Abstract
Abstract. Five mammalian members of the gp25L/ emp24/p24 family have been identified as major constituents of the cis-Golgi network of rat liver and HeLa cells. Two of these were also found in membranes of higher density (corresponding to the ER), and this correlated with their ability to bind COP I in vitro. This binding was mediated by a K(X)KXX-like retrieval motif present in the cytoplasmic domain of these two members. A second motif, double phenylalanine (FF), present in the cytoplasmic domain of all five members, was shown to participate in the binding of Sec23 (COP II). This motif is part of a larger one, similar to the F/YXXXXF/Y strong endocytosis and putative AP2 binding motif. In vivo mutational analysis confirmed the roles of both motifs so that when COP I binding was expected to be impaired, cell surface expression was observed, whereas mutation of the Sec23 binding motif resulted in a redistribution to the ER. Surprisingly, upon expression of mutated members, steady-state distribution of unmutated ones shifted as well, presumably as a consequence of their observed oligomeric properties.
The elucidation of the transport machinery of the secretory pathway has benefited from both biochemical and genetic approaches (Rothman and Wieland, 1996; Schekman and Orci, 1996). This has allowed for the identification of two types of transport vesicles that can be distinguished by their coat, coat protein (COP)1 I and II coatomer. Whereas COP I vesicles can be seen budding from membranes throughout the pathway, COP II vesicles are formed exclusively at ER exit sites (e.g., transitional elements) where they export packed and condensed cargo from the ER (Aridor and Balch, 1996; Bednarek et al., 1996). After budding, COP II vesicles quickly shed their coat (Bednarek et al., 1996) and are subsequently delivered to (or transformed into) a tubular vesicular network usually referred to as the ER-to-Golgi intermediate compartment (ERGIC) (Schweizer et al., 1990). This intermediate compartment is transported along microtubules towards the more centrally located Golgi apparatus where it forms part of the cis-Golgi network (CGN) (Saraste and Svensson, 1991; Presley et al., 1997). During this process, budding of COP I transport vesicles takes place, and these either bring cargo forward (Rothman and Wieland, 1996) and/or recycle components in a retrograde manner (Pelham, 1994; Bednarek et al., 1996). The COP I coatomer has a predominant concentration on and around the CGN, but can also be seen in later compartments such as the trans cisterna of the Golgi and the TGN (Oprins et al., 1993) as well as in the endocytic pathway (Whitney et al., 1995).
Recruitment of coatomer onto membranes argues for the presence of coat binding proteins and candidates for these have recently emerged, mainly through the realization that COP I coatomer directly binds to the ER retrieval motif, K(X)KXX, present in some resident proteins of the ER and the ERGIC. This motif, first identified in a viral protein, E3/19k, encoded by the human adenovirus 2, suffices to maintain reporter molecules in the ER through a constant retrieval from post-ER compartments. It was shown to be functional only if its second lysine was in a −3 position, and addition or deletion of amino acids COOH-terminal to this lysine abolished its ability to fully maintain proteins in the ER (Nilsson et al., 1989; Jackson et al., 1990). Remarkably, even the conservative substitution of lysines to either arginines or histidines led to loss of ER localization, and the same stringent criteria could be demonstrated in its ability to bind to COP I coatomer in vitro (Cosson and Letourneur, 1994). This strong functional correlation between ER retrieval and COP I coatomer binding placed COP I vesicles firmly onto the retrograde pathway and recently, several proteins with K(X)KXX- related motifs have been implicated as major coat binding receptors in vivo. Notably, ERGIC53 (p58 in rat), which is a major constituent of the ERGIC with lectin-like properties displays a functional K(X)KXX in its cytoplasmic domain (Tisdale et al., 1997). Antibodies to this part of the molecule effectively compete for coatomer binding in permeabilized cells, showing that this molecule serves as a major receptor for the coat. Furthermore, such tail antibodies also inhibit anterograde transport, implicating this protein as a facilitator of cargo transport.
In a similar fashion, members of the gp25L/emp24p/p24 (p24) family have also been implicated in bringing cargo forward from the ER and to bind coat proteins via their cytoplasmic domains. The two first members of this family were identified as a calnexin-associated integral membrane protein of the ER; (gp25L; Wada et al., 1991) and an endosome membrane protein in yeast (emp24; Singer-Kruger et al., 1993). Subsequent members were identified through genetic (erv25; Belden and Barlowe, 1996) and biochemical studies (CHOp24; Stamnes et al., 1995) and p23 (Blum et al., 1996; Sohn et al., 1996), as well as homology searches yielding a total of eight members in yeast and at least six in mammalian cells. Many of these display typical K(X)KXX motifs in their cytoplasmic domains, and genetic as well as biochemical studies have established links between these proteins and cargo export from the ER as well as being concentrated in COP II (Schimmöller et al., 1995; Elrod-Erickson and Kaiser, 1996; Belden and Barlowe, 1996) and I transport vesicles (Stamnes et al., 1995; Sohn et al., 1996). Another member, a putative ligand for the T1/ST2 receptor, was isolated as a cell surface protein (Gayle et al., 1996).
The function(s) of p24 members is yet to be demonstrated, but we have here undertaken a comparative study of five mammalian members and show that these proteins are highly abundant and reside, at steady state, in the CGN. Two of these members display typical K(X)KXX-like retrieval motifs and, as predicted, mediate binding of COP I coatomer in vitro. A second motif, composed of the two conserved phenylalanines conserved in all five members, mediates binding of Sec23 (COP II). Mutation of the retrieval-like motif redistributes mutated members as well as nonmutated ones to more distal parts of the secretory pathway including the cell surface. Mutation of the conserved double phenylalanine (FF) motif in only two members equally redistributed mutated and nonmutated family members to an ER-like location. This indicates a reliance on their association with each other to define their steady-state distributions.
Materials and Methods
Membrane Preparation, Subcellular Fractionation, Deglycosylation, Gel Filtration, Rate Zonal Centrifugation and NH2-terminal Sequencing
Membranes were isolated from rat liver and HeLa cells (Sonnichsen et al., 1994) as described previously. For analytical fractionations, rat liver was homogenized as described by Bergeron et al. (1982). The homogenates were subjected to successive centrifugation at 1570 g max for 10 min, the pellet washed once, and then both supernatants were pooled and centrifuged at 280,000 g max for 40 min (fraction multiple large platforms [MLP]). MLP fractions were then loaded onto a 0.5–2.5 M sucrose gradient and centrifuged at 80,000 g av for 18 h. Fractions were collected and analyzed for their content of GalT, calnexin, GMP25, and p58. GalT assays were as described previously (Bergeron et al., 1982). After SDS-PAGE and Western blotting, filters were incubated with antibodies (see below) to reveal corresponding p24 members as well as other endogenous markers. HRP– conjugated secondary antibodies (Tago Inc., Burlingame, CA) using the enhanced chemiluminescence system (Amersham, Buckinghamshire, UK) or 125I-labeled goat anti–rabbit (NEN, Mississauga, CA) were used to visualize immunoblots.
For rate zonal centrifugation, Golgi fractions were solubilized in 20 mM Hepes, pH 6.8, 50 mM NaCl, and 2% sodium cholate. After incubation for 1 h at 4°C, the lysate was centrifugated at 15,000 rpm for 10 min to remove insoluble debris, and then the supernatant was loaded onto a 5–35% sucrose gradient containing 20 mM Hepes, pH 6.8, 50 mM NaCl, and 0.3% sodium cholate, and then centrifugated at 35,000 rpm for 17 h using a SW40 rotor (Beckman Instruments, Palo Alto, CA). Fractions were then collected and subjected to SDS-PAGE and immunoblotting.
Deglycosylation experiments were performed according to the recommendations of the supplier (Boehringer Mannheim, Mannheim, Germany). For pronase treatment, fractions were incubated for 1 h at 4°C with 1 M KCl/0.25 M sucrose/4 mM imidazole (KCl-SI buffer), centrifuged at 15,000 g max for 5 min, and then resuspended in SI buffer. The fractions were then incubated with different amounts of pronase, with or without 0.1% Triton X-100, for 30 min at 0°C. The reaction was stopped by adding Pefebloc (Boehringer Mannheim) and EDTA (final concentrations, 5 and 10 mM, respectively) and boiled for 5 min. Sample buffer was then added, and the samples processed for SDS-PAGE and Western blotting.
For NH2-terminal sequencing, fractions were subjected to Triton X-114 extraction carried out by the method of Bordier (1981), and then subjected to SDS-PAGE and transferred onto polyvinyl idene difluoride (PVDF) membranes. Membranes were rinsed three times with 1 M NaCl, once with water, and then stained for 30 s with 0.1% Coommassie brilliant blue R in 50% methanol and destained. Protein bands of interest were cut out and subjected to NH2-terminal sequence analysis by automated Edman degradation according to the protocol of Hewick et al. (1981). Sequences were obtained by searching the following databases: GenBank, Swiss-Prot, and TIGR.
Cloning
For α2, overlapping expressed sequence tag (EST) nucleotide sequences were used to design the primers used for the synthesis of the PCR probe from reverse transcriptase (RT) rat liver RNA. Total RNA was extracted from rat liver using the method of Chomczynski and Sacchi (1987). Total RNA (4 μg) was used with the RT kit (GIBCO BRL, Gaithersburg, MD). Template cDNA and 100 pmoles of primers were used for the PCR reaction. The products were separated in a 3% agarose gel, and then the band corresponding to the predicted size was excised and electroeluted, followed by ethanol precipitation, and then cloned into the SmaI site of plasmid pTZ19R and sequenced using a (Applied Biosystem, Foster City, CA) sequencing kit. To clone α2, a human brain cDNA was diluted, amplified, and then the plates were rinsed with Luna-Bertani (LB) medium and the phage DNA was precipitated with ethanol. A PCR reaction was carried out using S and AS primer, and the positive pools screened using the RT-PCR fragment labeled by nick translation. One positive clone was then amplified, cut with EcoRI, and then a 1.3-kb fragment was subcloned into the EcoRI site of pTZ19R and sequenced.
For the other p24 family members, overlapping EST nucleotide sequences were used to design primers for nested PCRs. In each case, two sets of primers were designed and used to amplify full-length cDNAs using the Marathon cDNA amplification system (Clonetech Laboratories Inc., Palo Alto, CA). In all cases, a 3′ race was combined with a 5′ race to ensure that full-length cDNAs were obtained. The results of each pair was then used to design a forced-nested PCR to amplify full-length cDNAs from human placenta marathon-ready cDNA. All full-length cDNAs were designed to carry flanking BamHI sites to allow insertion into the pCMUIV expression vector (Nilsson et al., 1989) for sequencing and transient transfection.
Antibodies
Rabbit polyclonal antibodies were generated against synthetic peptides corresponding to internal sequences of each p24 family member. Peptides used were TPGLGMCVEVKDPC (α2), CVEVKDPEDKVILAREY (α2), DVEITGPNDKGIYKGDC (β1), CRLEGPDGKVLYKEM (γ3), CYVEDPQGNTIYRET (γ4), STLEFQVITGGHYDVDC (γ3/4), CKITDSAGHILYSKEDA (δ1), and RHLKSFFEAKKLV (cytoplasmic domain-α2). Peptides were coupled to keyhole limpet hemocyanin (KLH) or BSA carrier and then injected into rabbits together with Freunds Adjuvant (Sigma Chemical Co., Deisenhofer, Germany). Affinity purification was carried out essentially as described (Harlow and Lane, 1985). Rabbit polyclonal and mouse monoclonals against calnexin (Ou et al., 1993), KDEL receptor (Sonnichsen et al., 1994), myc (Evan et al., 1985), and vesicular stomatitis virus (VSV)-G tag (Kreis, 1986), and Sec23 (Paccaud et al., 1996) have been described, as have antibodies against COP I components (Sohn et al., 1996). A mAb against β-COP was used for indirect immunofluorescence (Allan and Kreis, 1986).
COP Binding to Cytoplasmic Domains
Synthetic peptides corresponding to the cytoplasmic domains of p24 family members: (CYLKSFFEAKKLV) (α2), (CYLKRFFEVRRVV) (β1), (CKSFFSDKRTTTTRVGS) (γ3), (CKSFFTEKRPISRAVHS) (γ4), (CYLRRFFKAKKLIE) (δ1); and to control proteins: erd2 (CDFFYLYITKVLKGKKLSLPA), E3/19k (CKYKSRRSFIDEKKMP), E3/19kk/ss (CKYKSRRSFIDESSMP), UDP/GT (CVVKSGDKPSLSARYVVRTGKKGKRD), GalNAc-T1 (MRKFAYC), and Man II (MKLSRQFTVFGC); as well as control peptides C1 (α2-internal peptide), C2 (β1-internal peptide), C3 (γ3/4-internal peptide), C4 (δ1-internal peptide); δ1 mutants (CAARRFFKAKKLIE) (CYLSSFFKAKKLIE) (CYLRRAAKAKKLIE) (CYLRRFFSSKKLIE) (CYLRRFFKASSLIE) (CYLRRFFKAKKAAE) (CYLRRFFKAKKLSS) (CYLRRAAKASSLIE) and E3/19kk/ss mutants; (CKYKSRRSFFDESSMP) (CKYFYRRSFIDESSMP) (CKYKSRRSAADESSMP) (CKYFYRRSFFDESSMP) were coupled to thiopropyl Sepharose beads via cysteines as described by Sohn et al. (1996). Briefly, 1.5 mg of each peptide was incubated with 0.5 ml of thiopropyl Sepharose in coupling buffer (50 mM Tris, 0.5 M NaCl, pH 7.3) overnight. Binding efficiency was estimated by spectrophotometry at 343 nm. Excess free binding groups were blocked by β-mercaptoethanol in blocking buffer (14 μl β-mercaptoethanol in 10 ml of 50 mM NaAc, 0.5 M NaCl, pH 4.5). After coupling, Sepharose beads were stored at 4°C in coupling buffer supplemented with NaN3. For binding, HeLa cells were washed twice in PBS and lysed with 50 mM Hepes, 90 mM KCl, 300 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, pH 7.3, supplemented with a protease inhibitor cocktail. Lysate was precleared by centrifugation for 15 min at ∼17,500 g (Sorval SS-34 at 12,000 rpm; DuPont de Nemours, Bad Homburg, Germany), and then allowed to react with Sepharose beads containing 5 nmol of coupled peptide for 2 h at 4°C. Beads were then washed four or five times with lysate buffer, followed by addition of 100 μl sample buffer, and then heated to 95°C to release bound components. This was followed by SDS-PAGE and Western blot analysis using rabbit polyclonal antibodies against different COP proteins. Bound antibodies were revealed by enhanced chemiluminescence.
Cell Culture, Transfection, and Immunofluorescence
HeLa cells stably expressing N-acetylglucosaminyltransferase (NAGT) I tagged with a myc epitope (Nilsson et al., 1993) were kept in DME (GIBCO BRL, Paisley, Scotland) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and Geneticin (G-418 sulphate) (Sigma Chemical Co., Deisenhofen, Germany) (450 μg/ml). Cells were transfected using the calcium phosphate technique as described previously (Pääbo et al., 1986), except that 30 μg plasmid DNA was used per 10 cm Petri dish of monolayer cells. At 72 h after transfection, cells were fixed and then processed for immunofluorescence as described previously (Nilsson et al., 1993; Peranen et al., 1993). Proteins were visualized by respective primary antibody followed by species-specific secondary antibodies conjugated to either FITC (Tago Inc.) or Texas red (Vector Labs, Inc., Peterborough, United Kingdom). Fluorescent signals were visualized with a 24-red-green-blue (RGB) charge-coupled device (CCD) (Hamamatsu Phototonics, Hamamatsu City, Japan) on an Axiovert 100TV (Carl Zeiss, Inc., Thornwood, NY) using a 100× lens and then recorded directly onto a disk using the Open Lab system (Improvision, Coventry, UK).
Electron Microscopy
Preparation of tissue for cryoimmune EM was carried out as previously described (Dahan et al., 1994). Briefly, male Sprague-Dawley rats (100– 125 g body weight) were fasted overnight and anaesthetized. Livers were perfusion-fixed with Ringer's lactate solution for 30 s, followed by freshly prepared 4% paraformaldehyde/0.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, at a flow rate of 10 ml/min for 10 min. Small pieces of liver were immersed in fixative for 1 h at 4°C, and then washed several times in 4% sucrose/0.1 M phosphate buffer. Liver pieces were then equilibrated, cryoprotected for 30 min–1 h with 2.3 M sucrose as described by Tokuyasu (1980), and then frozen directly in liquid nitrogen. Tissue sectioning, immunolabeling, and contrasting were carried out as described by Dahan et al. (1994), except that grids were floated on drops of 6 M GnHCl (in 50 mM Tris, pH 7.4) for 5 min before labeling. Primary antibodies and antibody– gold conjugates were diluted in PBS, pH 7.4, containing 2% BSA, 2% casein, and 0.5% ovalbumin.
Analysis of Gold Label.
Stubs from two animals were used for the quantitative analysis of labeling for α2. Whole hepatocytes were evaluated on micrographs (final magnification of 14,000×) for the labeling distribution of α2 within secretory compartments. 5,014 gold particles were scored in a total of 18 whole hepatocytes; gold particles were allocated to smooth ER, rough ER, and Golgi apparatus. Gold particle labeling within other cellular compartments (i.e., mitochondria, peroxisomes, lysosomes, multivesicular endosomes, and plasma membrane) was much lower than that observed over secretory apparatus compartments.
Results
Identification of Golgi Membrane Proteins
To identify abundant Golgi integral–membrane proteins, we purified rat hepatic membranes enriched for the Golgi marker, β1,4-galactosyltransferase (GalT) but relatively diminished in endosomal contamination (Dominguez et al., 1997; manuscript submitted for publication). Morphological analysis of the Golgi fraction revealed stacked saccules with a mottled lipoprotein content, especially in saccular distentions that are characteristic (Dahan et al., 1994) of the hepatic Golgi apparatus in situ (Fig. 1 A). In addition, associated tubular vesicular structures and fenestrated elements were evident in preparations evaluated by the random sampling methodology of Baudhuin et al. (1967).
Figure 1.
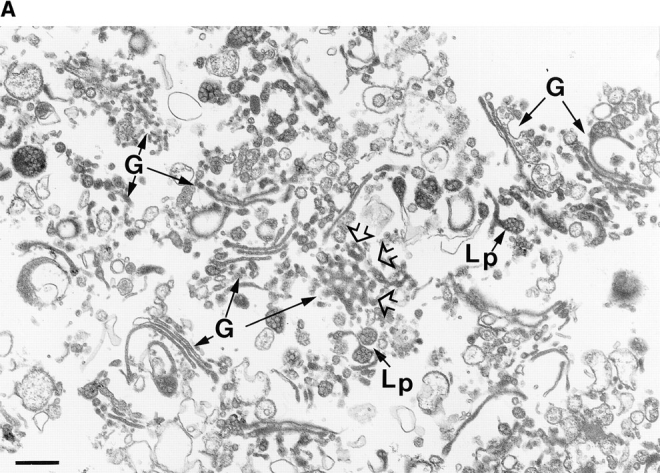
(A) Random view of the purified hepatic Golgi fraction. Random sampling of the fraction (160-fold purification over the homogenate for the Golgi marker GalT) visualized after cross sectioning of filtered fraction reveals an abundance of stacked Golgi saccules (G) with a mottled content (Lp), especially in saccular distensions identified previously in Golgi apparatus in situ as apoE-containing lipoprotein particles (Dahan et al., 1994). Fenestrated structures (open arrowheads) are also evident. Bar, 400 nm. (B) Identification and NH2-terminal sequence of Golgi integral membrane proteins. The membrane proteins of the Golgi fraction were extracted with Triton X-114, subjected to SDS-PAGE, transferred to PVDF membranes, stained by Coomassie brilliant blue, and then processed for NH2-terminal sequencing. Upper and lower case residues represent, respectively, certain and less certain amino acids. Repeated sequencing at five different regions of the broad band, indicated by the square bracket, has revealed only the indicated sequence. Four proteins of the p24 family were identified: Rat p23, the homologue to human p23; rat p24, the rat homologue of CHOp24; rat GMP25, the rat homologue to human GMP25, and a fourth previously uncharacterized family member, p26.
Separation of peripheral from integral membrane Golgi proteins was achieved by Triton X-114 partitioning according to Bordier (1981) and then the proteins were separated by SDS-PAGE (Fig. 1 B). The Triton X-114 detergent phase yielded 41 distinct major bands visible by Coomassie blue staining. These bands were sufficiently pure and abundant to allow their NH2-terminal sequencing (Fig. 1 B). We identified the resident Golgi protein α2,6-sialyltransferase (SialylT), a type II membrane protein marker of very low abundance (Weinstein et al., 1982, 1987), thereby indicating the sensitivity of the method, and apolipoprotein E, a major amphipathic cargo protein of hepatic Golgi apparatus that partitions only partly in the detergent phase. We also partially sequenced four low molecular weight proteins that were predicted members of the p24 family (Fig. 1 B). As estimated from their Coomassie blue staining, these proteins collectively represented a significant proportion of the total integral membrane protein complement of the fraction.
cDNA Cloning, Sequencing, and Proposed Nomenclature
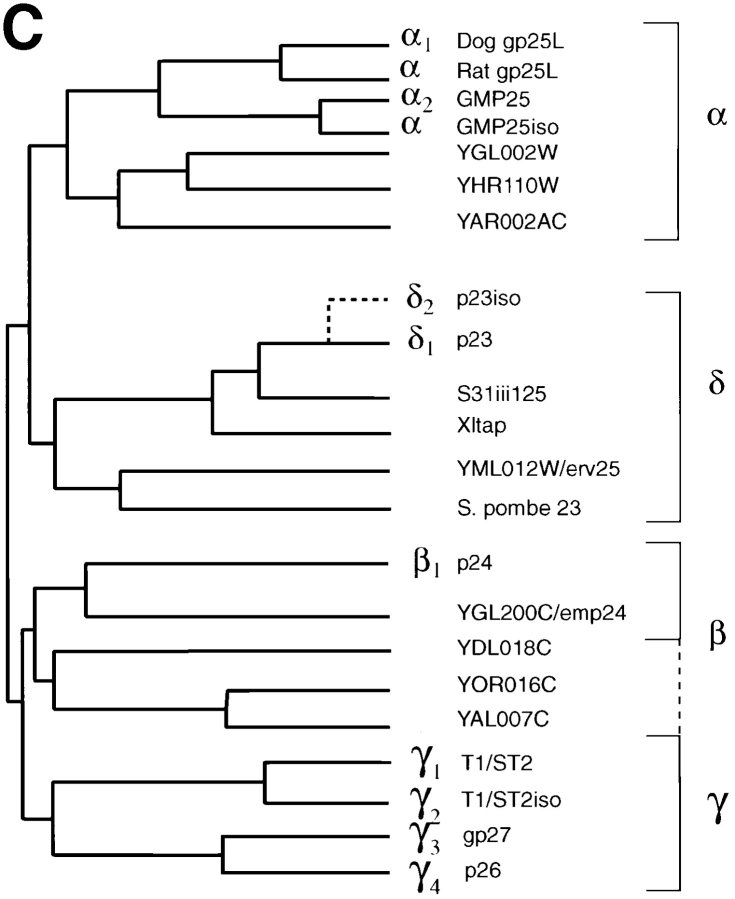
To clone the human cDNA of one of them (GMP25), oligonucleotide primers were synthezised based on overlapping sequences in two of the dbEST entries. These were used to generate a probe from rat liver RNA by RT-PCR. With this probe, screening of a human brain library led to the full-length sequence illustrated in Fig. 2 A, with the rat liver–deduced protein sequence corresponding to residues 1–43 of human GMP25. EST databases aided in the identification of the protein sequences in Fig. 1 B as constituents of the p24 family and also revealed a fifth member, p26. For the other family members, full-length cDNAs were isolated (Fig. 2 B) as described in Materials and Methods. Despite sharing a homology of only 20%, the same amino acids were found at similar positions for all five family members (Fig. 2 B, highlighted by shading) (Blum et al., 1996). Interestingly, most of the conserved amino acids cluster towards the membrane-spanning domain (Fig. 2 A, highlighted by shading) and, when modelled in a helical wheel, face one side. Searching the data base (Fig. 2 C) revealed that these sequences clustered into four major groups. Each group was represented by at least one member found in the purified hepatic Golgi fraction (Fig. 1 B). Two of the sequences (α2 and δ1) revealed the dilysine ER retrieval sequence (Jackson et al., 1990) at their carboxy termini (Fig. 2 B). Referring to their expected molecular weights or database accession numbers does not allow for easy comparison. We suggest they be referred to as p24 preceded by a single letter referring to the species as proposed by Fiedler and co-workers (1996), and subscripted by the Greek letter α, β, γ, or δ, followed by a number according to their position in the cluster tree and their relative time of publication. The human sequence of GMP25 is denoted with number 2 since this member does not relate well to the gp25L (cp24α1). We suggest, therefore, that the ones examined in this study are termed hp24α2 (α2), hp24β1 (β1), hp24δ1 (δ1), hp24γ3 (γ3), and hp24γ4 (γ4) (Fig. 2 C).
Figure 2.
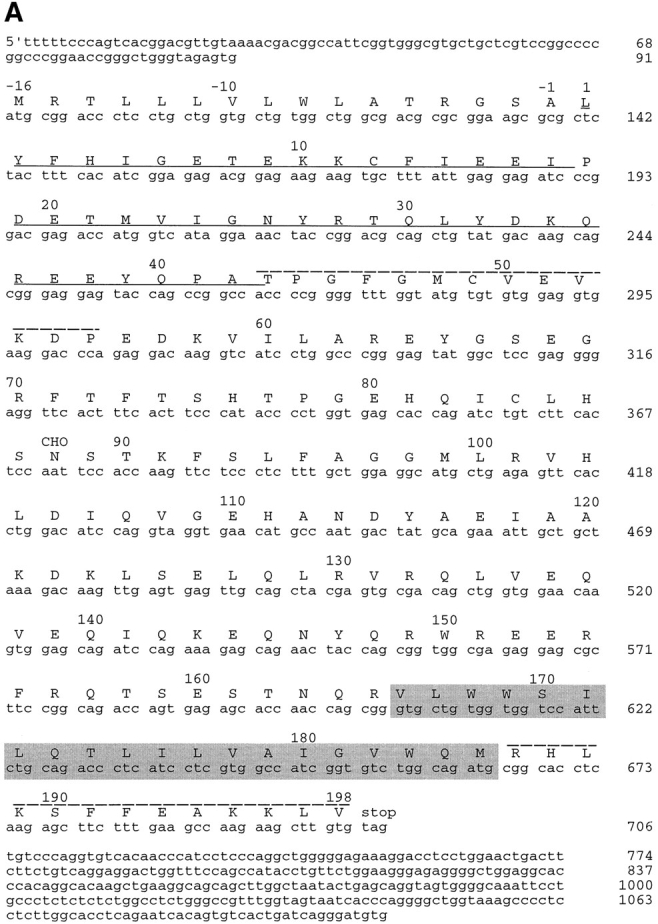
cDNA cloning and comparison with other family members. (A) cDNA sequence and the deduced amino acid sequence of GMP25. Residue +1 represents the first amino acid as determined from the NH2-terminal sequence analysis with the preceding amino acids representing the signal sequence. The underlined sequence corresponds to the sequence amplified by RT-PCR of rat liver total RNA that was then used as a probe to screen the human brain library. The putative transmembrane sequence as deduced from the algorithm of Rost et al. (1995) is highlighted by shading. CHO indicates the single predicted site of N-glycosylation. Peptide sequences to generate antibodies used for subcellular fractionation (residues 43–55) and immunolocalization on cryosections (residues 186–189) are indicated. (B) Comparison of coding sequences. Alignment of obtained sequences was performed using the Multialin Program. Amino acids found to be conserved are highlighted. Sequences to generate further peptide antibodies for immunofluorescence and subcellular fractionation studies in the family of membrane proteins are underlined. (C) Cluster tree. Sequence relationships among the deduced protein sequences was performed using the multiclusteral option of PIMA multisequence alignment based on the amino acid classification scheme described by Smith and Smith (1990). Branches of the deduced tree represented by the four major membrane proteins found in hepatic Golgi fractions are indicated on the right.
Membrane Orientation of α2
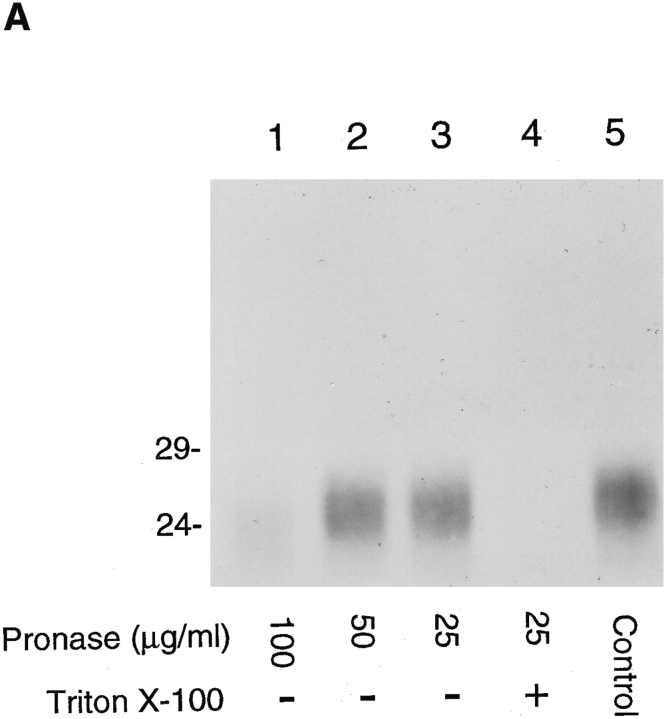
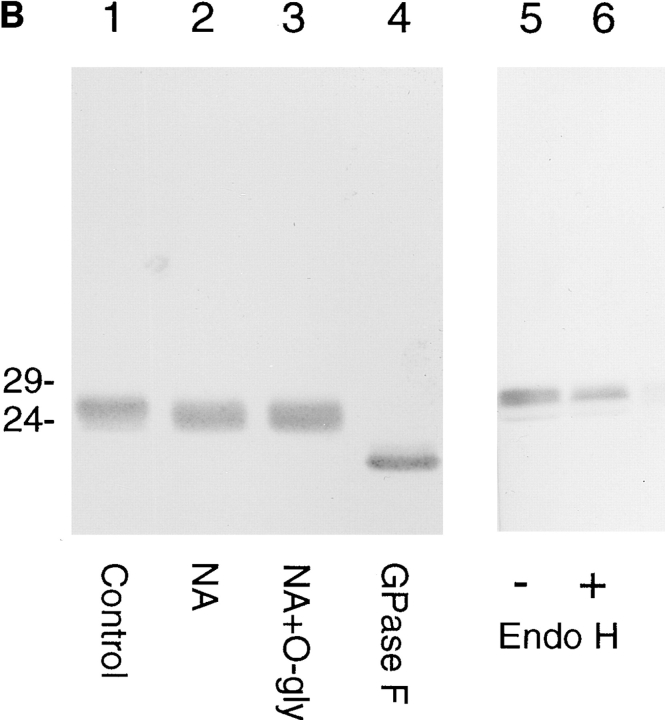
The amino acid sequence of α2 as well as the other family members suggests a type I integral membrane protein with a predicted cleaved signal sequence (von Heijne, 1986), the cleavage of which was confirmed from NH2-terminal sequencing (Fig. 1 B). Synthetic peptides deduced from the sequence of the α2 protein were used to raise antibodies. Protease protection assays and immunoblotting with affinity-purified antibody to residues 43–55 (Fig. 2 A) were used to probe the predicted intraluminal location of these residues of the α2 protein in purified hepatic Golgi fractions. This protein was sensitive to proteinase digestion only in the presence of detergent (Fig. 3 A), showing that the major portion of the molecule resides in the lumen. The sequence of the α2 protein predicted a single site of N-linked glycosylation at residue 88. Digestion with neuraminidase resulted in a slight shift in mobility on SDS-PAGE gels, consistent with terminal sialylation and a post-ER location for the α2 protein (Fig. 3 B). No additional molecular weight shift on SDS-PAGE was affected when samples were treated with both neuraminidase and O-glycosidase (Fig. 3 B), indicating that the protein was unlikely to be O-glycosylated. Treatment with peptide N-glycosyl amidase F (Gpase F) led to an increase in mobility of ∼3-kD consistent with a single site of N-glycosylation. The sensitivity to neuraminidase predicted an endoglycosidase H (endo H)-resistant glycoprotein, and this was confirmed (Fig. 3 B).
Figure 3.
Identification and orientation of the α2 sequence as a type I integral membrane glycoprotein. (A) Orientation of α2 by pronase digestion of hepatic Golgi fractions. 50 μg of Golgi fraction protein were subjected to proteolytic digestion. At a pronase concentration of 25 μg/ml, α2 was completely digested when 0.1% Triton X-100 was added, (lane 4) but unaffected in absence of detergent (lane 3). (B) Glycanase digestion of α2. 50 μg of Golgi fraction protein was untreated (lane 1, Control), treated with neuraminidase (lane 2, NA), neuraminidase and O-glycosidase (lane 3, NA+O+gly), or GpaseF (lane 4, GPase F). Treatment without (lane 5) or with endo H (lane 6) did not affect the mobility of the α2 protein. All immunoblots were with affinity-purified antibody to residues 43–55 of the α2 sequence (refer to Fig. 2 A).
Immunofluorescent Localization
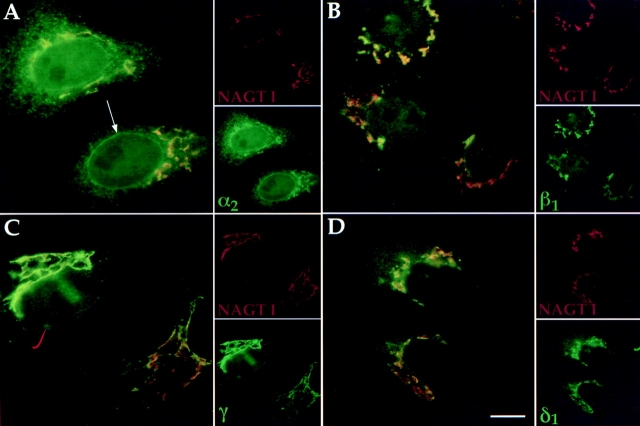
Conventional indirect immunofluorescence was attempted in HeLa cells stably expressing NAGT I with a myc epitope convenient for double labeling (Nilsson et al., 1993). Endogenous staining was weak, and transient expression was necessary for a stronger immunofluorescent signal. All five members were coexpressed, since transfection of individual members resulted in the formation of large, membranous structures of unknown origin (data not shown). It is presently unclear whether this was caused by high levels of expression, or if coexpression was required to circumvent formation of such structures. Upon coexpression, the α2 protein showed an apparent colocalization with the Golgi marker (Fig. 4 A) as well as an apparent nuclear envelope and cytoplasmic reticular pattern typical of the ER. For the β1 (Fig. 4 B), γ3/4 (Fig. 4 C; revealed by antibodies recognizing both forms), and δ1 (Fig. 4 D) members, apparent colocalization with NAGT I was found. Other Golgi markers such as SialylT, β-COP, and syntaxin 5 were also stained, and in all cases revealed an apparent colocalization for the majority of α2, β1, γ3/4 and δ1 immunofluorescence (data not shown).
Figure 4.
Immunofluorescence localization of p24 family members after transient transfection in HeLa cells. The different cDNAs of all hp24 family members were cotransfected into HeLa cells as described in Materials and Methods. At 72 h after transfection, cells were fixed and then processed for indirect immunofluorescence using corresponding and specific primary antibodies. Their localization was compared to the Golgi resident protein NAGT I (red), and, as can be seen, α2 (A, green), β1 (B, green), γ3/4 (C, green), and β1 (D, green) proteins colocalized (yellow) with NAGT I to the Golgi apparatus. An additional localization of α2 to the nuclear envelope is indicated by an arrow in A. Bar, 5 μm.
EM Localization of α2 in Rat Liver Cryosections
The requirement of transfection for clear colocalization necessitated additional approaches. Using affinity-purified antisera to residues 186–198 of the α2 sequence, localization in cryosections of rat liver was attempted. The results revealed antigenicity that was concentrated in smooth tubular membranes (Fig. 5, arrowheads) around Golgi apparatus and along the length of cisternal membranes of rough ER (Fig. 5, A and B, arrows). Quantitation of gold particle labeling within the secretory pathway compartments revealed 59% of labeling within smooth membranous networks, 35 within rough ER cisternae, and 6 within the Golgi apparatus (a total of 5,014 gold particles were counted in cryosections of 18 hepatocytes from livers of two rats). Remarkably, the labeling distribution within Golgi stacks and closely surrounding regions revealed that α2 protein was heterogenously distributed, i.e., being concentrated in membranous tubulovesicular profiles immediately surrounding the Golgi apparatus, and the first saccule on one side of a given Golgi stack (Fig. 5, B–D, see brackets), suggesting a CGN localization. Few Golgi apparatus profiles were labeled over central Golgi saccules.
Figure 5.
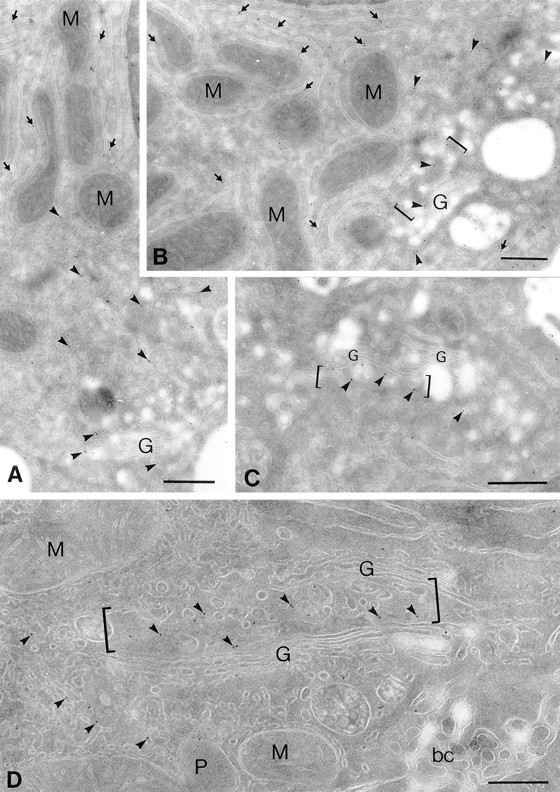
In situ distribution of the α2 protein in rat liver hepatocyte ultrathin cryosections. Cryosections of rat liver hepatocytes were immunolabeled with the rabbit anti-C–terminus antibody against the α2 sequence followed by goat anti– rabbit IgG 10-nm-gold conjugates. Representative profiles of labeling within secretory compartments in liver hepatocytes are shown. Profiles of flattened cisternae of the rough ER are labeled along their length (A and B, arrows); note the gold particles lining mostly the cytosolic surface of the rough ER cisternae where the COOH-terminal domain of the α2 sequence is expected. α2 labeling is distributed throughout tubular smooth membranous profiles (arrowheads) in the Golgi/bile canalicular region of hepatocytes; prominent gold particle labeling can be seen in tubulovesicular membranous profiles closely approaching one side of a given Golgi apparatus (B–D, brackets). Labeling within the Golgi apparatus for the α2 protein is similarly restricted to one of the saccules on one side of a Golgi stack, whereas most other saccules reveal negligible labeling. Mitochondria (M), peroxisomes (P), and bile canaliculus (bc) were largely devoid of labeling. Bars, 400 nm.
Subcellular Distribution
The EM immunolocalization of the α2 sequence suggested a lack of colocalization with Golgi markers. This was tested by subcellular fractionation. Analytical fractionation of rat liver MLP fractions (total membranes minus nuclei; Fig. 6) revealed a median density of the Golgi marker GalT of 1.122, with a distribution different to that of the ER marker calnexin, with a median density of 1.178. The distribution of the intermediate compartment marker p58 (median density of 1.149) was intermediate to that of GalT and calnexin. The overall distribution of the α2 protein was similar to that of p58. However, the α2 protein revealed a slightly greater median density (1.153) and a peak (as determined by quantitative Western blotting) displaced to higher density to that of p58. Hence, as predicted from the EM studies, the α2 protein was not in a compartment coincident with the GalT Golgi marker.
Figure 6.
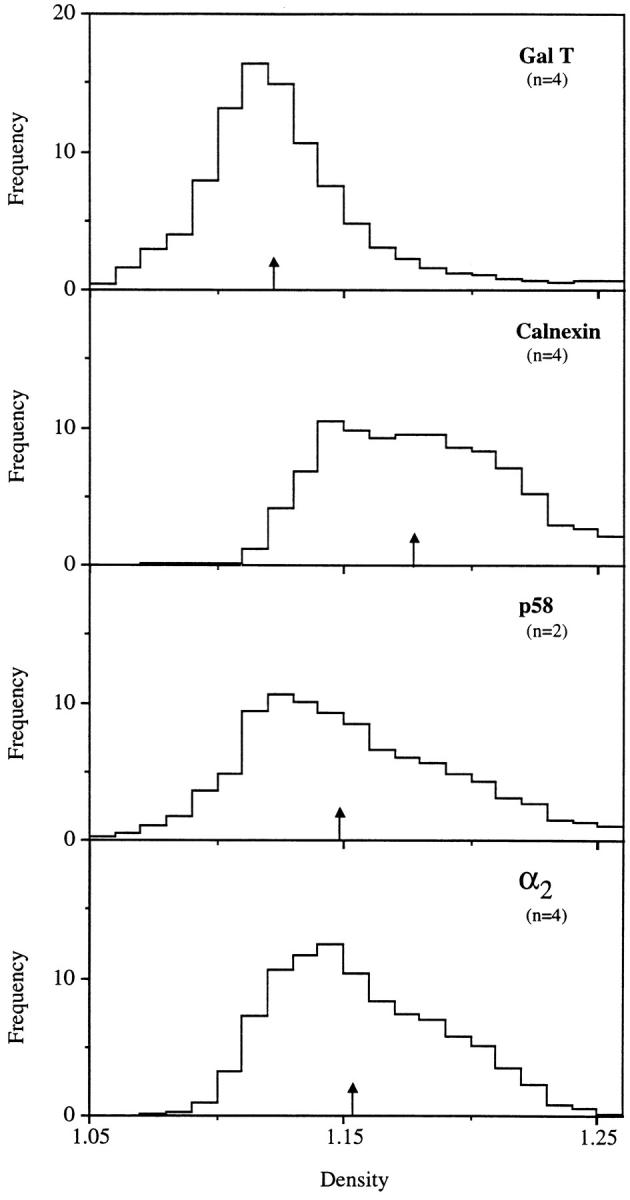
Analytical fractionation of the α2 protein in rat liver homogenates. The MLP fraction from rat liver homogenates was centrifuged on linear sucrose gradients as described in Materials and Methods. Equal volumes of each fraction were evaluated for their content of GalT as evaluated by enzyme assay, calnexin, p58, and α2 protein as recognized by immunoblots, and quantification was evaluated by densitometry. The number of separate fractionations (n) is indicated. Results were normalized according to the methodology of Beaufay et al. (1964).
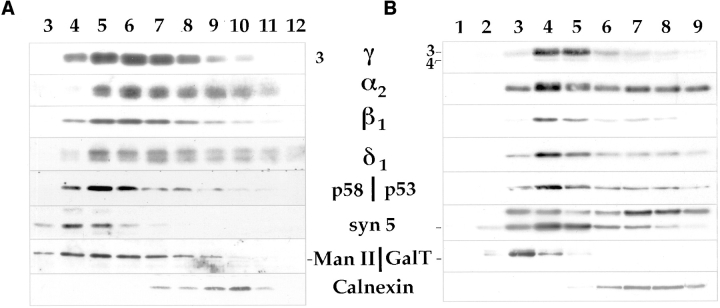
The subcellular distribution of all family members in rat liver membranes (Fig. 7, left) as well as in HeLa cells (Fig. 7, right), was compared. For rat liver homogenates, similar analytical gradients and a starting preparation of total membranes (MLP) similar to those used for the studies in Fig. 6 were used. Although overlapping, the distribution of the α2 and δ1 family members were displaced towards higher densities than the γ3 and β1 members. The distribution of p58, the intermediate compartment protein, was similar to the latter proteins, whereas syntaxin 5 (a Golgi resident target-soluble N-ethylmaleimide–sensitive factor attachment protein receptor (t-SNARE), Banfield et al., 1994) and the Golgi marker mannosidase II (Man II) were distributed towards lower densities. Calnexin was found in high density fractions. The heterogeneity in distribution of the p24 family members was confirmed in similar fractionation studies of HeLa cells (Fig. 7, right). This protocol used Nycodenz (Life Technologies, Eggenstein, Leopold Shafen, Germany) gradients instead of sucrose, and the use of a monoclonal antibody to GalT instead of Man II as Golgi marker and p53, the human equivalent to p58 as marker of the intermediate compartment. Taken together, a distinct pattern emerged in density gradients for both rat liver and HeLa cells for the α2 and δ1 family members. They revealed a distribution skewed more towards the ER marker than did the γ3, γ4, and β1 family members.
Figure 7.
Analytical fractional of all family members in homogenates from rat liver and HeLa cells. Total membranes from rat liver (A) or HeLa cells (B) were centrifuged on linear sucrose (rat) or Nycodenz (HeLa) gradients as described in Materials and Methods. Equal volumes of each fraction were determined for their content of calnexin, Man II (Rat) or GalT (HeLa), syntaxin 5, p58 (rat), p53 (HeLa), and different p24 family members, using specific antibodies generated to the peptides described in Fig. 2 A and with antibodies specific to the Golgi markers (Man II, Gal T), the intermediate compartment markers (p58, p53) or ER marker (calnexin). The high density end of the gradient is on the right.
COP I and II Coatomer Binding to Cytosolic Domains In Vitro
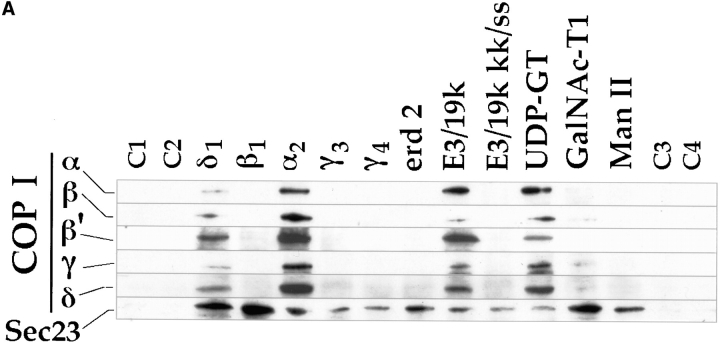
Both δ1 and α2 show COOH-terminal sequences similar to the previously identified ER retrieval motif known to bind COP I coatomer (Cosson and Letourneur, 1994), K(X)KXX. Whereas the latter displays sequences fitting this motif, the former has one additional amino acid known to partially impair its function. (Jackson et al., 1990). A third member, the β1, shows sequences reminiscent of the K(X)KXX motif, but with its critical lysines substituted to arginines. Substitution of these two lysines is known to eliminate the ability of the K(X)KXX motif to prevent reporter molecules from reaching late Golgi compartments (Jackson et al., 1990), and to effectively abolish binding to COP I coatomer (Cosson and Letourneur, 1994). The two additional members, γ3 and γ4, showed no sequences that could be related to known motifs. To investigate the ability of each member to bind COP I coatomer, synthetic peptides corresponding to their cytoplasmic domains were attached to thio-Sepharose, and then incubated with HeLa cytosol. Their binding was compared to that of peptides corresponding to the cytoplasmic domains of a protein of the CGN, i.e., the KDEL receptor (erd2) (Lewis and Pelham, 1990), as well as peptides corresponding to the type II cytosolic domains of the Golgi resident enzymes N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (Hagen et al., 1993; Homa et al., 1993) and Man II (Moremen and Robbins, 1991). As positive controls, peptides corresponding to the cytosolic domain of E3/19k (Cladaras and Wold, 1985) and UDP-glucuronyltransferase (UDP-GT) (Jackson et al., 1987) were included. These peptides contain the K(X)KXX motif. A mutant, E3/19k kk/ss, having the two lysines known to be critical for Golgi-to-ER retrieval (Jackson et al., 1990) and COP I coatomer binding (Cosson and Letourneur, 1994), mutated to serines was incorporated as a negative control.
Fig. 8 A shows that sequences corresponding to the cytoplasmic domains of δ1 and α2 associated with COP I components (as tested for α, β, β′, γ, and δ COP subunits) as did those of E3/19k and UDP-GT. Little, if any, association of COP I subunits was observed to the other peptides under the conditions used. Interestingly, a small but significant binding of COP I components was observed to the peptide corresponding to the cytoplasmic domain of GalNAc-T1 that might be explained by the presence of two basic amino acids immediately following the methionine (MRK-) of this type II transmembrane protein. Whereas a similar motif composed of double arginine (RR) has been shown to be sufficient for Golgi-to-ER retrieval of proteins of type II topology (Schutze et al., 1994), binding of such a motif to COP I subunits has yet to be established.
Figure 8.
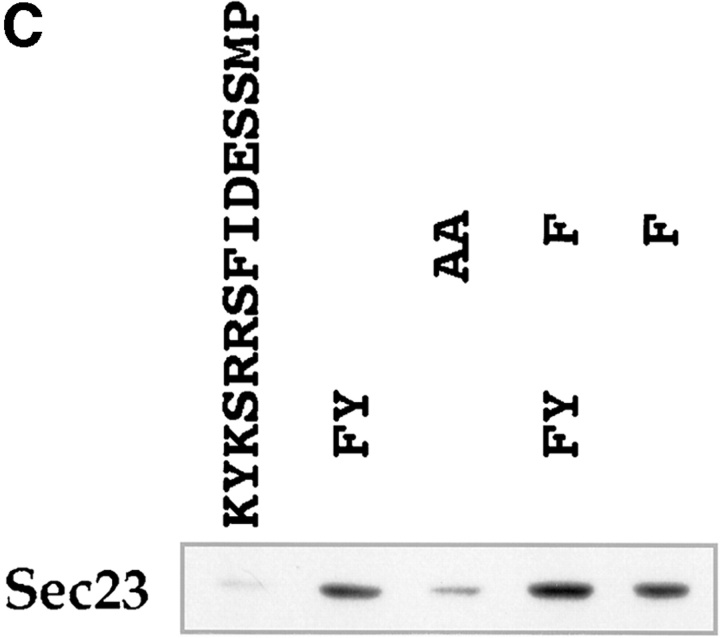
Binding of COP I and II proteins to cytoplasmic domains. Peptides corresponding to different cytoplasmic domains were coupled to thiopropyl Sepharose beads as described in Materials and Methods, and then assayed for their ability to bind COP I and II. (A) Binding of coat proteins to the cytoplasmic domains of α2, β1, δ1, γ3, γ4, erd2, E3/19k, E3/19k kk/ss (KK mutated to SS), UDP-GT, GalNac-T1, Man II, and control peptides C1-4. Binding of COP I components (tested for were α, β, β′, γ, and δ-COP) to α2 and δ1 was comparable to E3/19k and UDP-GT. The human homologue of Sec23 (hSec23) of the COP II coatomer showed specific binding to all cytoplasmic tails, but not to the four control peptides C1-4. Note the additional binding of the Sec23 to β1 and δ1 as well as to GalNac-TI. (B) Binding of coat proteins as in A but using the cytoplasmic domain of δ1 where pair-wise amino acid substitutions reveal a decrease of COP I binding upon KK-to-SS substitution, whereas Sec23 of COP II is reduced upon FF-to-AA substitution. (C) Introduction of aromatic residues into the mutated E3/19k peptide increases binding of Sec23.
The surprising finding of these experiments was the failure of COP I subunits to bind β1, γ3, and γ4. This would have been predicted from earlier studies (Fiedler et al., 1996; Sohn et al., 1996) that showed significant binding to both β1 and to a mutated version of the δ1 where its COOH-terminal sequence had been changed from KKLIE to SSLIE. Therefore, we examined the influence of using fresh versus frozen cell and tissue lysates under different buffer conditions, but were unable to reveal any detectable binding to the other members (data not shown). However, upon prolonged exposures, a weak but significant binding to β1 could be observed, and this was further increased when supplying purified COP I coatomer (data not shown). Under these conditions, no binding to γ3 and γ4 was observed, suggesting a weak but reproducible binding of COP I coatomer to β1.
The unexpected finding that COP I coatomer could bind cytoplasmic domains as partial complexes (Cosson and Letourneur, 1994), rather than a complete unit, was recently extended by Fiedler and co-workers who showed that two subcomplexes of COP I coatomer either bound to a putative but conserved FF motif present in the cytoplasmic domain in most p24s members, or to the K(X)KXX-like motif. This was suggested to provide the mechanistic explanation for how COP I coatomer could be present on both forward (coated by COP I subunits β, γ, and ζ) and retrograde (coated by COP I sub units α, β′, and ε) moving transport vesicles (Fiedler et al., 1996; Orci et al., 1997). Under no conditions were we able to reproduce subcomplex formation of the COP I coatomer (ζ and ε not assayed for). Rather, our findings are more in line with those observed by Sohn and co-workers (1996) (Fig. 3 A, lanes 1 and 5), who showed clear binding of all COP I subunits to δ1 (ζ and ε not assayed for). However, given that a blotting assay has clear technical limitations in quantitative terms, we do not exclude the possibility of individual protein members of the COP I coatomer complex to exhibit some degree of variation in their binding to different cytoplasmic domains of the p24s.
Hence, COP I binding in this study appeared restricted to the presence of a K(X)KXX-like retrieval motif. Under the same experimental conditions, cytoplasmic domains were also tested for their possible binding of COP II (by testing for the presence of human Sec23). Unexpectedly, all cytoplasmic domains bound Sec23, albeit to different extents (Fig. 8 A). Most Sec23 binding was seen to the β1 and δ1 proteins, but also to GalNAc-TI and Man II. The binding of Sec23 was specific. Four control peptides (C1–4) of similar lengths and charge compositions to those of the cytosolic domains of the respective α2, β1, γ3, and δ1 family members failed to bind under the conditions used. At least in vitro, this would separate COP II (Sec23) away from COP I in terms of selectivity of the different family members. However, the observation that the δ1 protein strongly binds both COP I and II coatomers predicts a competitive recruitment of COP I and II for this family member. This was investigated in greater detail whereby peptides corresponding to the cytoplasmic domain of δ1 were mutated so that amino acids encoding this domain were substituted two by two. Where possible, hydrophobic residues were substituted to alanines and hydrophilic ones to serines. The result of such an experiment is shown in Fig. 8 B. Both COP I and II binding was examined and as expected, when changing the KKLIE to SSLIE, COP I binding was reduced significantly. This is consistent with the observations of Sohn and co-workers (1996). However, only a slight, if any, decrease of COP I binding was observed when the FF motif was mutated to double arginine (AA), and this was unexpected since Sohn and co-workers (1996) showed a greater dependency on this motif for binding to all COP I subunits (ζ and ε not assayed for). Instead, we detected the influence of this motif in modulating COP I binding, since changing both the FF and the double lysine (KK) to AA and double serine (SS), respectively, completely abolished binding to COP I (Fig. 8 B, lane 9). In contrast, binding of Sec23 was greatly reduced upon changing the FF to AA but was completely unaltered when changing downstream sequences that included the KK. An influence of upstream sequences on binding Sec23 was also observed, suggesting that the FF motif is part of a larger one. This was examined in greater detail using the mutant, E3/19k kk/ss that binds Sec23 at relatively low levels but no COP I. This cytoplasmic domain (outlined in Fig. 8 C) was altered in such a way that a FF motif was introduced in two places either alone or together. Also, an existing F was altered to A to see if this reduced the original binding of Sec23. Fig. 8 C shows that changing this F to A did not result in a decrease in the already relatively low level of Sec23 binding, whereas introduction of an additional F next to the preexisting one increased Sec23 binding significantly. Furthermore, when introducing FY upstream of this F residue resembling those found in δ1, Sec23 binding increased even further. Therefore, we tentatively conclude from these studies that whereas the KK residues present in δ1 and α2 mediate binding of COP I subunits, the FF residues, presumably part of a larger motif (see Discussion) allow for the binding of Sec23. However, a minor influence of the latter residues on COP I binding was also detected.
In Vivo Mutational Analysis and Oligomeric Properties of p24s
Since only two out of the five chains bound COP I in relatively high amounts, and this coat is implicated in retrograde transport (Cosson and Letourneur, 1994), eliminating the Golgi-to-ER retrieval motif of the α2 and δ1 family members may alter their distribution. The α2 and δ1 sequences were accordingly mutated with two S residues replaced for the two K residues at the positions of their putative retrieval motifs. All 5 cDNAs were cotransfected as in the above experiment. Using antibodies specific to each family member, both mutated forms of α2 and δ1 showed a clear redistribution to the cell surface (shown is α2, Fig. 9 A). Unexpectedly, the wild-type β1, γ3/4 proteins (Fig. 9 B, γ3/4 proteins as revealed by peptide antisera recognizing both forms) were also redistributed as a consequence. This shows that the K(X)KXX-like motifs of the α2 and δ1 proteins are required for the intracellular steady-state distribution of all five p24 family members. Since the FF motif was shown to mediate binding to Sec23 (COP II) and this coatomer complex was needed for exit from the ER (Barlowe et al., 1994), this motif was mutated in both α2 and δ1 to AA. Upon cotransfection, the mutated p24s (Fig. 9 C, shown is α2) were now found to distribute to the ER together with the other three nonmutated members (Fig. 9 D, γ3/4 proteins). Hence, the proteins may show heterotypic interactions in vivo and this was further examined in vitro.
Figure 9.
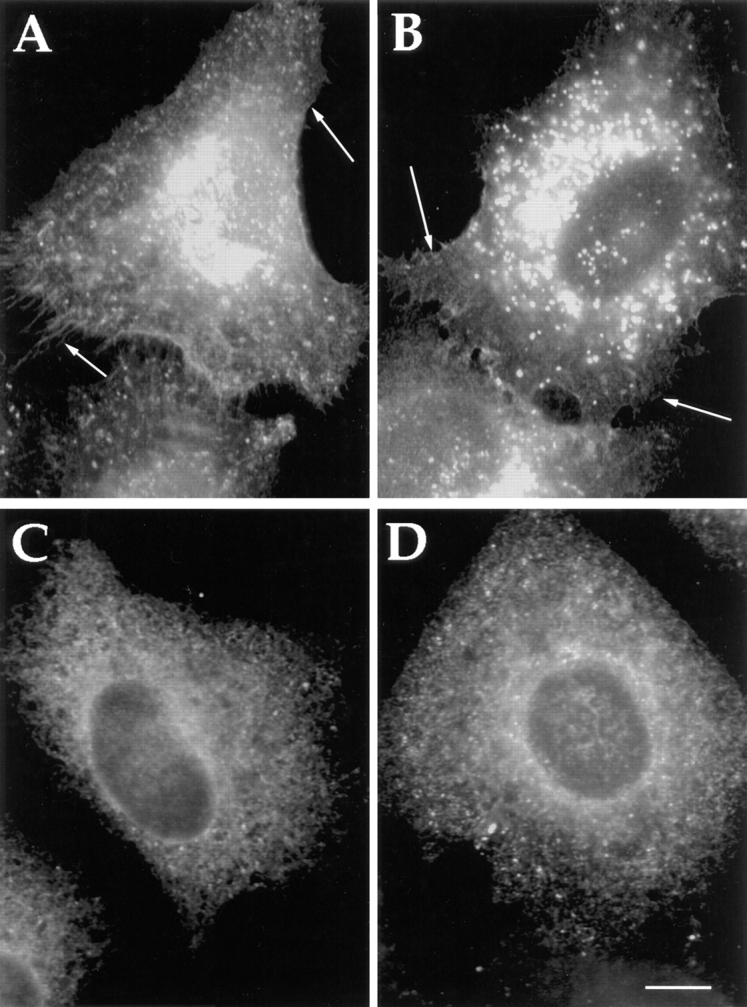
Altered distribution of family members after mutation of K(X)KK(X) on FF motifs of the cytosolic domains of α2 and β1 proteins. Cotransfection of all five p24s in where either the KK (A and B) or the FF (C and D) motif has been altered to SS or AA, respectively, in both α2 and δ1. Altered redistribution and apparent cell surface staining can be seen with the KK/SS mutants (shown is α2)(A). Remarkably, the unmutated members (shown is γ3/4) (B) also reveals redistribution to the cell surface. Similarly, an apparent ER staining can be seen with the FF/AA mutants (shown is α2)(C). This also leads to the apparent redistribution of the unmutated members to the ER (shown is γ3/4)(D). Bar, 5 μm.
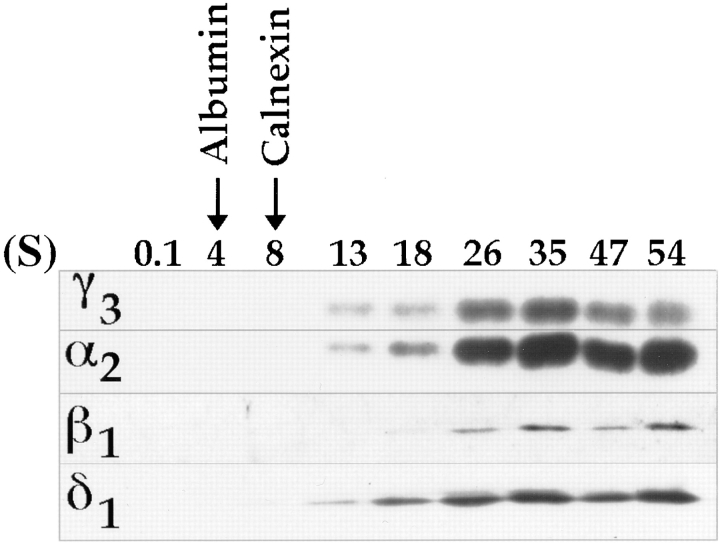
Highly purified rat liver Golgi membranes were subjected to detergent solubilization followed by velocity gradient centrifugation and fractionation. Precipitated proteins were then subjected to SDS-PAGE and Western blotting to reveal the different p24s. As can be seen in Fig. 10, the p24s appear to cosediment as large oligomeric complexes (peaking near 35S as evaluated using the McEwen method [1966]), well away from the membrane protein, calnexin (a resident ER protein contaminant of the fraction), and the soluble cargo molecule, serum albumin.
Figure 10.
Sedimentation density gradient centrifugation showing oligomeric behavior of p24s. Golgi fraction was solubilized with sodium cholate containing buffer, loaded onto a sucrose gradient, and subjected to centrifugation as described in Materials and Methods. Fractions were collected and evaluated for their content in α2, β1, δ1, and γ3 by Western blotting followed by ECL. All four show a similar peak distribution around 35S.
Discussion
The observation that p24 family members redistribute as a consequence of the ER retrieval motif K(X)KXX(X) was demonstrated by immunofluorescence upon coexpression of all five members. This redistribution also affected nonmutated family members, suggesting heterotypic interactions among the p24 family members. Such interactions were further demonstrated in vitro by subjecting detergent-solubilized material to velocity gradient centrifugation. The α2, β1, δ1, and γ3 members cosedimented as large complexes (evaluated to peak at a sedimentation value of ∼35S by the method of McEwen [1966]). This corresponds to very large complexes of ∼1–2 million D, consistent with values obtained for these proteins upon gel filtration (data not shown). In yeast, the homologous proteins of β and δ (emp24 and erv25, respectively) have been demonstrated by chemical cross-linking to be in a complex when present in COP II vesicles (Belden and Barlowe, 1996).
KK to SS mutations resulting in a redistribution of the p24 family members to the cell surface can be readily explained by the importance of these residues for retrieval (Nilsson et al., 1989; Jackson et al., 1990) and recruitment of COP I coatomer (Cosson and Letourneur, 1994; Letourneur et al., 1994). The latter was shown to be recruited in vitro. Both α2 and δ1 showed clear COP I binding in what appeared to be a KK-restricted manner, and in comparable amounts to the positive controls, E3/19K and UDP-GT, which have both been shown to display functional K(X)KXX retrieval motifs (Nilsson et al., 1989). This KK-restricted binding was shown by mutating corresponding amino acids in the E3/19K as well as in δ1. In the case of the latter, the clear reduction of COP I binding upon the changing KK-to-SS motif shows that even though this member does not display a perfect K(X)KXX retrieval motif, COP I binding is still, at least in part, mediated by this motif. The one additional amino acid present in the δ1 member shifts the last K of the KK motif from a crucial −3 to a −4 position. This is known to greatly affect retrieval of this motif when present in a reporter molecule. Therefore, it is not surprising that δ1 binds relatively less than does α2 (compare α2 and δ1, Fig. 8 A). However, binding of COP I to δ1 was not completely abolished upon changing its KK motif to SS, suggesting that upstream sequences would mediate binding as well. However, this could only be shown indirectly. When both KK and FF were substituted to SS and AA, respectively, COP I binding was reduced even further or effectively eliminated. However, when substituted alone, such upstream sequences (including the FF motif) showed very little effect on COP I binding. This is in stark contrast to that observed by Sohn and co-workers (1996), who showed a complete elimination of COP I binding when changing the FF motif to AA. At present, we have no explanation for this discrepancy. What is apparent though, both from this study and that of Sohn and co-workers (1996), is the lack of subcomplexes of COP I coatomer binding to cytoplasmic domains of p24s.
The β1 and γ chains failed to bind COP I in appreciable amounts. Furthermore, mutation of the KK motif of the δ1 member would have been expected to only reduce binding of some of the COP I subunits but not all. Fielder et al. (1996) found clear binding of COP I subunits to the cytoplasmic domains of all p24 family members tested. As well, they observed that only a subset of COP I subunits (α, β′, and ε) bound in a KK-restricted manner, whereas the other subunits (β, γ, and ζ) bound in a FF-dependent manner. It has been shown that the COP I complex can be split into either separate subunits or apparent subcomplexes when using relatively high salt concentrations (0.3– 0.5 M). This was observed by Cosson and Letourneur (1994) who compared the binding of COP I under either low or high ionic strength to peptides displaying the K(X)KXX motif. At high ionic strength, binding was only observed for α, β′, and possibly ε, whereas at lower ionic strength, binding of β and γ was also observed. Evidence that the COP I complex is sensitive to high ionic strength was also shown by Lowe and Kreis (1995), who subjected the COP I complex to either 1 M NaCl, 0.5 M Tris-HCl, or 0.25 M MgCl2. This resulted in a breakdown of the COP I complex into either subcomplexes or individual subunits in vitro. However, under the conditions used in this study (0.4 M salt), we were unable to split the COP I complex. Rather, it appeared as a single complex and this persisted upon altering buffer conditions including salt concentrations and amounts of cell extracts supplied in the assay (data not shown). Under all conditions assayed for, COP I appeared to bind as a complex relying mostly on the presence of a KK motif. Recent studies of Lowe and Kreis (1996) have also shown that COP I coatomer is present as a single complex in the cytosol with all subunits represented equally in the complex (only the ζ subunit of COP I has been observed to be in excess as a monomer). However, although we failed to split the COP I complex in vitro, we do not preclude the existence of such subcomplexes in vivo. It is possible that the COP I complex could be split by posttranslational modifications such as phosphorylation, and that this would reveal binding to the FF motif as observed by Fiedler et al. (1996). However, it remains unclear why we failed to detect abundant binding of COP I to either the KK-to-SS mutated δ1 or to the other p24s not displaying K(X)KXX-like motifs. Perhaps only upon splitting the COP I complex into subcomplexes would β, γ, and ζ bind to the FF motif. However, if this was the case, it is difficult to explain why Sohn et al. (1996) detected binding of not only β, γ, and ζ, but also α and β′ to hp24δ1 where its KK motif had been mutated to SS. Clearly, only by revealing the presence of COP I complexes in vivo, can we address the significance of subcomplexes, and examine to what extent such subcomplexes correlate with function (e.g., anterograde and retrograde COP I vesicle-mediated transport).
Other resident proteins of the exocytic pathway reveal motifs similar to the KK motif, although in different configurations and topologies. For example, the GalNAcT-1, which showed a weak binding of COP I, has in its cytoplasmic domain residues that in this type II protein, could be expected to serve as a retrieval signal (Schutze et al., 1994). Future work will reveal if there exists a correlation among resident proteins of the exocytic pathway in their extent of retrieval and their relative ability to bind COP I.
Whereas the KK-to-SS mutation led to a more distal distribution (including the cell surface) of the p24 family members, mutation of FF-to-AA redistributed the same proteins to a more ER-like location. As for the KK-to-SS mutations, the FF-to-AA mutations were only on the α2 and δ1 family members. Yet, all family members redistributed to an ER-like location, consistent with their suggested association as heterooligomers in vivo. FF motifs in the cytosolic domains of this family of membrane proteins have previously been implicated in exit from the ER (Fiedler et al., 1996). However, rather than binding COP I directly, we show the FF motif is involved in binding of the COP II coatomer subunit sec23, as mutation of this motif in δ1 greatly reduced the binding of Sec23. This helps to explain the shift of their steady-state distributions to the ER upon expression, as decreased binding of COP II would reduce their ability to be exported from the ER (Barlowe et al., 1994; Campbell and Schekman, 1997). Binding of Sec23 was observed in all cytoplasmic domains tested. This suggested a nonlinear motif, since there was no apparent similarity between these domains, except for the presence of one or more aromatic residues. However, the binding to Sec23 is unlikely to be the result of nonspecific association to an aromatic residue because no binding was detected to the four control peptides used in the study, which all contained at least one aromatic residue. We conclude that Sec23 binding involves aromatic residues, but that these must be presented in a favorable way. Sequences upstream of the FF motif but not downstream showed significant influence on the binding of Sec23 to δ1. These show a high degree of conservation in all p24s, suggesting that FF is part of a larger motif. For example, the strong endocytosis and putative AP2 binding motif, F/YXXXXF/Y, can be modeled onto sequences corresponding to the cytoplasmic domains of β1, δ1, and the γ chains. However, the γ chains showed comparable levels of Sec23 binding to α2, suggesting that other amino acids are influencing Sec23 binding as well (e.g., the two basic amino acids preceding the FF motif: compare KSFF (α2, γ3, and γ4) with RRFF (β1 and δ1). Further mutational studies using E3/19k showed that Sec23 binding could be increased if an additional aromatic residue to mimic the FF motif was introduced. When this F/YXXXXF/Y motif was created, Sec23 binding was increased even further. At this stage, the importance of a F/YXXXXF/Y motif in some of the p24s can only be speculated upon, but it raises some exciting parallels to the endocytic pathway where this motif appears to mediate or at least greatly influence binding of AP2 to endocytosed membrane proteins (Johnson et al., 1990; Trowbridge, 1991; Kornfeld, 1992; Milgram et al., 1996; Rapoport et al., 1997).
When coexpressed, the five p24 family members colocalized with the medial Golgi marker NAGT I as judged by indirect immunofluorescence. By EM immunolabeling, the α2 member was largely (60%) located to small tubular networks approaching one side of the Golgi stack with the stacked saccules accounting for only a minimum (6%). By analytical fractionation, the majority of this family was found in compartments whose density was distinct to that of Golgi markers. Taken together, these observations suggest that at steady state, all family members are found to their greatest extent in the CGN. Since the α2 family member was terminally glycoslated in rat liver Golgi fractions (the γ3 member has also been found to be N-glycosylated and endo H resistant in HeLa cells [Füllekrug, J., and T. Nilsson, unpublished data]), then this predicts they are subjected to retrograde transport from compartments as late as the trans-Golgi. The slightly different distributions of the p24s in compartment(s) between the ER and stacked saccules of the Golgi apparatus as revealed by subcellular fractionation and indirect immunofluorescence likely reflects the sum of the opposing anterograde and retrograde signals to which they are subjected and their relative affinities for each other (see above).
The p24s have been suggested to serve as cargo receptors bringing cargo out of the ER. This is supported by the observation that β (emp24) and δ (erv25) are found enriched in COP II vesicles in yeast. Furthermore, the deletion of these membrane proteins decreased the rate of transport of selected ER-derived cargo (Schimmöller et al., 1995; Stamnes et al., 1995; Elrod and Kaiser, 1996). Consistent with β (emp24) and δ (erv25) being concentrated in COP II vesicles, in yeast we observed a higher binding of Sec23 to their human counterparts. Despite the generation of polyclonal antibodies to nine different peptides of the five p24s examined in this study, none were immunoprecipitating. Future coimmunoprecipitation studies using appropriate antibodies during pulse-chase protocols will clarify the roles of these family members in cargo delivery, recycling, and their oligomerization in mammalian cells. The steady-state concentration of the p24s in the CGN suggests additional roles. For example, their high abundance in Golgi fractions along with the intermediate compartment marker p53/58 that has also been shown to associate with COP I coatomer predicts these three proteins (α2, δ1, and p53/58) represent the major COP I coatomer binding proteins of the cell. Indeed, the high proportion of all integral membrane proteins accounted for by the p24 family members in the hepatic Golgi fraction (Fig. 1) suggests that along with p53/58, they represent the major integral membrane proteins of the secretory pathway between the ER and the Golgi apparatus. An explanation for their relative abundance, oligomeric behavior, and steady-state distribution in the CGN could, therefore, be that these serve to form and maintain membrane structures between the ER and Golgi apparatus, and that their relative abilities to bind COP I and II serves to position them in this part of the pathway as well as to perhaps stabilize the membranes in which they reside by binding directly to COP I (Kreis and Pepperkok, 1994). Such a structural role is testable.
Footnotes
We thank the following colleagues for kindly sharing reagents, even before their publication: J. Rothman (Sloan-Kettering, NY) and F. Wieland (University of Heidelberg, Heidelberg, Germany) for gifts of p24 and p23 antibodies; T. Kreis (University of Geneva, Geneva, Switzerland) and C. Harter (University of Heidelberg ) for COP I reagents; H.-P. Hauri (University of Basel, Basel, Switzerland) and J. Saraste (University of Bergen, Bergen, Norway) for sharing p53 and p58 antibodies; H.-D. Söling (University of Göttingen, Göttingen, Germany), for the KDEL receptor antibody; N. Hui (Imperial Cancer Research Fund, London, England) for syntaxin 5 antibodies and J. Ostermann (Vanderbilt University School of Medicine, Nashville, TN) for purified coatomer I. Also, we wish to acknowledge F. Parlati (Genetics Group, Biotechnology Research Institute, Montreal, Canada) for help in cloning; B. Storrie (Blacksburg) and M. Otter (Cell Biology Programme, European Molecular Biology Laboratory [EMBL], Heidelberg, Germany), for critically reading the manuscript. EST's were obtained from the Institute for Genomic Research database TIGR and from GenBank. We would also like to thank the support facilities at EMBL along with A. Bell, P. Cameron, and the Sheldon Biotechnology center at McGill for antibody production, peptide sequencing, and DNA sequencing.
This work was supported by a McGill major Hydro-Québec fellowship to M. Dominguez, a Deutsche Forschungsgemeinschaft to J. Füllekrug, a Swiss National Science Fundation grant (31-43366-95) and Jules Thorn Charitable Overseas Trust to J.-P. Paccaud, and the Medical Research Council of Canada and Glaxo Wellcome Inc. to J.J.M. Bergeron.
M. Dominguez and K. Dejgaard contributed equally to this work.
Address all correspondence to T. Nilsson, Cell Biology Programme, European Molecular Biology Laboratory, Meyerhofstrasse 1, 69012 Heidelberg, Germany. Tel.: (49) 622-138-7294. Fax: (49) 622-138-7512. E-mail: nilsson@embl-heidelberg.de
1. Abbreviations used in this paper: CGN, cis-Golgi network; COP, coat proteins; ECL, enhanced chemiluminescence; endo H, endo glycosidase H; ERGIC, ER-to-GOLGI intermediate compartment; GalNAc-T1, N-acetylgalactosylaminyltransferase I; GalT, β1,4-galactosyltransferase; Man II, α1,2 mannosidase II; MLP, multiple large platform; NAGT I, N-acetylglucosaminyltransferase I; RT, reverse transcriptase; Sialyl, α2,6-sialytransferase; UDP-GT, UDP-glucuronyltransferase.
References
- Allan VJ, Kreis TE. A microtubule-binding protein associated with membranes of the Golgi apparatus. J Cell Biol. 1986;103:2229–2239. doi: 10.1083/jcb.103.6.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Balch WE. Principles of selective transport: coat complexes hold the key. Trends Cell Biol. 1996;6:315–320. doi: 10.1016/0962-8924(96)10027-1. [DOI] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warren G, Pelham HR. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Baudhuin P, Evrard P, Berthet J. Electron microscopic examination of subcellular fractions. J Cell Biol. 1967;32:405–416. doi: 10.1083/jcb.32.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaufay H, Jacques P, Baudhuin P, Sellinger OZ, Berthet J, De Duve C. Tissue fractionation studies. 18. Resolution of mitochondrial fractions from rat liver into three distinct populations of cytoplasmic particles by means of density equilibration in various gradients. Biochem J. 1964;92:184–205. doi: 10.1042/bj0920184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek SY, Orci L, Schekman R. Traffic COPs and the formation of vesicle coats. Trends Cell Biol. 1996;6:468–473. doi: 10.1016/0962-8924(96)84943-9. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Erv25p, a component of COP II-coated vesicles, forms a complex with emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J Biol Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- Bergeron JJ, Rachubinski RA, Sikstrom RA, Posner BI, Paiement J. Galactose transfer to endogenous acceptors within Golgi fractions of rat liver. J Cell Biol. 1982;92:139–146. doi: 10.1083/jcb.92.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Feick P, Puype M, Vandekerckhove J, Klengel R, Nastainczyk W, Schulz I. Tmp21 and p24A, two type 1 proteins enriched in pancreatic microsomal membranes, are members of a protein family involved in vesicular trafficking. J Biol Chem. 1996;271:17183–17189. doi: 10.1074/jbc.271.29.17183. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Campbell JL, Schekman R. Selective packaging of cargo molecules into endoplasmic reticulum-derived COP II vesicles. Proc Natl Acad Sci USA. 1997;94:837–842. doi: 10.1073/pnas.94.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cladaras C, Wold WS. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985;140:28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Dahan S, Ahluwalia JP, Wong L, Posner BI, Bergeron JJ. Concentration of intracellular hepatic apolipoprotein E in Golgi apparatus saccular distensions and endosomes. J Cell Biol. 1994;127:1859–1869. doi: 10.1083/jcb.127.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson MJ, Kaiser CA. Genes that control fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Gayle MA, Slack JL, Bonnert TP, Renshaw BR, Sonoda G, Taguchi T, Testa JR, Dower SK, Sims JE. Cloning of a putative ligand for the T1/ST2 receptor. J Biol Chem. 1996;271:5784–5789. doi: 10.1074/jbc.271.10.5784. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Van Wuyckhuyse B, Tabak LA. Purification, cloning, and expression of a bovine UDP-GalNAc: polypeptide N-acetyl-galactosaminyltransferase. J Biol Chem. 1993;268:18960–18965. [PubMed] [Google Scholar]
- Harlowe, E., and D. Lane. 1985. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 519–552.
- Hewick RM, Hunkapiller MW, Hood LE, Dreyer WJ. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981;256:7990–7997. [PubMed] [Google Scholar]
- Homa F L, Hollander T, Lehman DJ, Thomsen DR, Elhammer AP. Isolation and expression of a cDNA clone encoding a bovine UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1993;268:12609–12616. [PubMed] [Google Scholar]
- Jackson MR, McCarthy LR, Harding D, Wilson S, Coughtrie MW, Burchell B. Cloning of a human liver microsomal UDP-glucuronosyltransferase cDNA. Biochem J. 1987;242:581–588. doi: 10.1042/bj2420581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KF, Chan W, Kornfeld S. Cation-dependent mannose 6-phosphate receptor contains two internalization signals in its cytoplasmic domain (published erratum appears in Proc. Natl. Acad. Sci. USA.1991. 88: 1591) Proc Natl Acad Sci USA. 1990;87:10010–10014. doi: 10.1073/pnas.87.24.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S. Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO (Eur Mol Biol Organ) J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE, Pepperkok R. Coat proteins in intracellular membrane transport. Curr Opin Cell Biol. 1994;6:533–537. doi: 10.1016/0955-0674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. A human homologue of the yeast HDEL receptor. Nature. 1990;348:162–163. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. In vitro assembly and disassembly of coatomer. J Biol Chem. 1995;270:31364–31371. doi: 10.1074/jbc.270.52.31364. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. In vivoassembly of coatomer, the COP I coat precursor. J Biol Chem. 1996;271:30725–30730. doi: 10.1074/jbc.271.48.30725. [DOI] [PubMed] [Google Scholar]
- McEwen CR. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1966;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Milgram SL, Mains RE, Eipper BA. Identification of routing determinants in the cytosolic domain of a secretory granule-associated integral membrane protein. J Biol Chem. 1996;271:17526–17535. doi: 10.1074/jbc.271.29.17526. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Robbins PW. Isolation, characterization, and expression of cDNAs encoding murine α-mannosidase II, a Golgi enzyme that controls conversion of high mannose to complex N-glycans. J Cell Biol. 1991;115:1521–1534. doi: 10.1083/jcb.115.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Pypaert M, Hoe MH, Slusarewicz P, Berger EG, Warren G. Overlapping distribution of two glycosyltransferases in the Golgi apparatus of HeLa cells. J Cell Biol. 1993;120:5–13. doi: 10.1083/jcb.120.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprins A, Duden R, Kreis TE, Geuze HJ, Slot JW. β-COP localizes mainly to the cis-Golgi side in exocrine pancreas. J Cell Biol. 1993;121:49–59. doi: 10.1083/jcb.121.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Söllner T, Rothman JE. Bidirectional transport by distinct populations of COP I-coated vesicles. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- Ou WJ, Cameron PH, Thomas DY, Bergeron JJ. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- Pääbo S, Weber F, Nilsson T, Schaffner W, Peterson PA. Structural and functional dissection of an MHC class I antigen-binding adenovirus glycoprotein. EMBO (Eur Mol Biol Organ) J. 1986;5:1921–1927. doi: 10.1002/j.1460-2075.1986.tb04445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccaud J-P, Reith W, Carpentier J-L, Ravazzola M, Amherdt M, Schekman R, Orci L. Cloning and functional characterisation of mammalian homologues of the COP II component Sec 23. Mol Biol Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. About turn for the COPs? . Cell. 1994;79:1125–1127. doi: 10.1016/0092-8674(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Peranen J, Rikkonen M, Kaariainen L. A method for exposing hidden antigenic sites in paraformaldehyde-fixed cultured cells, applied to initially unreactive antibodies. J Histochem Cytochem. 1993;41:447–454. doi: 10.1177/41.3.8429208. [DOI] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rapoport I, Miyazaki M, Boll W, Duckworth B, Cantley LC, Shoelson S, Kirchhausen T. Regulatory interactions in the recognition of endocytic sorting signals by AP-2 complexes. EMBO (Eur Mol Biol Organ) J. 1997;16:2240–2250. doi: 10.1093/emboj/16.9.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Saraste J, Svensson K. Distribution of the intermediate elements operating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmöller F, Singer-Kruger B, Schroder S, Kruger U, Barlowe C, Riezman H. The absence of emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO (Eur Mol Biol Organ) J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Franson JAM, Matter K, Kreis TE, Ginsel L, Hauri H. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B, Frank R, Crausaz F, Riezman H. Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J Biol Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- Smith RF, Smith TF. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci USA. 1990;87:118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms JB, Wieland FT. A major transmembrane protein of Golgi-derived COP I-coated vesicles in coatomer binding. J Cell Biol. 1996;5:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Fullekrug J, Nguyen P, Van, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Craighead MW, Hoe MH, Lampen N, Geromanos S, Tempst P, Rothman JE. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc Natl Acad Sci USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale EJ, Plutner H, Matteson J, Balch WE. p53/58 binds COP I and is required for selective transport through the early secretory pathway. J Cell Biol. 1997;137:581–593. doi: 10.1083/jcb.137.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Trowbridge IS. Endocytosis and signals for internalization (published erratum appears in Curr. Opin. Cell Biol. 1991. 3:1062) Curr Opin Cell Biol. 1991;3:634–641. doi: 10.1016/0955-0674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucl Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJD, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- Weinstein J, de Souza e Silva U, Paulson JC. Sialylation of glycoprotein oligosaccharides N-linked to asparagine. Enzymatic characterization of a Gal β1 to 3(4)GlcNAc α2 to 3 sialyltransferase and a Gal β1 to 4GlcNAc α 2 to 6 sialyltransferase from rat liver. J Biol Chem. 1982;257:13845–13853. [PubMed] [Google Scholar]
- Weinstein J, Lee EU, McEntee K, Lai P H, Paulson JC. Primary structure of β-galactoside α2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J Biol Chem. 1987;262:17735–17743. [PubMed] [Google Scholar]