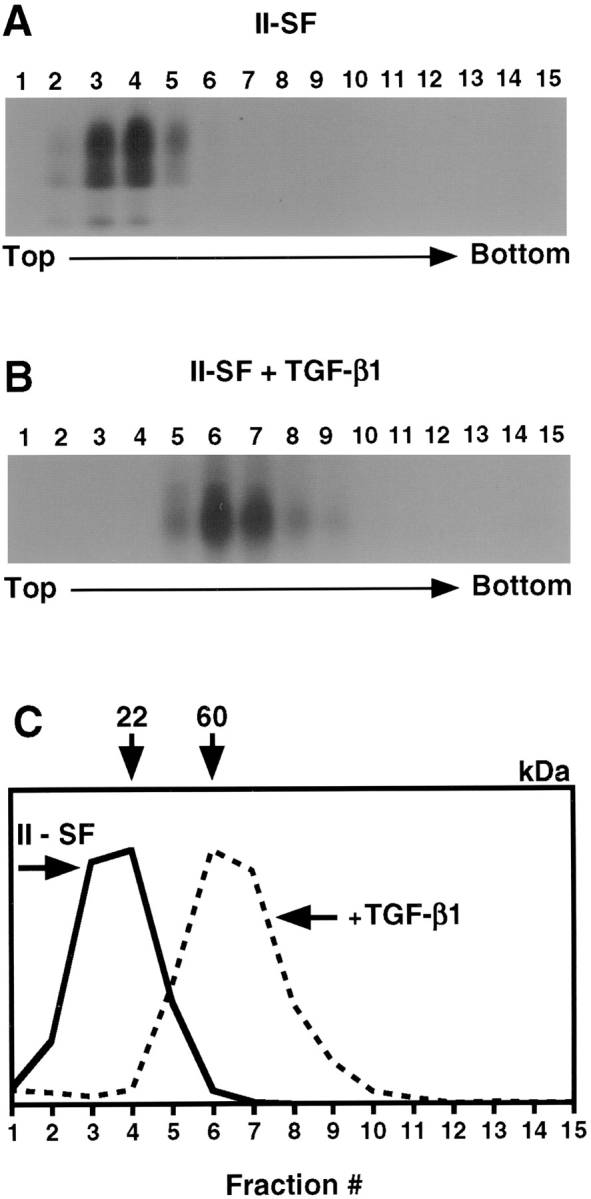
The ectodomain of TβRII is a monomer and dimerizes upon TGF-β1 binding. (A) COS7 cells were transfected with II-SF. 48 h after transfection, they were incubated in starvation medium and labeled with 35S-Express, and the media collected from the cells were concentrated and centrifuged on 5–15% sucrose gradients without detergent (see Materials and Methods). Fractions were immunoprecipitated with α-HA, and immunoprecipitates were analyzed by SDS-PAGE. (B) Unlabeled media from transfected COS7 cells were collected and cross-linked to iodinated TGF-β1, as described under Materials and Methods. Samples were loaded on sucrose gradients and processed as for A. (C) A graphical representation of the quantitation of bands, as for Figs. 1 and 6.
Analogous experiments were conducted on the soluble form of TβRI (I-SF), described under Materials and Methods. The results are depicted in Fig. 8. Metabolically labeled I-SF migrated with a peak at fractions 3–6 (Fig. 8), consistent with a monomer of molecular mass around 24 kD. The broad peak suggests that some I-SF migrated faster, and there could be some I-SF dimers present. These results demonstrate that the exoplasmic domains of both TβRI and TβRII are mainly monomeric and are not sufficient for dimerization by themselves. II-SF, however, can form dimers upon TGF-β1 binding.
Figure 8.
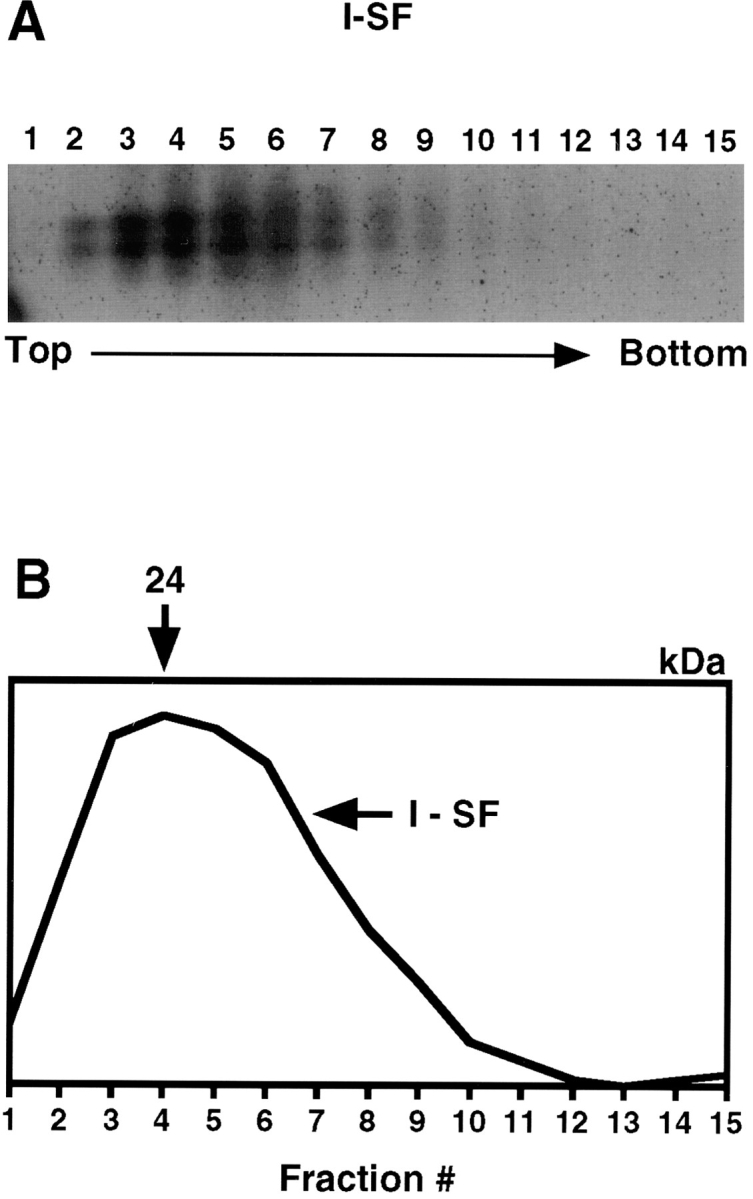
The ectodomain of TβRI is monomeric. (A) COS7 cells were transfected with I-SF and metabolically labeled, and the media collected from the cells were centrifuged on 5–15% sucrose gradients without detergent as described in Fig. 7 A. Immunoprecipitation and SDS-PAGE were as in Fig. 7. (B) Graphical representation of the quantified bands seen in A.
Discussion
TGF-β elicits a broad spectrum of effects, ranging from control of cell growth and extracellular matrix composition to differentiation and cell death (37). TGF-β signal transduction requires two cell surface receptors, TβRI and TβRII, which are serine-threonine kinases (9, 24). A substantial amount of evidence attests to interactions between TβRI and TβRII upon ligand binding (3, 7, 27, 43, 44). Homomeric interactions of TGF-β receptors, however, have not been investigated in detail, especially for TβRI. Although there are clear indications that homomeric interactions of type I and type II TGF-β receptors are important functionally for signal transduction and regulation, there has thus far been no physical evidence for TβRI homooligomerization, and the cellular site of TβRII homooligomerization is not known (6, 12, 27, 28, 40). In the current studies, we combined several complementary approaches to investigate this issue: (a) sedimentation velocity studies to determine the size of metabolically labeled receptor complexes; (b) coimmunoprecipitation experiments on coexpressed, differentially tagged forms of TβRI to explore homooligomer formation; and (c) immunofluorescence copatching studies on cells coexpressing differentially tagged TβRI forms to establish the existence of receptor homodimers at the surface of intact cells.
Sedimentation velocity experiments on metabolically labeled TβRI demonstrated that the receptors migrate as a complex whose size corresponds to a dimer. This complex was observed in the ER of several cell types, from naturally expressing L6 (Fig. 1) or Mv1Lu cells to transiently expressing COS7 cells (Fig. 2). This indicates that the dimer-sized complex formed is not affected by the level of expression. The fact that the complex size fits a TβRI dimer does not necessarily point to the existence of such dimers since some other cellular protein of a similar size can be associated with the receptor. This issue can be resolved by the use of other methods (coimmunoprecipitation and immunofluorescence copatching) to demonstrate that TβRI does indeed form homodimers and to establish independently the existence of TβRI dimers at the cell surface. The latter experiments require the expression of epitope-tagged receptors at levels high enough for good fluorescence visualization, and they were therefore performed in COS7 cells. The use of the overexpressing COS cells is justified in view of the similar complex size found in centrifugation studies for TβRI expressed transiently in COS cells or naturally in the other cell types. Furthermore, similar results were obtained in the copatching studies on COS7 cells expressing either high (∼200,000 receptors/cell at the surface) or low (∼20,000) receptor levels (determined as described in reference 12).
The coimmunoprecipitation studies (Fig. 4) using tagged TGF-β type I receptors clearly demonstrate physical association between TβRI polypeptides. Thus, the complexes formed are relatively stable and withstand, at least partially, the detergent solubilization and immunoprecipitation conditions. Together with the centrifugation velocity studies, which point to a dimer-sized complex, these results suggest that the complex is indeed a homodimer of TβRI.
Coimmunoprecipitation experiments do not provide a measure for the percentage of receptors that reside in oligomers since only part of the cell population (∼40%, as determined by immunofluorescence) coexpresses the two differently tagged receptors. Immunofluorescence copatching studies circumvent this problem by selecting only coexpressing cells for analysis; they also provide an independent means to determine the size of the receptor oligomers and, most importantly, do so at the surface of live cells. By coexpressing HA- and myc-tagged TβRI, we have observed that ∼50% of the patches were dyed yellow (Fig. 5), implying that the majority of TβRI at the surface of COS7 cells is homodimeric. The reasoning is as follows: For monomers, each type of tagged receptor would be swept into separate patches by the respective antibody, yielding no copatching. In the case of a dimer, only half of the dimers would be composed of receptors carrying different tags; these will be swept mostly into copatches, while the great majority of uniformly tagged dimers will reside in either green or red patches. For trimers and tetramers, the expected percentage of complexes carrying different tags is 75 and 87.5%, respectively. Clearly, the results of the copatching experiments are in good agreement with the receptors being dimeric. They provide an independent proof for the homodimeric nature of the dimer-sized complex observed in the centrifugation experiments.
The copatching experiments, as well as the coimmunoprecipitation studies, show that TβRI dimers are unaffected by the presence of TGF-β1 (Figs. 4 and 5); this is not surprising since TβRI does not bind TGF-β1 in the absence of TβRII. Interestingly, the dimeric structure of TβRI is not disrupted by pretreatment of the cells with DTT under conditions known to eliminate TGF-β1 binding to these receptors (Figs. 4 and 5). These findings demonstrate that the homooligomeric complex of the type I receptors can withstand whatever structural alteration is mediated by the DTT treatment, and which interferes with the ability of TβRI to be cross-linked to TGF-β1 in the presence of TβRII (reference 4; Rodriguez, C., R. Lin, P.E. Scherer, and H.F. Lodish, unpublished results).
Sedimentation velocity studies analogous to those performed on TβRI were conducted on TβRII (Fig. 6). These experiments establish that TβRII appears in dimer-sized complexes, which are formed in the ER. Together with previous studies using immunofluorescence copatching (12) and coimmunoprecipitation (6, 12), which demonstrate that TβRII chains interact with each other, we can conclude that TβRII appears both in the ER and at the cell surface as a dimer. The copatching studies (12) indicate that the dimeric nature of TβRII is not altered after TGF-β1 binding. Moreover, the inherent tendency of TβRII to form dimers is independent of expression level, as indicated by the similar results obtained in sedimentation velocity studies performed on naturally expressing cell lines and on overexpressing COS cells.
To explore the role of the exoplasmic domain in promoting homodimerization, the soluble exoplasmic domains of TβRI and TβRII secreted by COS7 cells were analyzed by sedimentation velocity experiments (Figs. 7 and 8). In both cases, the receptors migrated primarily as monomers, indicating that these domains are insufficient to promote dimerization of either receptor. TGF-β1 induced dimerization of II-SF (Fig. 7), which binds ligand (24), in agreement with a report that TGF-β2 and TGF-β3 also promote dimerization of the ectodomain (20). The ligand-mediated homomeric interactions of II-SF, however, are disrupted by the presence of nonionic detergent, indicating that they are significantly weaker than in wild-type TβRII (data not shown). Accordingly, the intact TβRII is dimeric in the absence of ligand and does not require ligand binding for dimerization. An immediate conclusion is that the cytoplasmic and/or transmembrane domains of TβRII must have a major contribution to the homomeric interactions. This conclusion is in accord with the report (6) that the cytoplasmic domains of TβRII overexpressed in 293 cells can form homooligomers. There is, however, evidence that the ectodomains of both receptors also contribute to the homomeric interactions since chimeras composed of the cytoplasmic and transmembrane regions of TβRI or II and the ectodomain of the Epo receptor required dimerization by Epo to interact (27, 28). Taken together, it seems likely that several domains scattered throughout the receptors contribute to the homomeric interactions. In the case of TβRII, it appears that ligand binding can enhance the interactions between the ectodomains.
In conclusion, we have combined several methods to characterize homooligomer formation of TβRI and TβRII. The findings demonstrate that both receptors exist as homodimers that form in the ER and persist at the cell surface. The roles of homooligomerization and its involvement in the mechanism(s) of TGF-β signaling should be further explored. It appears, however, that for TβRI the homodimerization has a critical role in signal transduction (27, 40), and for TβRII it is involved in modulating signaling (28). The variety of biological responses mediated by TGF-β ligands raises the possibility that an intricate equilibrium between various homo- and heterocomplexes, which can be affected by the binding of various ligands, plays a role in regulating those responses.
Footnotes
1. Abbreviations used in this paper: α-HA, mouse monoclonal antibody which recognizes a specific epitope of the HA protein; α-myc, mouse monoclonal antibody which recognizes a specific c-myc sequence; α-IIC, a polyclonal rabbit antiserum raised against the COOH-terminal 16 amino acids of TβRII; I-SF, soluble HA-tagged TβRI ectodomain; II-SF, soluble HA-tagged TβRII ectodomain; ECL, enhanced chemiluminescence; Endo H, endoglycosidase H; Epo, erythropoietin; GαM, goat IgG anti– mouse IgG; HA, influenza hemagglutinin; Octyl-POE, n-octyl-polyoxyethylene; TGF-β, transforming growth factor β; TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor.
We thank Professor Z. Kam (Weizmann Institute, Israel) for his invaluable help with image analysis, and Orit Gutman (Tel Aviv University) and Haya Yankelev (Whitehead Institute) for expert technical assistance.
This research was supported in part by a grant from the Israel Science Foundation administered by the Israel Academy of Sciences and Humanities (to Y.I. Henis), by a project grant from the Israel Cancer Research Fund (Y.I. Henis), and by National Institutes of Health Grants CA63260 (to H.F. Lodish) and DK02290 (to R.G. Wells).
Lilach Gilboa and Rebecca G. Wells contributed equally to this work.
Address all correspondence to Harvey F. Lodish, The Whitehead Institute, Nine Cambridge Center, Cambridge, MA 02142. Tel.: (617) 258-5216. Fax: (617) 258-6768. E-mail: lodish@wi.mit.edu
References
- 1.Attisano L, Wrana JL. Signal transduction by members of the transforming growth factor-β superfamily. Cytokine Growth Factor Rev. 1996;7:327–339. doi: 10.1016/s1359-6101(96)00042-1. [DOI] [PubMed] [Google Scholar]
- 2.Attisano L, Wrana JL, Lopez-Casillas F, Massague J. TGF-β receptors and actions. Biochim Biophys Acta. 1994;1222:71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 3.Bassing CH, Yingling JM, Howe DJ, Wang T, He WW, Gustafson ML, Shah P, Donahoe PK, Wang X-F. A transforming growth factor β type I receptor that signals to activate gene expression. Science. 1994;263:87–89. doi: 10.1126/science.8272871. [DOI] [PubMed] [Google Scholar]
- 4.Cheifetz S, Massague J. Isoform-specific transforming growth factor-β binding proteins with membrane attachments sensitive to phosphatidylinositol-specific phospholipase C. J Biol Chem. 1991;266:20767–20772. [PubMed] [Google Scholar]
- 5.Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 6.Chen R-H, Derynck R. Homomeric interactions between type II transforming growth factor-β receptors. J Biol Chem. 1994;269:22868–22874. [PubMed] [Google Scholar]
- 7.Chen R-H, Moses HL, Maruoka EM, Derynck R, Kawabata M. Phosphorylation-dependent interaction of the cytoplasmic domains of the type I and type II transforming growth factor-β receptors. J Biol Chem. 1995;270:12235–12241. doi: 10.1074/jbc.270.20.12235. [DOI] [PubMed] [Google Scholar]
- 8.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franzen P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin CH, Miyazono K. Cloning of a TGFβ type I receptor that forms a heteromeric complex with the TGFβ type II receptor. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- 10.Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. 726 pp.
- 11.Henis YI, Gutman O, Loyter A. Sendai virus envelope glycoproteins become laterally mobile on the surface of human erythrocytes following fusion. Exp Cell Res. 1985;160:514–526. doi: 10.1016/0014-4827(85)90198-3. [DOI] [PubMed] [Google Scholar]
- 12.Henis YI, Moustakas A, Lin HY, Lodish HF. The types II and III transforming growth factor-β receptors form homo-oligomers. J Cell Biol. 1994;126:139–154. doi: 10.1083/jcb.126.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki M, Moustakas A, Lin HY, Lodish HF, Carr BI. Growth inhibition by transforming growth factor β (TGF-β) receptor type I is restored in TGF-β-resistant cells after expression of TGF-β receptor type II cDNA. Proc Natl Acad Sci USA. 1993;90:5359–5363. doi: 10.1073/pnas.90.11.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kam, Z., H. Chen, J.W. Sedat, and D.A. Agard. 1992. Analysis of three- dimensional image data: display and feature tracking. In Electron Tomography: Three-dimensional Imaging with the Transmission Electron Microscope. J. Frank, editor. Plenum Press, New York. 237–256.
- 15.Kam Z, Jones MO, Chen H, Agard DA, Sedat JW. Design and construction of an optimal illumination system for quantitative wide-field multi-dimensional microscopy. Bioimaging. 1993;1:71–81. [Google Scholar]
- 16.Kolodziej PA, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 18.Kurkela R, Vuolas L, Vihko P. Preparation of F (ab′)2fragments from monoclonal mouse IgG1 suitable for use in radioimaging. J Immunol Methods. 1988;110:229–236. doi: 10.1016/0022-1759(88)90108-1. [DOI] [PubMed] [Google Scholar]
- 19.Laiho M, Weis FM, Boyd FT, Ignotz RA, Massague J. Responsiveness to transforming growth factor-β restored by complementation between cells defective in TGF-β receptors I and II. J Biol Chem. 1991;266:9108–9112. [PubMed] [Google Scholar]
- 20.Letourneur O, Goetschy JF, Horisberger M, Grutter MG. Ligand-induced dimerization of the extracellular domain of the TGF-β receptor type II. Biochem Biophys Res Commun. 1996;224:709–716. doi: 10.1006/bbrc.1996.1088. [DOI] [PubMed] [Google Scholar]
- 21.Lin HY, Lodish HF. Receptors for the TGF-β superfamily: multiple polypeptides and serine/threonine kinases. Trends Cell Biol. 1993;3:14–19. doi: 10.1016/0962-8924(93)90195-7. [DOI] [PubMed] [Google Scholar]
- 22.Lin HY, Moustakas A. TGF-β receptors: structure and function. Cell Mol Biol. 1994;40:337–349. [PubMed] [Google Scholar]
- 23.Lin HY, Wang X-F, Ng-Eaton E, Weinberg RA, Lodish HF. Expression cloning of the TGF-β type II receptor, a functional transmembrane serine/threonine kinase. Cell. 1992;68:775–785. doi: 10.1016/0092-8674(92)90152-3. [DOI] [PubMed] [Google Scholar]
- 24.Lin HY, Moustakas A, Knaus P, Wells RG, Henis YI, Lodish HF. The soluble exoplasmic domain of the type II transforming growth factor (TGF)-β receptor. A heterogeneously glycosylated protein with high affinity and selectivity for TGF-β ligands. J Biol Chem. 1995;270:2747–2754. doi: 10.1074/jbc.270.6.2747. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Casillas F, Cheifetz S, Doodey J, Andres JL, Lane WS, Massague J. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF–β receptor system. Cell. 1991;67:785–795. doi: 10.1016/0092-8674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF-β signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 27.Luo K, Lodish HF. Signaling by chimeric erythropoietin-TGF-β receptors: homodimerization of the cytoplasmic domain of the type I TGF-β receptor and heterodimerization with the type II receptor are both required for intracellular signal transduction. EMBO (Eur Mol Biol Organ) J. 1996;15:4485–4496. [PMC free article] [PubMed] [Google Scholar]
- 28.Luo K, Lodish HF. Positive and negative regulation of type II TGF-β receptor signal transduction by autophosphorylation on multiple serine residues. EMBO (Eur Mol Biol Organ) J. 1997;16:1970–1981. doi: 10.1093/emboj/16.8.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 30.Massague J. Receptors for the TGF-β family. Cell. 1992;69:1067–1070. doi: 10.1016/0092-8674(92)90627-o. [DOI] [PubMed] [Google Scholar]
- 31.Massague J. TGFβ signaling: receptors, transducers, and Mad proteins. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 32.Massague J, Weis-Garcia F. Serine/threonine kinase receptors: mediators of transforming growth factor β family signals. Cancer Surv. 1996;27:41–64. [PubMed] [Google Scholar]
- 33.Massague J, Attisano L, Wrana JL. The TGF-β family and its composite receptors. Trends Cell Biol. 1994;4:172–178. doi: 10.1016/0962-8924(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell EJ, Lee K, O'Connor-McCourt MD. Characterization of transforming growth factor-β (TGF-β) receptors on BeWo choriocarcinoma cells including the identification of a novel 38-kDa TGF-β binding glycoprotein. Mol Biol Cell. 1992;3:1295–1307. doi: 10.1091/mbc.3.11.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moses HL, Yang EY, Pietenpol JA. TGF-β stimulation and inhibition of cell proliferation: new mechanistic insights. Cell. 1990;63:245–247. doi: 10.1016/0092-8674(90)90155-8. [DOI] [PubMed] [Google Scholar]
- 36.Moustakas A, Lin HY, Henis YI, Plamondon J, O'Connor MD, McCourt, Lodish HF. The transforming growth factor β receptors types I, II, and III form hetero-oligomeric complexes in the presence of ligand. J Biol Chem. 1993;268:22215–22218. [PubMed] [Google Scholar]
- 37.Roberts, A.B., and M.B. Sporn. 1990. The transforming growth factor- betas. In Peptide Growth Factors and Their Receptors. M.B. Sporn and A.B. Roberts, editors. Springer-Verlag, Heidelberg. 419–472.
- 38.Seed B, Aruffo A. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X-F, Lin HY, Ng-Eaton E, Downward J, Lodish HF, Weinberg RA. Expression cloning and characterization of the TGF-β type III receptor. Cell. 1991;67:797–805. doi: 10.1016/0092-8674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 40.Weis-Garcia F, Massague J. Complementation between kinase-defective and activation-defective TGF-β receptors reveals a novel form of receptor cooperativity essential for signaling. EMBO (Eur Mol Biol Organ) J. 1996;15:276–289. [PMC free article] [PubMed] [Google Scholar]
- 41.Wells RG, Yankelev H, Lin HY, Lodish HF. Biosynthesis of the type I and type II TGF-β receptors. Implications for complex formation. J Biol Chem. 1997;272:11444–11451. doi: 10.1074/jbc.272.17.11444. [DOI] [PubMed] [Google Scholar]
- 42.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 43.Wrana JL, Attisano L, Carcamo J, Zentella A, Doodey J, Laiho M, Wang X-F, Massague J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 44.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]