Abstract
Abstract. Reversible phosphorylation plays an important role in pre-mRNA splicing in mammalian cells. Two kinases, SR protein-specific kinase (SRPK1) and Clk/Sty, have been shown to phosphorylate the SR family of splicing factors. We report here the cloning and characterization of SRPK2, which is highly related to SRPK1 in sequence, kinase activity, and substrate specificity. Random peptide selection for preferred phosphorylation sites revealed a stringent preference of SRPK2 for SR dipeptides, and the consensus derived may be used to predict potential phosphorylation sites in candidate arginine and serine-rich (RS) domain–containing proteins. Phosphorylation of an SR protein (ASF/SF2) by either SRPK1 or 2 enhanced its interaction with another RS domain–containing protein (U1 70K), and overexpression of either kinase induced specific redistribution of splicing factors in the nucleus. These observations likely reflect the function of the SRPK family of kinases in spliceosome assembly and in mediating the trafficking of splicing factors in mammalian cells. The biochemical and functional similarities between SRPK1 and 2, however, are in contrast to their differences in expression. SRPK1 is highly expressed in pancreas, whereas SRPK2 is highly expressed in brain, although both are coexpressed in other human tissues and in many experimental cell lines. Interestingly, SRPK2 also contains a proline-rich sequence at its NH2 terminus, and a recent study showed that this NH2-terminal sequence has the capacity to interact with a WW domain protein in vitro. Together, our studies suggest that different SRPK family members may be uniquely regulated and targeted, thereby contributing to splicing regulation in different tissues, during development, or in response to signaling.
Pre-mRNA splicing in mammalian cells takes place in a multi-component complex known as the spliceosome, which contains two classes of splicing factors: U1, U2, U5, and U4/6 small nuclear ribonucleoprotein particles (snRNPs)1 and non-snRNP splicing factors (for review see Krämer, 1996). A series of interactions between pre-mRNA and snRNPs during spliceosome assembly are critical for splice-site selection, and most importantly, for establishing a catalytic core for the splicing reaction to occur in the spliceosome. However, most of these interactions are mediated by non-snRNP factors. Among the best characterized non-snRNP factors are the superfamily of arginine/serine-rich (RS) domain-containing splicing factors (for review see Fu, 1995). The family is composed of “classic” SR proteins that are characterized by one or two RNA recognition motifs at the NH2 terminus, an RS domain at the COOH terminus, and other RS domain–containing polypeptides. RNA recognition motifs are responsible for binding to RNA, whereas RS domains appear to function as protein–protein interaction modules to mediate various spliceosome assembly steps. Thus, some RS domain–containing proteins may mediate complex assembly on RNA, whereas others may function as adaptor molecules to bring complexes together in spliceosome assembly. Because RS domain–containing proteins play a critical role in selecting and pairing functional splice sites, many splicing factors in this family are not only essential for constitutive splicing, but can also affect alternative splicing (for review see Fu, 1995; Manley and Tacke, 1996; Valcárcel and Green, 1996).
All RS domain–containing proteins are probably posttranslationally modified by phosphorylation, and reversible phosphorylation has been shown to play an important role in splicing. For example, no splicing complexes could be detected in nuclear extract treated with the phosphatase PP1, indicating that phosphorylation is essential for early steps of spliceosome assembly, and SR proteins appear to be the major targets of the PP1 treatment (Mermoud et al., 1994). Consistent with these findings, phosphorylation was recently shown to promote specific protein–protein interactions between U1 70K and ASF/SF2, both of which contain an RS domain (Xiao and Manley, 1997). On the other hand, dephosphorylation is critical for splicing after spliceosome assembly. When nuclear extracts were treated with phosphatase inhibitors, spliceosome assembly was not affected, but splicing was blocked. Splicing in the stalled spliceosome could then be rescued by phosphatase PP1 or PP2A (Mermoud et al., 1992). Moreover, incorporation of nonhydrolyzable γ-S-ATP in the RS domain–containing U1 70K blocked splicing after spliceosome assembly (Tazi et al., 1993). Together, these studies strongly indicate that a phosphorylation–dephosphorylation cycle is important for RNA splicing. Additionally, PP1 was also shown to affect splice-site selection in vitro (Cardinal et al., 1994). Because SR proteins are known to affect splice-site selection both in vitro and in vivo, and PP1 clearly caused dephosphorylation of these splicing factors in nuclear extract, it is likely that the PP1 effect on alternative splicing is mediated through changes in the phosphorylation state of SR proteins (Mermoud et al., 1994).
Two families of kinases, SR protein-specific kinase (SRPK) and Clk/Sty, have been identified that phosphorylate RS domain–containing splicing factors. Our lab identified and cloned human SRPK1 in the pursuit of an activity that mediates splicing factor redistribution in the cell cycle (Gui et al., 1994a ). SRPK1 is a kinase highly specific for RS domain–containing splicing factors because it recognizes only arginine (not lysine) and phosphorylates only serine (not threonine) in its substrates (Gui et al., 1994b ). Clk/Sty was initially cloned as a cdk-like kinase by PCR (Johnson and Smith, 1991; Ben-David et al., 1991) as well as a dual specificity kinase in an expression screening (Howell et al., 1991). In fact, three highly related Clk/Sty kinases are expressed in mammalian cells (Hanes et al., 1994). Clk/Sty was later found to interact with RS domain– containing splicing factors in the yeast two-hybrid system, and to efficiently phosphorylate these splicing factors in vitro (Colwill et al., 1996a ). Interestingly, Clk/Sty itself contains several SR or RS repeats at the NH2 terminus, which appear to contribute to its high affinity binding with RS domain–containing substrates. SRPK1 and Clk/Sty share 32% identity in their kinase domains and both contain a signature amino acid sequence referred to as the LAMMER motif (Colwill et al., 1996a ). A systematic comparison between the two kinases revealed that SRPK1 displays high specific activity towards RS domain–containing splicing factors, whereas Clk/Sty has broader substrate specificity (Colwill et al., 1996b ). These observations suggest that these two kinases may have distinct functions in pre-mRNA splicing. Clearly, splicing is likely to be regulated by multiple kinases and phosphatases, given the complexity of the splicing reaction and the diversity of splicing factors involved.
In this paper, we report the cloning and characterization of a new SRPK family member named SRPK2 that was discovered based on its sequence similarity to SRPK1. A series of biochemical experiments demonstrate that SRPK1 and 2 are very similar with respect to their enzymatic activity and substrate specificity. Both kinases promoted specific protein–protein interactions between RS domain–containing splicing factors and their overexpression induced the redistribution of splicing factors from nuclear speckles to nucleoplasm, indicating that both kinases may be involved in the regulation of spliceosome assembly in vivo. However, these two kinases are differentially expressed in various human tissues and each kinase contains unique sequence features that may contribute to their specific function and/or regulation in vivo. Therefore, mammalian cells may have evolved multiple kinases to regulate RNA splicing, and these SR protein kinases may themselves be targets for regulation.
Materials and Methods
cDNA Cloning of SRPK2
A database search revealed multiple expression seqence tag (EST) clones that are homologous but not identical to SRPK1. One such cDNA clone (Genbank/EMBL/DDBJ accession number H00135) was obtained from Research Genetics, Inc. (Huntsville, AL), and sequencing analysis showed that the clone encodes a serine/threonine kinase and displays 78% identity to SRPK1 in their kinase domains. DNA probes derived from the clone were used to screen a human fetal brain cDNA library in the Lambda ZAP II vector (Stratagene, La Jolla, CA). From 5 × 105 plaques, 31 positive clones were obtained. Restriction analysis showed that all the clones are derived from different regions of a single cDNA, and thus, the longest clone was sequenced in both strands.
Expression of SRPK2 and Its Substrates
The HpaI–NheI fragment encoding the full-length SRPK2 was cloned into the pAcG2T vector (PharMingen, San Diego, CA) to express the kinase as a glutathione-S-transferase (GST) fusion protein by baculovirus according to manufacturer's instructions. GST-SRPK2 was purified on glutathione-Sepharose 4B beads (Pharmacia Biotechnology Inc., Piscataway, NJ). ASF/SF2 and its mutant derivatives were expressed and purified from bacteria (Gui et al., 1994b ; Colwill et al., 1996b ). GST-ASF/SF2 and U2AF65 were also made in bacteria. Other RS domain–containing proteins (SRp20, GST-SRp40, GST-SC35, SRp55) were expressed by baculovirus as described (Gui et al., 1994a ).
Northern Blotting Analysis
A human multiple tissue Northern blot was purchased from Clontech Laboratories Inc. (Palo Alto, CA). Soas2, HBL100, Bosc23, SF763, MG63, and Hela cells were cultured in DME (Life Technologies, Gaithersburg, MD) plus 10% heat-inactivated FCS (Hyclone, Logan, UT). Lan5 cells were grown in DME-F12 plus 10% FCS. THP1, U937, Weri1, and Lnz308 cells were cultured in RPMI 1640 (Irvine Scientific, Santa Ana, CA) plus 10% FCS. Total RNA from 6 × 105 cultured cells was extracted using RNAexol (BioChain, San Leandro, CA); 15–20 μg of total RNA was separated in 1% agarose gel containing 2.2 M formaldehyde and then blotted onto Hybond-N+ nylon membrane (Amersham Corp., Arlington Heights, IL). After baking the membrane at 80°C for 2 h, the membrane was stained with methylene blue to visualize RNA molecular markers (Life Technologies) and destained in H2O. The blots were prehybridized at 68°C for 1 h in 10 ml ExpressHyb Hybridization solution (Clontech Laboratories Inc.), and then hybridized with random primed probes as specified in Fig. 8. After hybridization for 1–2 h, the blots were washed at 50°C for 15 min sequentially in 0.1% SDS plus 0.5, 0.25, and 0.1× SSC before exposure to X-ray film. For normalization, each blot was stripped by boiling in 0.5% SDS for 5 min and reprobed for glyceraldehyde-3-phosphate dehydrogenase.
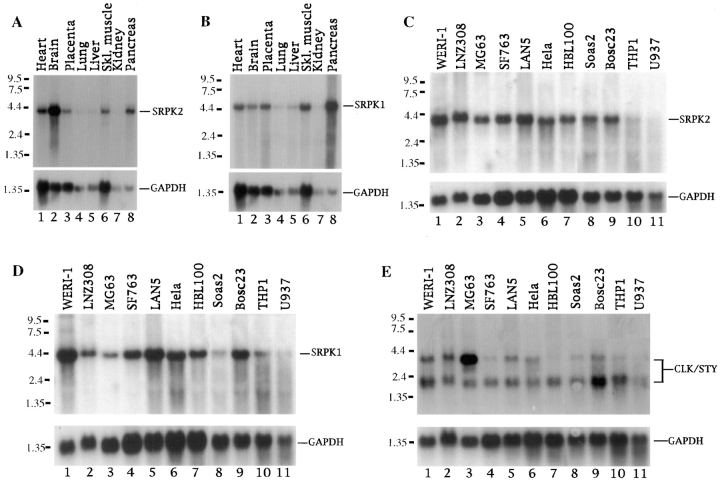
Figure 8.
Northern blotting analysis of SR protein kinases (indicated in the right of each panel) in multiple human tissues (A and B) and cell lines (C–E).
In Vitro Kinase Assay
The kinase activity of purified SRPK1 and 2 was normalized using bacterial ASF/SF2 as a substrate and then used to phosphorylate other substrates as described (Gui et al., 1994b ). Each purified substrate was normalized (using BSA as a standard) to 0.1 μg for testing phosphorylation in Fig. 2 B or to 1 μg in Fig. 2 C. To determine whether bacterial ASF/SF2 could gain the mAb104 phosphoepitope upon phosphorylation by SRPK2, 0.1 μg of GST-ASF/SF2 was incubated with purified SRPK2 in the presence of 1 mM ATP under kinase assay conditions followed by Western blotting with mAb104 (culture supernatant). The blot was developed by enhanced chemiluminescence (ECL; Pierce, Rockford, IL). To compare the relative specific activities of SRPK1 and 2, full-length SRPK1 and 2 cDNAs were fused with a FLAG-tag sequence at their NH2 termini, and cloned into the pSP73 vector for in vitro transcription/translation in the TNT system (Promega Corp., Madison, WI). The translated products were immunoprecipitated using the M2 anti-FLAG monoclonal antibody followed by a kinase assay on beads as previously described (Colwill et al., 1996b ). After SDS-PAGE, 35S-labeled kinases and 32P-labeled ASF/SF2 were quantitated by phosphoimaging (Molecular Dynamics Inc., Sunnyvale, CA).
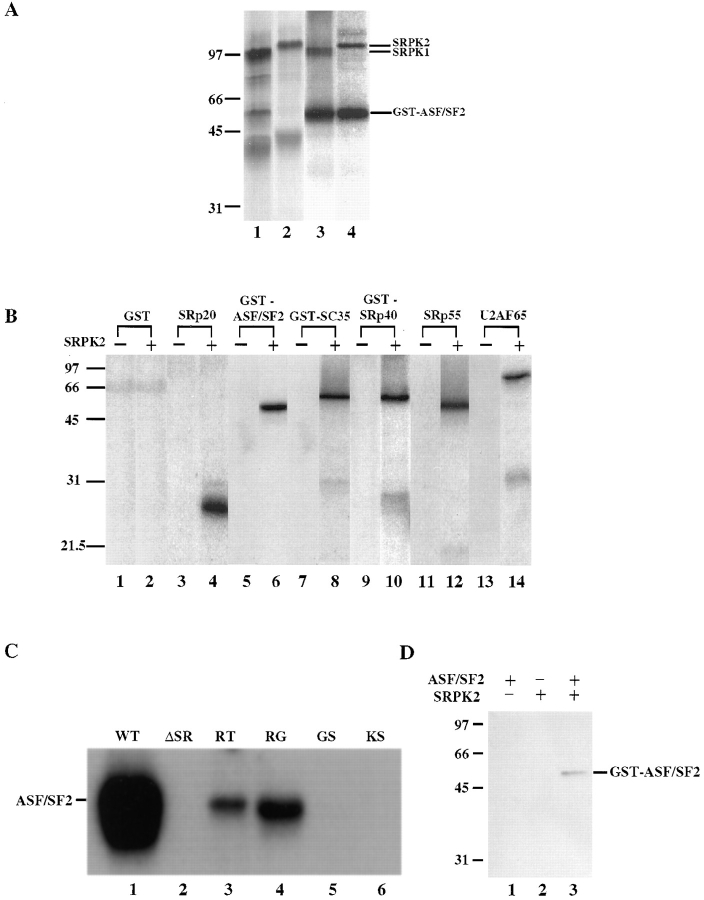
Figure 2.
Biochemical characterization of SRPK2. (A) In vitro translated SRPK1 (lane 1) and SRPK2 (lane 2) were immunoprecipitated with anti-FLAG antibody followed by kinase assay on beads using bacterial GST-ASF/SF2 as substrate (lanes 3 and 4). In vitro translated and precipitated kinases (35S-labeled) and phosphorylated GST-ASF/SF2 (32P-labeled) are indicated on the right. (B) Phosphorylation of purified RS domain–containing proteins (∼0.1 μg each) by SRPK2. (C) Phosphorylation of a panel of ASF/SF2 mutants (∼1 μg each) by SRPK2. These mutants were described by Caceres and Krainer (1993) and used in our previous characterization of SRPK1 (Gui et al., 1994b ) and Clk/Sty (Colwill et al., 1996b ). Residual phosphorylation in the RT and RG mutant proteins likely occurred outside the mutagenized region as discussed previously (Gui et al., 1994b ). (D) SRPK2-mediated phosphorylation restored the mAb104 phosphoepitope on bacterial GST-ASF/SF2.
Determination of Phosphorylation Consensus by Peptide Selection
Rationale for peptide library design and synthesis were as previously described (Songyang et al., 1994). In the current study, a serine-oriented and arginine-directed peptide library (see Fig. 4 A) was used. For the kinase reaction, purified SRPK2 (100 U, as defined in Gui et al., 1994a ) was incubated at 25°C for 1 h with 1 mg of degenerate peptide mixture in a 300-μl reaction volume in the presence of 100 mM ATP, trace labeled with [γ- 32P]ATP (∼6 × 105 cpm), 1 mM DTT, 10 mM MgCl2, and 50 mM Tris, pH 7.5. After the reaction, free ATP was removed by DEAE-Sephacel chromatography, and phosphopeptides were quantitatively separated from the unphosphorylated mixture on a ferric chelation column as previously described (Songyang et al., 1994). Peptide sequencing and data analysis were also as described (Songyang et al., 1994).
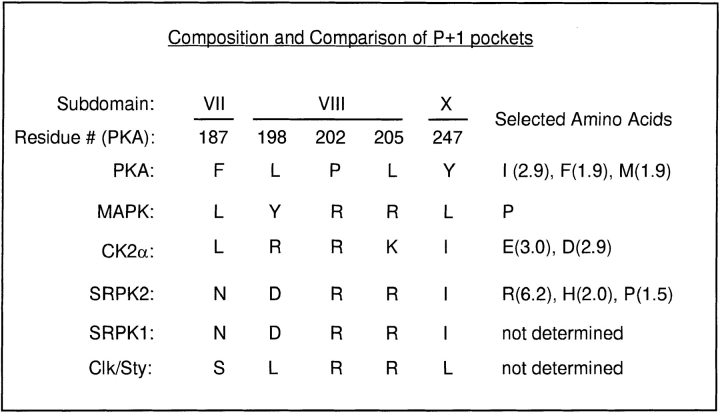
Figure 4.
Composition and comparison of P+1 pockets. The pocket composition for PKA, MAPK, and CK2α and preferred amino acids in the P+1 position of their substrates are from Songyang et al. (1996). The assignment of amino acids that form the P+1 pockets for SRPK1 and 2 is based on the sequence alignment shown in Fig. 1 B, and the selected amino acids for SRPK2 are from Fig. 3 B. The deduced P+1 pocket for Clk/Sty is also based on a similar sequence alignment with kinases whose tertiary structures have been determined (not shown). The subdomain locations of individual amino acids in the P+1 pocket and their positions corresponding to those of PKA are indicated.
Protein–Protein Interaction Determined by the GST Pulldown Assay
The assay was performed essentially as described (Xiao and Manley, 1997). Bacterially expressed GST-ASF/SF2 was kinased in vitro by baculovirus-expressed GST-SRPK1 or 2 as described (Gui et al., 1994b ), except that the ATP concentration was raised to 1 mM, and the reaction was incubated at 30°C for 4 h. Unphosphorylated GST-ASF/SF2 was prepared in a parallel sample lacking ATP. Treated proteins were desalted on G25 columns equilibrated in NETN (20 mM Tris-Cl, pH 8.0, 100 mM NaCl, 0.5 mM EDTA, 0.5% NP-40; Xiao and Manley, 1997), and rebound to glutathione-Sepharose. Beads were washed twice with NETN containing 1 M NaCl, and then three times with NETN. Bead aliquots containing 1–2 μg GST or GST-ASF/SF2 were incubated in 200 μl NETN with 3 μl in vitro translated [35S]methionine U1 70K for 30 min at 4°C. Beads were washed three times in NETN, treated with RNase and washed once, and then boiled in SDS-PAGE sample buffer. After SDS-PAGE, U1 70K was detected by autoradiography.
Indirect Immunofluorescence Microscopy
Full-length SRPK2 (the HpaI–NheI fragment) was subcloned into the NcoI–XbaI sites in the pUHD10-3 vector to create pUHD–SRPK2, in which a FLAG tag was fused with SRPK2 at its NH2 terminus. This vector system is used in cells expressing a tetracycline-controlled transactivator (tTA), and thus, gene expression is constitutive in the absence of tetracycline (Gossen and Bujard, 1992). 1 d before transfection, 5 × 104 tTA HeLa cells were seeded in 6-cm plates containing two or three 1.8-cm round coverslips coated with 0.1% gelatin. 10 μg of pUHD-SRPK2 plasmid DNA was transfected per plate by the calcium–phosphate method and the transfected cells were washed three times with PBS 16–24 h later. After culturing in fresh medium for another 16–24 h, cells were processed for immunostaining at room temperature as described (Fu and Maniatis, 1990). Briefly, cells were washed twice in PBS, and then fixed in 2% formaldehyde plus 0.2% Triton X-100 for 10 min. Fixed cells were permeablized in 1% Triton X-100 for 10 min, washed three times in PBS, and then blocked for 30 min in 20% FBS plus 0.5% Tween 20. Cells were stained for 1 h with the primary monoclonal M2 anti-FLAG (IgG1; 1:1,200 dilution from the 2 mg/ml stock purchased from Eastman Kodak Co. (Rochester, NY) and B1C8 (IgM; 1:50 dilution from culture supernatant; a gift from B. Blencowe and P. Sharp, Massachusetts Institute of Technology, Cambridge, MA) or mAb104 (IgM; undiluted culture supernatant), washed three times with PBS plus 0.1% NP-40, and developed for 1 h with secondary rhodamine-conjugated goat anti–mouse IgG and FITC-conjugated goat anti–mouse IgM (Southern Biotechnology, Birmingham, AL). Finally, the coverslips were mounted in an antiquenching solution (25 mg/ ml 1,4-diazabicyclo [2.2.2] octane and 70% glycerol in PBS) for examination under a Zeiss Axiophot fluorescence microscope.
Results
Identification and Cloning of SRPK2
We previously described the cloning and characterization of SRPK1, a serine kinase highly specific for the RS domain present in many splicing factors (Gui et al., 1994a ,b; Colwill et al., 1996b ). To gain further understanding of its biological functions, we conducted chromosomal mapping. Unexpectedly, probes derived from SRPK1 hit multiple loci in both mice and humans (our unpublished results), suggesting the possibility of multiple related kinases. These observations prompted us to search for SRPK1- related expression sequence tags in databases. We found eight clones displaying extensive homology to various regions within the kinase domain of SRPK1. Sequencing analysis revealed two kinases, one of which is SRPK1 and the other (represented by the clone H00135) is highly related, but not identical to SRPK1. We then screened a human fetal brain cDNA library using probes derived from H00135 and isolated a full-length clone, which contains an open reading frame encoding a novel kinase of 686 amino acids (Fig. 1 A).
Figure 1.
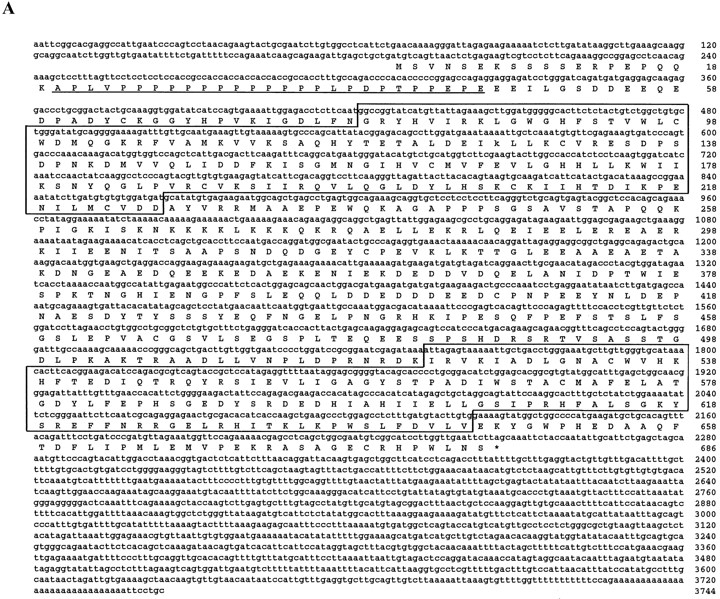
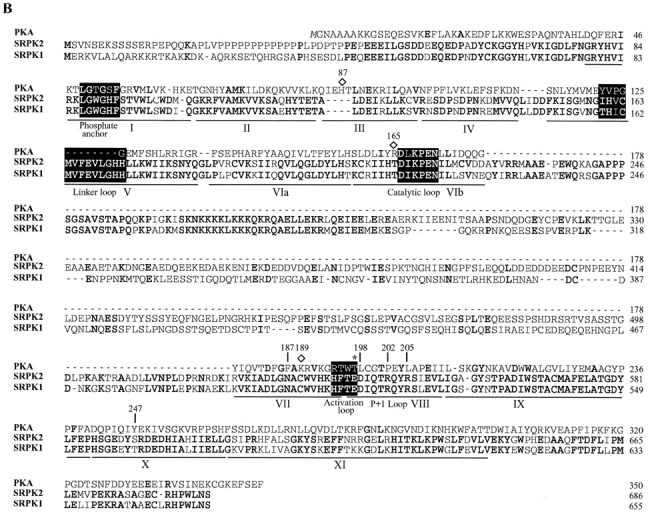
Sequence of SRPK2. (A) Nucleotide and deduced amino acid sequences of SRPK2 (GenBank/EMBL/DDBJ accession number U88666). Underlined is a proline-rich sequence and boxed are kinase catalytic domains. (B) Sequence comparison with SRPK1 and PKA. Kinase domains are underlined and marked according to Hanks and Quinn (1991). Identical amino acids between SRPK1 and 2 and conserved amino acids in PKA are in bold. Gaps (indicated by dashes) were introduced to optimize the alignment. Amino acids that form the P+1 pocket are numbered according to the PKA nomenclature. Functional loops are highlighted in black boxes. An asterisk indicates Thr197 in PKA and the corresponding amino acids in SRPK1 and 2, and diamonds label the amino acids that form ion pairs with Thr197 in PKA tertiary structure as previously described (Taylor and Radzio-Andzelm, 1994).
Sequence comparison with SRPK1 revealed several features of this new kinase: first, the new kinase displays 77% identity and 90% similarity to SRPK1 over their entire kinase domains (kinase domains are boxed in Fig. 1 A, and individual domains are underlined and marked in Fig. 1 B). Second, like SRPK1, the kinase domains of this new kinase are divided into two halves by a large spacer sequence (Fig. 1), a structural feature rare among serine/ threonine kinases, but common among tyrosine kinases (Hanks and Quinn, 1991). Interestingly, one-third of the spacer region is also highly conserved between the two kinases (Fig. 1 B). Comparison with cAMP-dependent protein kinase A (PKA; Fig. 1 B) indicates that the spacers are inserted between two β sheets (β8 and β9; Taylor and Radzio-Andzelm, 1994), suggesting that the spacers may function as an autonomous domain in these kinases (see below). Third, both SRPK1 and the new kinase have a small deletion between kinase domain II and III, and a small insertion in the linker loop within kinase domain V (Fig. 1 B). These differences may contribute significantly to the activity and specificity of these kinases (see Discussion for further details). Based on these structural similarities as well as on many shared biochemical properties with SRPK1 described below, we named the new kinase SRPK2.
SRPK2 also contains unique features that may be indicative of its function and/or regulation in vivo. Most notably, SRPK2 contains a proline-rich sequence APLVPPPPPPPPPPPPPLPDPTPPEP at the NH2 terminus (underlined in Fig. 1 A). This motif contains the binding consensus core for Src Homology 3 (SH3) domain–containing proteins (PXXP, X-any amino acid, P-proline; Ren et al., 1993), and for a subclass of WW domain–containing proteins that bind to proteins containing PPLP cores (underlined above) usually in the context of polyprolines (Chan et al., 1996; Sudol, 1996a ; Bedford et al., 1997). Indeed, a recent two-hybrid screen using a WW domain–containing protein as bait identified a mouse cDNA fragment, WBP6, which is highly homologous to SRPK2 (Bedford et al., 1997; see Discussion). Accordingly, this NH2-terminal proline–rich motif of SRPK2 may function as a targeting signal to interact with substrates and/or regulators.
SRPK2 Is Specific for RS Domain–containing Splicing Factors
The extensive sequence homology between SRPK1 and 2 suggests that they may have similar enzymatic activity and substrate specificity. To test this possibility, we prepared both kinases by in vitro transcription/translation followed by immunoprecipitation, and SR protein-kinase activity was assayed on beads using bacterially expressed ASF/SF2 as substrate. As shown in Fig. 2 A, both SRPK1 and 2 phosphorylated ASF/SF2 with a similar specific activity, demonstrating that SRPK2 is an active kinase for this SR protein. We then tested phosphorylation of other RS domain–containing splicing factors using purified baculovirus-expressed SRPK2. As shown in Fig. 2 B, SRPK2 efficiently phosphorylated SRp20, ASF/SF2, SC35, SRp40, and SRp55, as well as U2AF65. SRPK2 could not phosphorylate other common kinase substrates such as histone H1 or myelin basic protein (data not shown). Therefore, like SRPK1, SRPK2 is specific for RS domain–containing splicing factors.
To determine whether SRPK2-mediated phosphorylation took place in the RS domain and to further characterize the sequence requirement for SRPK2-mediated phosphorylation, we tested phosphorylation of a panel of ASF/ SF2 mutants. Previous studies demonstrate that, although both SRPK1 and Clk/Sty phosphorylate this splicing factor in its RS domain, they have different sequence requirements (Colwill et al., 1996b ). For example, changing serines to threonines in all RS dipeptides in ASF/SF2 abolished phosphorylation by both kinases, indicating that both proteins are serine-specific kinases. However, changing arginines to lysines abolished phosphorylation by SRPK1, but had little effect on phosphorylation by Clk/ Sty, indicating that SRPK1, but not Clk/Sty, requires arginine for substrate recognition and phosphorylation. As shown in Fig. 2 C, SRPK2-mediated phosphorylation of ASF/SF2 was abolished when the RS domain was deleted (ΔRS), showing that phosphorylation occurs in this domain. Replacing serines by threonines (RT) or glycines (RG) in all RS dipeptides in ASF/SF2 diminished phosphorylation (some residual phosphorylation likely takes place at unmutagenized serines outside the RS repeats; Gui et al., 1994b ), and mutation of arginines in the RS dipeptides, KS and GS, resulted in a complete loss of phosphorylation of ASF/SF2 by SRPK2. Therefore, SRPK2 has the same substrate specificity as SRPK1, displaying a stringent requirement for both arginine and serine in phosphorylating RS domain–containing splicing factors.
Previous studies also showed that a specific phosphorylated epitope is present in all standard SR proteins purified from mammalian cells and is recognized by the monoclonal antibody mAb104 (Zahler et al., 1992). This phosphoepitope can be restored to bacterially expressed ASF/ SF2 by both SRPK1- and Clk/Sty-mediated phosphorylation (Gui et al., 1994b ; Colwill et al., 1996b ). To verify if this is also true for SRPK2, bacterially expressed GST-ASF/SF2 was phosphorylated by SRPK2 and analyzed by Western blotting using mAb104. Fig. 2 D shows that ASF/ SF2 did regain the mAb104 specific phosphoepitope upon phosphorylation by SRPK2, and the phosphorylation was also accompanied by a mobility shift in SDS-PAGE as expected (data not shown, see Fig. 5 A). Taken together, these data demonstrate that SRPK1 and 2 have identical biochemical properties in phosphorylating RS domain–containing splicing factors in vitro, and support the idea that SRPK2 is one of the kinases responsible for phosphorylating those splicing factors in vivo.
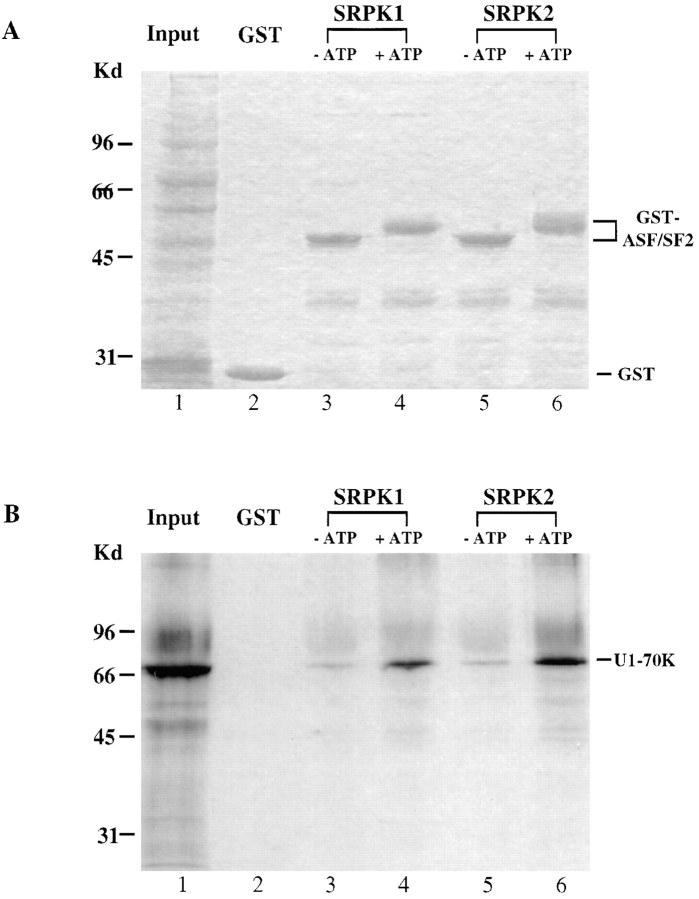
Figure 5.
In vitro protein–protein interaction enhanced by SRPK-mediated phosphorylation. Bacterial GST-ASF/SF2 was phosphorylated by SRPK1 (lane 4) or SRPK2 (lane 6) or mock-phosphorylated (lanes 3 and 5). In vitro translated U1 70K (lane 1, total input in each binding assay) was incubated with beads coupled with GST or treated GST-ASF/SF2. Bound proteins were resolved in a 10% SDS-PAGE followed by autoradiography. (A) Coomassie blue staining of total (lane 1) and bound proteins (lanes 2–6). Note that phosphorylated GST-ASF/SF2 displayed a marked mobility shift. (B) An autoradiograph of the same protein gel. U1 70K was not retained by GST (lane 2), but bound to some extent to mock-phosphorylated GST-ASF/SF2 (lanes 3 and 5) and efficiently to phosphorylated GST-ASF/SF2 by SRPK1 (lane 4) and SRPK2 (lane 6).
Selection of Preferred Substrates
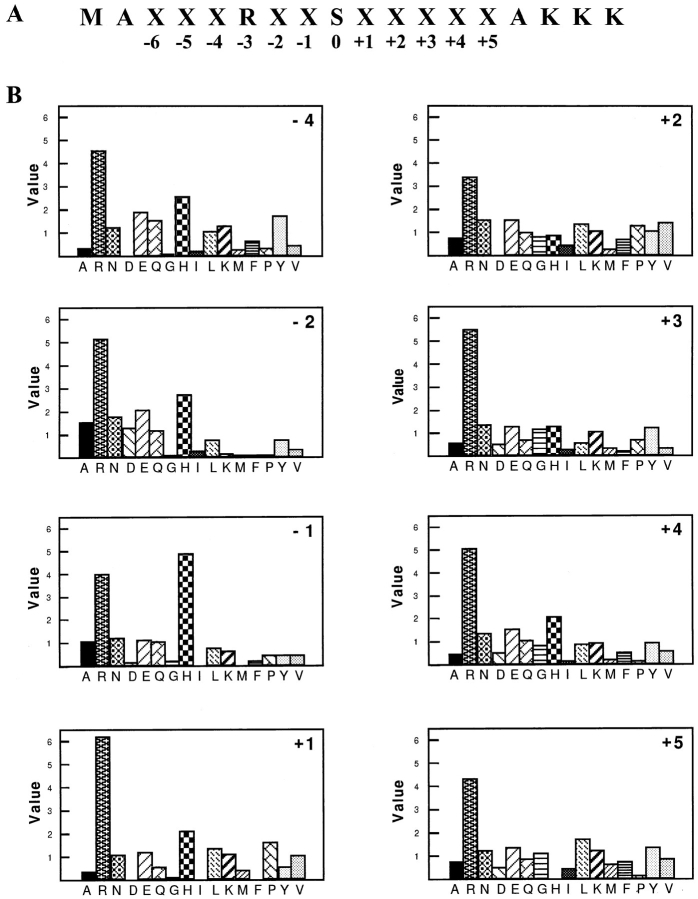
The above biochemical characterization demonstrates that the SRPK family members are highly specific for RS domain–containing splicing factors. To further define the substrate specificity and understand the structural basis for phosphorylation-site selection by the SRPK family of kinases, we took a peptide selection approach that has been developed to determine the substrate specificity of protein kinases (Songyang et al., 1994, 1996). The method involves phosphorylating a random peptide library using a purified kinase followed by quantitative separation and isolation of phosphopeptides, and then sequencing of the selected phosphopeptide pool. Amino acid(s) enriched in a given position will indicate a preference for this amino acid at that position in substrate recognition and phosphorylation. Because the SRPK family members phosphorylate RS domain–containing splicing factors, they are likely to be arginine-directed kinases (Songyang et al., 1996). Therefore, we chose a biased library in which serine was placed at position 0, and arginine at position −3 (Fig. 3 A). Purified SRPK2 was used to phosphorylate the peptide library. After quantitative removal of unphosphorylated peptides by iron-chelating chromatography, SRPK2-phosphorylated peptides were sequenced. Preferred amino acids at each position are presented in Fig. 3 B. Although the experiment was done using SRPK2, it is reasonable to assume that the data are representive of the SRPK family of kinases because both SRPK1 and 2 are highly related, displaying very similar kinase activity and substrate specificity as described above.
Figure 3.
Substrate specificity of SRPK2 determined by peptide selection. (A) The serine-oriented and arginine-directed peptide library used in this study. M = Met, A = Ala, K = Lys, and X = any amino acid except Ser, Thr, Cys, and Trp. The exclusion of Ser and Thr in the library was to ensure phosphorylation at the single serine residue, and omission of Cys and Trp was to avoid problems with sequencing and oxidation. Positions relative to the serine residue are indicated. (B) Each panel shows the relative abundance of amino acids at each position, which is indicated in the upper right corner.
As shown in Fig. 3 B, SRPK2 generally prefers arginine, but not lysine, near the phosphorylation site, which is consistent with the fact that both SRPK1 and 2 phosphorylate SR proteins, but not the KS mutant of ASF/SF2 (Fig. 2). Besides arginine, SRPK2 also selected histidine NH2-terminal of the serine. These observations suggest that the kinase may require positive charges on both sides of the serine, indicating that arginine-serine-arginine (RSR) is the best substrate for the kinase. Based on these selection data in combination with the mutagenesis results, it appears that phosphorylation mediated by the SRPK family of kinases requires three criteria: (a) SR and RS dipeptides are preferred; (b) A basic environment around the SR or RS dipeptide is critical; and (c) Lysine is not allowed, especially at the −2 position (Fig. 2 C and Fig. 3 B). These criteria explain why the SRPK family of kinases could not phosphorylate most common kinase substrates including histone H1 and myelin basic proteins (many contain lysines and hydrophobic amino acids around phosphorylation sites). Further, one may use these criteria to predict potential SRPK phosphorylation sites in a candidate protein substrate (see Discussion).
There are two notable exceptions to RSR as the best substrate in the selection. First, proline was also preferentially selected at the +1 position (compared to other positions). This finding explains the previous observation that SRPK1 could phosphorylate the SPRY peptide in ASF/ SF2 in vitro (Colwill et al., 1996b ). However, a synthetic SPRY peptide was not a good substrate for SRPK1 (Colwill et al., 1996b ), likely due to the lack of surrounding basic amino acids. Second, both polar (N and Q) and acidic (D and E) amino acids are significantly selected at the −2 position. This observation implies that the kinase may phosphorylate an SRSR-containing peptide, regardless of whether the first serine is unmodified (polar) or phosphorylated (acidic; note that serine was not included in randomized positions in the library so that its selection was not detected). Therefore, the SRPK family of kinases may be capable of phosphorylating both unphosphorylated and partially phosphorylated SR proteins.
Structural Basis for Substrate Specificity
Perhaps the most striking feature of the SRPK family of kinases is the strong selection of arginine at the P+1 position, which is in contrast to most protein kinases characterized so far (Songyang et al., 1994, 1996). Previous structural studies have revealed a pocket (called the P+1 pocket) in the kinase catalytic core that is specific for selecting the amino acid at the P+1 position in the substrate. Previous studies have established that the composition of the P+1 pocket is highly predictive of substrate specificity (Songyang, Z., unpublished results). To understand the mechanism for the P+1 selection by the SRPK family members, we aligned SRPK sequences with a number of kinases whose tertiary structures have been determined, and therefore the composition of their P+1 pockets is known (the sequence alignment of SRPK1 and 2 with PKA is shown in Fig. 1 B). The sequence comparison revealed identical P+1 pockets for SRPK1 and 2, which is consistent with identical substrate specificity between the two kinases, and provides a rationale for the selection of arginine at the P+1 position, as described below.
At positions marked in Fig. 1 B and summarized in Fig. 4, the P+1 pockets for PKA and many other kinases (their sequences not shown, see Hanks and Quinn, 1991) are composed of hydrophobic residues, explaining the preference of hydrophobic residues at the P+1 position in both selected and natural substrates (Songyang et al., 1994). In contrast, previous modeling predicted that the arginine in mitogen-activated protein kinase (MAPK) at the PKA 205 equivalent position sits in the bottom of the P+1 pocket and directly interacts with a secondary amine group in the substrate (Songyang et al., 1994). Proline was strongly selected by MAPK at the P+1 position because it is the only natural amino acid that has a secondary amine. For a similar reason, perhaps, proline was selected (although to a lesser extent) by SRPK2 as the kinase also has an arginine at this position (Arg550 in SRPK2, see Fig. 1 B and Fig. 4). Casein kinase IIα (CKIIα) has a highly basic P+1 pocket including an arginine at the PKA equivalent position 198, which is consistent with its selection for an acidic residue at the P+1 position in the substrate as explained earlier (Songyang et al., 1996). In contrast, the P+1 pocket of both SRPK1 and 2 contains conserved polar (Asn532 in SRPK2, which corresponds to Phe187 in PKA) and acidic (Asp543, which corresponds to Leu198 in PKA) residues. This acidic amino acid in the P+1 pocket of these kinases may be responsible for strong selection of arginine in the P+1 position in the substrate.
As described in the Introduction, Clk/Sty is capable of phosphorylating SR proteins; however it can also phosphorylate many other substrates which do not contain any RS motif (Colwill et al., 1996a ,b). Sequence alignment revealed that the P+1 pocket of Clk/Sty (Fig. 4) is similar to that of MAPK, explaining why Clk/Sty can phosphorylate the SPRY peptide more efficiently than SRPK1 (Colwill et al., 1996b ). Further, a polar serine (Ser327 in Clk/Sty) at the PKA equivalent position 187 may be partially responsible for the selection of arginine at the P+1 position. This may be functionally significant as this serine residue is conserved among all three Clk/Sty family members from both mouse and human (Hanes et al., 1994). One may further speculate that the phosphorylation specificity of Clk/ Sty for RS domain–containing splicing factors may be regulated by its own phosphorylation, thereby converting this polar amino acid (Ser327) to a negatively charged residue (phosphoserine). Clk/Sty may be autophosphorylated (Johnson and Smith, 1991; Ben-David et al., 1991) or phosphorylated by other kinases, but the phosphorylation site(s) remains to be determined and the postulated function and regulation of the serine residue in the P+1 pocket in the determination of substrate specificity will have to be proven by mutagenesis.
The SRPK Family of Kinases Mediates In Vitro Interactions between SR Domain–containing Splicing Factors
The biochemical data strongly suggest a role of SRPK1 and 2 in splicing. Recently, it was demonstrated that Clk/ Sty–mediated phosphorylation of ASF/SF2 facilitates its interaction with another RS domain–containing splicing factor, U1 70K (Xiao and Manley, 1997). To determine whether the SRPK family of kinases has a similar function, we tested the effect of SRPK1- and 2-mediated phosphorylation on protein–protein interactions by the GST-pulldown assay. Bacterially produced ASF/SF2 was efficiently phosphorylated by either kinase, resulting in a marked mobility shift in SDS-PAGE (Fig. 5 A), and phosphorylated ASF/SF2 interacted with in vitro translated U1 70K much more efficently than the mock-phosphorylated protein (Fig. 5 B). GST alone did not bring down any U1 70K, demonstrating the specificity of the assay. These data support a role for the SRPK family of kinases in spliceosome assembly by facilitating specific protein–protein interactions among spliceosomal components.
Localization of SRPK1 and SRPK2 and Evidence for Their Interaction with Splicing Factors In Vivo
It is known that splicing factors are concentrated in a speckled pattern in the nucleus, yet splicing takes place cotranscriptionally on nascent transcripts (for review see Spector, 1993). Thus, splicing factors must be recruited to sites of transcription and splicing (Jimenez-Garcia and Spector, 1993; Huang and Spector, 1996; Misteli and Spector, 1997). Previous studies showed that kinases, such as SRPK1 (Gui et al., 1994a ) and Clk/Sty (Colwill et al., 1996a ), were able to induce the redistribution of splicing factors from nuclear speckles to the nucleoplasm, indicating that these kinases may play an active role in the recruitment process. To provide further evidence that the SRPK family members interact with their substrates in vivo and mediate the localization of splicing factors in the nucleus, we determined the localization of both SRPK1 and 2 by peptide tagging or fusing to green fluorescent protein (GFP), and examined the effect of overexpression of these kinases on the localization of endogenous splicing factors.
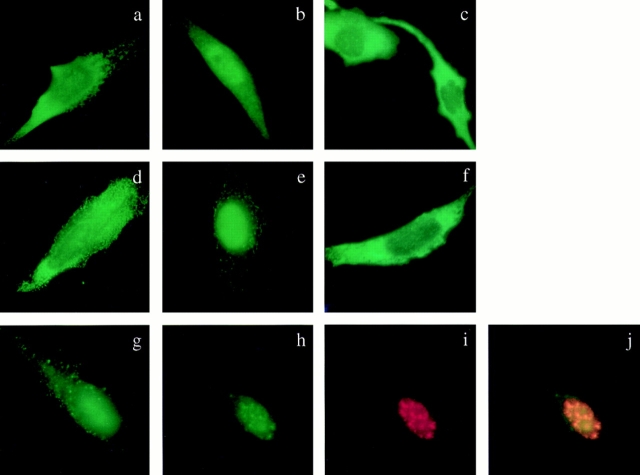
Previous studies showed that Clk/Sty is localized in the nucleus, consistent with its function in splicing (Colwill et al., 1996a ). As shown in Fig. 6, we also detected both SRPK1 and 2 in the nucleus. Surprisingly, both these kinases are also found in the cytoplasm. Among transfected cells, some displayed predominant cytoplasmic (Fig. 6, a and d), whereas others displayed nuclear (Fig. 6, b and e) localization. This heterogeneous pattern could be attributed to the level of the expressed kinases, the physiological state of the cell, or fixation and permeabilization conditions during immunolocalization. To examine the localization of the expressed kinases without fixation and permeabilization, we directly visualized GFP-SRPK1 (Fig. 6 c) and GFP-SRPK2 (Fig. 6 f) in living cells. The cytoplasmic signal appears to be predominant (Fig. 6, c and f), although the expressed kinases are also clearly visible in the nucleus (see below for further details). Therefore, it is likely that the endogenous kinases are present in both cellular compartments.
Figure 6.
Localization of SRPK1 and SRPK2 in transfected HeLa tTA cells. FLAG-tagged SRPK1 (a and b) and SRPK2 (d and e) are localized in both the cytoplasm (represented in a and d) and the nucleus (represented in b and e). Predominant cytoplasmic signal was seen with GFP-SRPK1 (c) and GFP-SRPK2 (f), although some punctate nuclear population was visible (see c, for example). When viewed in different focal planes, signals in both the cytoplasm (g) and the nucleus (h) were evident, and the nuclear population (h) colocalized with the B1C8 antigen (i), a marker for nuclear speckles. A merged image appears yellow (j).
The localization of the SRPK family of kinases in the cytoplasm is interesting because many kinases with a nuclear function are regulated by nuclear translocation. In fact, Dsk1, an SRPK family member from fission yeast, was also found to localize predominantly in the cytoplasm in interphase, but in the nucleus during mitosis (note that the yeast nuclear envelope does not break down in mitosis; Takeuchi and Yanagida, 1993). Thus, only a fraction of these kinases may be required for splicing in the nucleus in interphase, but a maximal level of such kinases may be necessary for the reorganization of the splicing machinery during mitosis, as suggested previously (Gui et al., 1994a ). Our studies, however, do not preclude the possibility that the SRPK family of kinases has a function in the cytoplasm. We further observed that SRPK1 and more clearly SRPK2 are not diffusely localized in the cytoplasm (Fig. 6, a, d, and g). Instead, they appear to be attached to some structure, which may function by anchoring these kinases in the cytoplasm. We are currently determining the sequence requirements for the localization of both kinases in the cytoplasm as well as in the nucleus.
Upon a close inspection of transfected cells, a punctate nuclear population of both kinases was occasionally seen in both fixed and living cells (see Fig. 6 c, for an example). When focused on different focal planes (compare Fig. 6, g and h), SRPK2 was clearly colocalized with B1C8 (Fig. 6, h–j), a novel RS domain–containing splicing factor (Blencowe and Sharp, personal communication) previously characterized as one of the markers for nuclear speckles (Blencowe et al., 1994). Similarly, we also detected colocalization of GFP-SRPK1 with SC35 in nuclear speckles (data not shown). These observations suggest that at least some fraction of SRPK1 and 2 interacts with their substrates in the nucleus.
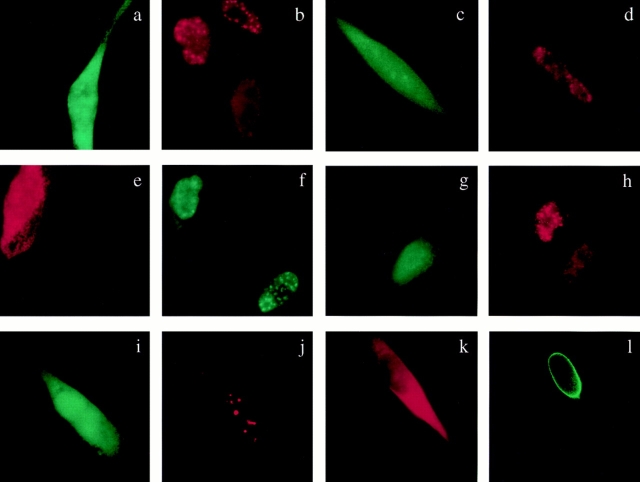
In most SRPK-transfected cells, images of nuclear speckles appear diffused, a situation reminiscent of overexpression of wild-type Clk/Sty (Colwill et al., 1996a ). In fact, this effect may reflect the movement of splicing factors mediated by the SRPK and Clk/Sty families of kinases in the nucleus (for review see Fu, 1995). As shown in Fig. 7, overexpression of both SRPK1 (Fig. 7, a and b) and SRPK2 (Fig. 7, e–h) effectively induced the redistribution of endogenous splicing factors from nuclear speckles to nucleoplasm. Splicing factors, such as the B1C8 antigen (Fig. 7 b) and SR proteins detected by mAb104 (data not shown), are concentrated in nuclear speckles in untransfected cells, but became diffusely localized in cells overexpressing SRPK1. Similar results were also obtained with SRPK2 (Fig. 7, e–h). In contrast, overexpression of a mutant SRPK1, which contains a point mutation (K109M) at the ATP-binding site and was previously shown to be an inactive kinase (Colwill et al., 1996b ), had little effect on the distribution of endogenous SR proteins (Fig. 7, c and d). These data are consistent with our published results that application of purified SRPK1 onto permeabilized cells in vitro induced the disassembly of the speckled nuclear domains (Gui et al., 1994a ), and strongly suggest that SRPK family members are directly involved in releasing SR proteins from nuclear speckles to the nucleoplasm. This may represent an essential step in recruiting splicing factors to nascent transcripts for cotranscriptional splicing suggested by Spector and his colleagues (Jimenez-Garcia and Spector, 1993; Huang and Spector, 1996; Misteli and Spector, 1997).
Figure 7.
Specific induction of redistribution of endogenous splicing factors by overexpression of SRPK1 and SRPK2. The B1C8 antigen (b, d, and f) and SR proteins (h) stained with mAb104 are concentrated in nuclear speckles of untransfected cells. Overexpression of FLAG-tagged SRPK1 (a) and SRPK2 (e and g) induced the redistribution of both the B1C8 antigen (b and f) and SR proteins (h) from speckles to the nucleoplasm. Overexpression of inactive SRPK1 had little effect on SR protein localization (c and d). Reduced signal in kinase-expressed cells likely reflects some loss of solubilized splicing factors during fixation and permeabilization. In contrast, overexpression of FLAG-tagged SRPK2 (i and k) had no effect on the structural integrity of PML-positive PODs detected by 5E10 (j) or the nuclear envelope stained with anti-lamine antibodies (l).
Finally, other nuclear structures, such as the PML oncogenic domains (Dyck et al., 1994) detected by the anti-PML mAb5E10 (Stuurman et al., 1992) and nuclear envelope detected by anti-lamine A/C antibodies (Gui et al., 1994a ), were not affected by overexpression of either SRPK1 or 2 (Fig. 7, i–l). Therefore, the SRPK family of kinases appears to specifically mediate the redistribution of splicing factors in the nucleus.
SR Protein Kinases Are Differentially Expressed
Current data suggest that both the Clk/Sty and SRPK families of kinases are involved in the regulation of spliceosome assembly by mediating the localization and interaction of splicing factors in mammalian cells. It is, however, unlikely that they are completely redundant kinases, as each kinase may have a more defined spectrum of substrates. Alternatively, these kinases may be differentially expressed and/or regulated. To explore these possibilities, we examined the expression of these SR protein kinases by Northern blotting analysis in multiple human tissues as well as in many commonly used experimental cell lines. As shown in Fig. 8, SRPK1 and 2 are expressed as single 4.4- and 4.3-kb transcripts, respectively, in multiple human tissues. SRPK1 is highly expressed in pancreas whereas SRPK2 is highly expressed in brain. Both kinases are moderately expressed in other tissues including heart and skeletal muscle and the expression of both is low in lung, liver, and kidney. These results clearly show that SRPK1 and 2 are differentially expressed, which may contribute to the tissue specific regulation of both constitutive and alternative splicing.
In 11 experimental human cell lines examined, SRPK1 and 2 are ubiquitously expressed, although with notable differences (Fig. 8, C and D). Compared to their expression in HeLa cells after normalizing against GAPDH, SRPK1 is expressed at a higher level than SRPK2 in Weri-1 and Lan5, but much lower in Soas2 cells, and both kinases exhibit a low level of expression in two leukemia cell lines, THP1 and U937. Clk/Sty is expressed as two transcripts (3.2 and 1.8 kb) resulting from alternative splicing as characterized previously (Duncan et al., 1995). The 1.8-kb transcript corresponds to the primary cDNA for the active kinase, whereas the 3.2-kb mRNA results from a partially spliced RNA, and therefore does not encode a functional kinase (Duncan et al., 1995). Interestingly, the 1.8-kb message is ubiquitously expressed, whereas the expression of the 3.2-kb RNA is cell specific (Fig. 8 E, lane 3, for example), indicating that Clk/Sty may be regulated by alternative splicing in a cell specific fashion. These results will guide us in choosing a cellular model to study the regulation of splicing by specific kinases in the future. Together, these data demonstrate that SR protein kinases are differentially expressed. Thus, various SR protein kinases may be regulated at multiple levels by differential expression, alternative splicing, nuclear translocation, phosphorylation activation, or suppression, etc. These multiple regulation pathways in combination with their distinct substrate specificity likely contribute to the complexity of regulation of alternative splicing in mammalian cells and tissues, in response to signaling, or during development.
Discussion
Expression of Multiple SR Protein Kinases in Mammalian Cells
Two families of SR protein kinases (Clk/Sty and SRPK) are now well characterized. The Clk/Sty family has three members (Hanes et al., 1994) and the SRPK family has two. Chromosome mapping suggests that the SRPK1 locus is situated on mouse chromosome 17 and human chromosome 6, and SRPK2 on mouse chromosome 5 and human chromosome 7, respectively (Wang, H.-Y., J.B. Bermingham, W. Lin, K.C. Arden, and X.-D. Fu, manuscript in preparation). In addition, probes derived from the conserved kinase domains detected additional loci in mouse chromosome X and human chromosome 8 and 18 (our unpublished results), indicating that these loci may encode additional SRPK family members or pseudogenes. Consistent with this prediction, a cDNA fragment (named WBP6) was recently isolated from a mouse library in a two-hybrid screen, which resembles the human SRPK2 (Bedford et al., 1997). The isolated partial cDNA has the coding capacity for proline-rich and kinase domains nearly identifical to the corresponding domains in SRPK2. However, the partial clone lacks NH2 terminal sequence, and the reported NH2 terminal part is completely different from that of SRPK2. Thus, it remains unclear whether the isolated cDNA represents the mouse homologue of human SRPK2 or another highly related kinase.
Kinases responsible for phosphorylation of RS domain– containing splicing factors may not be confined to the two families described above. Previous work showed that PKA and protein kinase C can phosphorylate ASF/SF2 in vitro (Colwill et al., 1996a ), although the functional significance of these phosphorylation events remains unclear. In addition, the NimA kinase selects arginines on both sides of serine, suggesting that this kinase may also be involved in phosphorylating SR proteins (Songyang et al., 1996). Consistent with this possibility, a catalytically mutant form of this kinase has been shown to colocalize with splicing factors in nuclear speckles (Lu et al., 1996). However, unlike SRPK2, NimA stringently requires a phenylalanine at the −3 position (Songyang et al., 1996), indicating that the kinase may, at most, phosphorylate only a subset of SR proteins. NimA functions as a cell cycle regulator (Lu and Hunter, 1995), but whether or not it also has a role in splicing and is capable of phosphorylating some SR proteins remain to be verified in future studies.
Structural Basis for the Activity and Specificity of the SRPK Family of Kinases
Recombinant SRPK1 and 2 are highly active and specific, indicating that these kinases may have an open conformation for substrate recognition and catalysis. Although the crystal structure of these kinases remains to be elucidated, comparison with PKA revealed a basis for substrate specificity in the P+1 pocket as described in Results. In addition, the insertion in the linker loop within kinase domain V (Fig. 1 B) may contribute to the separation between the small and large lobes, and thus, to an open conformation of the SRPK family of kinases for substrate recognition and orientation. In PKA, Thr197 in the activation loop is consititutively phosphorylated upon synthesis and the phosphate on Thr197 is highly resistant to hydrolysis by phosphatase (Steinberg et al., 1993). According to the PKA crystal structure, this phosphothreonine forms ion pairs with His87 in the small lobe, and Arg165 and Lys189 in the large lobe, and these interactions are postulated to be critical for the conformation of the active site in the enzyme (see discussion by Taylor and Radzio-Andzelm, 1994). The salt bridge between His87 and phosphothreonine197 is at least partially responsible for the closed conformation of PKA (Knighton et al., 1991), and the interaction is broken for the open conformation of the enzyme (Zheng et al., 1993). As discussed earlier regarding the structural features of Clk/Sty and SRPK1 (Colwill et al., 1996a ), these two families of kinases have a completely different set of amino acids in the corresponding positions. As illustrated in Fig. 1 B, Thr197 in PKA is replaced by a glutamic acid residue in both SRPK1 and 2; it is unclear which amino acid corresponds to His87 as the region in kinase domain III contains a small, but critical deletion in both SRPK1 and 2; and Arg165 and Lys189 are replaced by threonine and cysteine, respectively. These differences suggest that SRPK1 and 2 as well as Clk/Sty may have a very different conformation than PKA at their active sites, which may explain why these two families of kinases are both highly active and selective in phosphorylating RS domain–containing splicing factors.
Proline-rich Domain in SRPK2
Characterization of the second SRPK family member not only illustrates the complexity of regulation of splicing factors by multiple kinases, but also implies potential regulation of the kinase itself. One interesting structural feature of SRPK2 is its NH2 terminal proline–rich domain, which contains several consensus binding sites for an SH3 domain (Ren et al., 1993). To explore potential regulation of splicing by signaling, we are currently testing whether SRPK2 is associated with any SH3 domain–containing proteins. The proline-rich sequences are also frequent binding targets for WW domain–containing proteins (Chen and Sudol, 1995; Chan et al., 1996; Macias et al., 1996; Sudol, 1996b ). Indeed, it was recently shown that a formin-binding protein interacts with a SRPK2-like kinase (WBP6) from mouse in a two-hybrid screen (Bedford et al., 1997). Whether this association is of functional significance remains to be investigated. The WW domain is present in a growing number of structural, regulatory, and signaling molecules (see an automatic update of WW domain–containing proteins under the following electronic address: http://www.brok.embl-heidelberg.de/Modules/ww-gif.html; Sudol et al., 1996a). Some splicing factors, such as Prp40 from yeast, also contain this domain (Kao and Siliciano, 1996). Therefore, it will be interesting to investigate whether the NH2 terminal proline–rich domain in SRPK2 interacts with WW domain proteins, especially WW domain–containing splicing factors. Such interactions may contribute to a unique substrate specificity, regulation and/or cellular targeting of the kinase in mammalian cells.
Implication of Nuclear and Cytoplasmic Localization of the SRPK Family of Kinases
The SRPK family of kinases is characterized by a large spacer sequence dividing conserved catalytic kinase domains into two halves. Structural modeling of both kinases suggests that the spacers constitute a unique domain, which may function autonomously. This structural arrangement is rare among serine/threonine kinases, but common in tyrosine kinases. For example, upon PDGF binding on the cell surface, the PDGF receptor becomes phosphorylated in its spacer, which then serves as a platform for signal relay by interacting with adaptor molecules (for review see Ullrich and Schlessinger, 1990). By such an analogy, the spacer sequences may also influence the biological function of SRPKs in mammalian cells.
A clue to the role of these spacer sequences comes from the observation that deletion of the spacer from Dsk1, a SRPK family member from fission yeast, resulted in exclusive nuclear localization of the mutant kinase in interphase cells (Takeuchi and Yanagida, 1993). A similar phenomenon was also seen with both SRPK1 and 2 (our unpublished results). Thus, the spacer sequences may be involved in the regulation of nuclear translocation of these SR protein kinases.
As shown above, SRPK1 and 2 are present in the nucleus and cytoplasm. The localization of these kinases in the nucleus is consistent with their putative function in splicing. The significance of their presence in the cytoplasm is unclear, but might reflect additional functions of these kinases in the cytoplasm. Alternatively, a fraction of these kinases may be restricted to the cytoplasm because oversupply of these kinases can completely alter the organization of the splicing machinery in the nucleus, and therefore, may be detrimental to normal cell physiology. We observed that both SRPK1 and 2 are localized in the cytoplasm in a nondiffused fashion, although the underlying cytoplasmic structure remains to be characterized. These findings, taken together with the observations that SRPK1 and 2 become exclusively localized in the nucleus after their spacers are removed (our unpublished observations), suggest that these kinases may be anchored in the cytoplasm through their spacer sequences.
Rules to Predict Preferred Phosphorylation Sites for the SRPK Family of Kinases
Our peptide selection studies have provided a structural basis to explain the stringent substrate specificity observed with the SRPK family of kinases. This information can also be used to predict potential kinase substrates. As described in Results, these kinases appear to prefer phosphorylating an SR or RS dipeptide in a basic environment enriched with arginine or histidine, but not lysine. This rule applies to all proven substrates tested so far in our lab. For example, all standard SR proteins satisfy such a rule and they can be efficiently phosphorylated by either SRPK1 or 2 in vitro. We also tested phosphorylation of other RS domain–containing splicing factors, such as human U2AF65 (Zamore et al., 1992) and U2AF35 (Zhang et al., 1992), and their homologues from Drosophila (dU2AF50 and dU2AF38; Kanaar et al., 1993; Rudner et al., 1996) and fission yeast (Prp2; Potashkin et al., 1993). All these RS domain–containing splicing factors contain multiple SR or RS dipeptides and are phosphorylated by SRPK1 (our unpublished observations).
It should be emphasized that not all RS domain–containing proteins implicated in splicing are substrates for SRPK1 or 2. For example, Clk/Sty contains 10 SR or RS repeats at the NH2 terminus (Ben-David et al., 1991). However, it could not be phosphorylated in vitro by SRPK1 (our unpublished result), and inspection of its RS domain indicates that none of those dipeptides satisfies the criteria for recognition and phosphorylation by SRPK1 and 2 as described above. In addition, Snp1p, a U1 70-K homologue from yeast, contains a number of dispersed SR or RS dipeptides (Smith and Barrell, 1991). Again, none of those dipeptides is surrounded by arginine or histidine, and Snp1p could not be phosphorylated by SRPK1. In contrast, the B1C8 antigen, which was initially identified by an antinuclear matrix monoclonal antibody (Blencowe et al., 1994), was found to be a good substrate for SRPK1 during its biochemical purification, and recent cloning and characterization show that it is an RS domain–containing splicing factor (Blencowe et al., 1998). Finally, we have recently cloned and characterized a novel splicing factor, which is a U2AF35-related protein called Urp in a two- hybrid screen using SRPK1 as bait (Tronchere et al., 1997). Urp contains a typical RS domain at the COOH terminus and the protein was an excellent substrate for SRPK1. Therefore, the stringent substrate specificity and the sequence requirement defined by peptide selection may be used to identify additional RS domain–containing proteins that may have a role in splicing.
How SR Protein Kinases May Affect Splicing
It was recently reported that Clk/Sty facilitates SR protein–protein interactions and prevents nonspecific protein–RNA interactions (Tacke and Manley, 1997; Xiao and Manley, 1997). In this paper, we show that the SRPK family of kinases has an analogous function in promoting SR protein–protein interactions. Thus, it is conceivable that multiple kinases are involved in splicing in mammalian cells. However, the function of these SR protein kinases in splicing is just beginning to be understood. Previous studies using phosphatase inhibitors indicate that phosphorylation of SR proteins is required for spliceosome assembly, and thus, SR protein kinases can be considered as activators for SR proteins (Mermoud et al., 1994). However, subsequent dephosphorylation is also important for splicing as blocking dephosphorylation by phosphatase inhibitors prevented splicing from occurring in the fully assembled spliceosome (Mermoud et al., 1994). In agreement with this finding, we observed that too much SRPK1 is also inhibitory to splicing, likely by interfering with dephosphorylation in a later step of the splicing reaction (Gui et al., 1994a ). These observations are consistent with the idea that the level of SR protein kinases in the nucleus must be regulated.
SR proteins are known to affect splice-site selection both in vitro and in vivo, and, as such, it seems likely that SR protein kinases might affect alternative splicing. Earlier studies demonstrated that SR proteins generally stimulate the selection of the proximal 5′ and 3′ splice sites if two competing splice sites contain identical splicing signals (Fu et al., 1992). This situation, however, is unlikely to be the case in most natural alternative splicing systems. In the SV-40 T/t alternative splicing system, for example, different SR proteins were shown to prefer different splice sites (Zahler et al., 1993). Therefore, selection of a specific splice site may be dictated by combinatory actions and competition of mutiple SR proteins and other splicing factors. For this reason, it will be difficult to predict which direction a specific alternative splicing event will follow after the activation of multiple SR proteins by a specific SR protein kinase. A specific alternative splicing event may be regulated by the expression of a unique spectrum of SR proteins, which may in turn be regulated by kinases with distinct activity and substrate specificity. The identification of additional SR protein kinases will ultimately contribute to an understanding of regulatory events governing alternative splicing.
Footnotes
We thank Susan Taylor of University of California at San Diego (UCSD; La Jolla, CA) (UCSD) for many useful discussions on kinase structure and function, and Marius Sudol (Mount Sinai School of Medicine, New York) for many stimulating discussions. We are grateful to B. Blencowe and P. Sharp (Massachusetts Institute of Technology, Cambridge, MA) for B1C8 antibody, L. De Jong (University of Amsterdam) for the 5E10 anti-PML antibody, D. Black (University of California, Los Angeles, Los Angeles, CA) for Weri-1 and Lan5 cell lines, C. Glass (UCSD) for U937 and THP1 cell lines, M. Kamps (UCSD) for HBL100 and HT1080 cell lines, and J. Wang (UCSD) for Bosc23 and Soas2 cell lines. The authors thank members of the Fu lab for cooperation and suggestions during the course of this study, especially S. Chandler for help in SRPK2 expression and purification, and L. Feng for help in immunohistochemistry.
J.A. Dyck and J.M. Yeakley are supported by fellowships from Damon Runyon-Walter Winchell Foundation and National Institutes of Health (NIH), respectively. X-D. Fu is a Leukemia Society of American Scholar. This work was supported by a grant from NIH (GM52872) to X-D. Fu.
Address all correspondence to Xiang-Dong Fu, Division of Cellular and Molecular Medicine, Department of Medicine, University of California, San Diego, La Jolla, CA 92093-0651. Tel.: (619) 534-4937. Fax: (619) 534-8549. E-mail: xdfu@ucsd.edu
1. Abbreviations used in this paper: GFP, green fluorescent protein; GST, glutathione-S-transferase; MAPK, mitogen-activated protein kinase; PKA, protein kinase A; RS, arginine and serine-rich; RSR, arginine-serine-arginine; SH3, src homology 3; snRNP, small nuclear ribonucleoprotein particle; SRPK, SR protein-specific kinase; tTA, tetracycline-controlled transactivator.
References
- Bedford MT, Chan DC, Leder P. FBP WW domains and the ABL SH3 domains bind to a specific class of proline-rich ligands. EMBO (Eur Mol Biol Organ) J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David Y, Letwin K, Tamock L, Bernstein A, Pawson T. A mammalian protein kinase with potential for serine/threonine and tyrosine phosphorylation is related to cell cycle regulators. EMBO (Eur Mol Biol Organ) J. 1991;10:317–325. doi: 10.1002/j.1460-2075.1991.tb07952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., R. Issner, J.A. Nickerson, and P.J. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. In press. [DOI] [PMC free article] [PubMed]
- Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO (Eur Mol Biol Organ) J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B, Cohen PT, Lamond AI. Protein phosphatase 1 can modulate alternative 5′ splice site selection in a HeLa splicing extract. FEBS Lett. 1994;35:276–280. doi: 10.1016/0014-5793(94)00973-2. [DOI] [PubMed] [Google Scholar]
- Chan DC, Bedford MT, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO (Eur Mol Biol Organ) J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Sudol M. The WW of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO (Eur Mol Biol Organ) J. 1996a;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakaly JM, Gish GD, Caceres JF, Pawson T, Fu X-D. SRPK1 and Clk/Sty protein kinases show distinct substrate specifities for serine/arginine-rich splicing factors. JBiol Chem. 1996b;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Duncan PI, Howell BW, Marius RM, Drmanic S, Douville EM, Bell JC. Alternative splicing of STY, a nuclear dual specificity kinase. J Biol Chem. 1995;270:21524–21531. doi: 10.1074/jbc.270.37.21524. [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine-serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994a;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Tronchere H, Chandler SD, Fu X-D. Purification and characterization of a kinase specific for the serine-and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes J, von der Kammer H, Klaudiny J, Scheit KH. Characterization by cDNA cloning of two new human protein kinases. Evidence by sequence comparison of a new family of mammalian protein kinases. J Mol Biol. 1994;244:665–672. doi: 10.1006/jmbi.1994.1763. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Howell BW, Afar DE, Lew J, Douville EM, Icely PL, Gray DA, Bell JC. STY, a tyrosine-phosphorylating enzyme with sequence homology to serine/threonine kinases. Mol Cell Biol. 1991;11:568–572. doi: 10.1128/mcb.11.1.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Johnson KW, Smith KA. Molecular cloning of a novel human cdc2/CDC28-like protein kinase. J Biol Chem. 1991;266:3402–3407. [PubMed] [Google Scholar]
- Kanaar R, Roche SE, Beall EL, Green MR, Rio DC. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science. 1993;262:569–573. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- Kao HY, Siliciano PG. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton DR, Zheng J, Ten LF, Eyck, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hunter T. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell. 1995;81:413–424. doi: 10.1016/0092-8674(95)90394-1. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AI. Ser/Thr-specific protein phosphophatases are required for both catalytic steps pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AE. Regulation of mammalian splicesome assembly by a protein phosphorylation mechanism. EMBO (Eur Mol Biol Organ) J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Serine/threonine phosphatase I modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. . Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci USA. 1993;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V, Barrell BG. Cloning of a yeast U1 snRNP 70K protein homologue: functional conservation of an RNA-binding domain between humans and yeast. EMBO (Eur Mol Biol Organ) J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. A structural basis for the substrate specifities of protein ser/thr kinases: primary sequence preference of casein kinase I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5 and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Steinberg RA, Cauthron RD, Symcox MM, Shuntoh H. Autoactivation of catalytic (C alpha) subunit of cyclic AMP-dependent protein kinase by phosphorylation of threonine 197. Mol Cell Biol. 1993;13:2332–2341. doi: 10.1128/mcb.13.4.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, De Graof A, Floore A, Josso A, Humbel B, De Jong L, Van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996a;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- Sudol M. The WW module competes with the SH3 domain? . Trends Biochem Sci. 1996b;21:161–163. [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module-the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Yanagida M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol Biol Cell. 1993;4:247–260. doi: 10.1091/mbc.4.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Radizo-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunnel C, Luhrmann R. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature. 1993;363:283–286. doi: 10.1038/363283a0. [DOI] [PubMed] [Google Scholar]
- Tronchere H, Wang J, Fu X-D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Knighton DR, Xuong HH, Taylor SS, Sowadski JM, Ten LF, Eyck Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]