Abstract
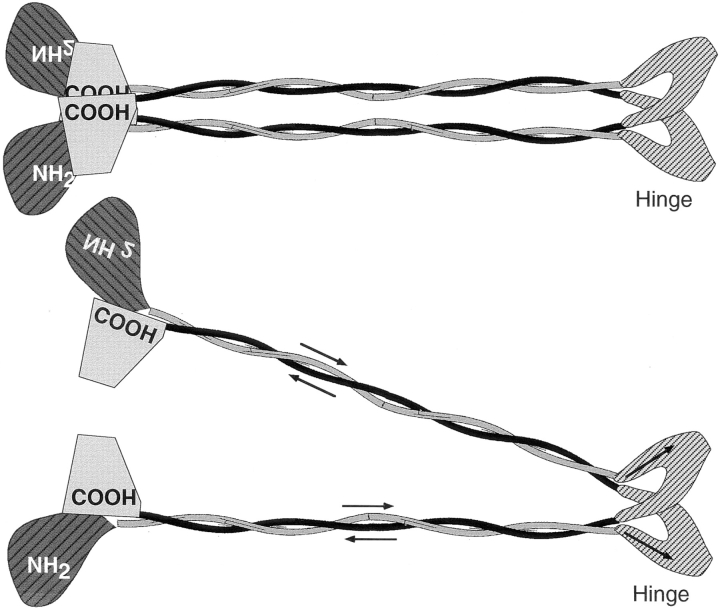
Structural maintenance of chromosomes (SMC) proteins function in chromosome condensation and several other aspects of DNA processing. They are large proteins characterized by an NH2-terminal nucleotide triphosphate (NTP)-binding domain, two long segments of coiled coil separated by a hinge, and a COOH-terminal domain. Here, we have visualized by EM the SMC protein from Bacillus subtilis (BsSMC) and MukB from Escherichia coli, which we argue is a divergent SMC protein. Both BsSMC and MukB show two thin rods with globular domains at the ends emerging from the hinge. The hinge appears to be quite flexible: the arms can open up to 180°, separating the terminal domains by 100 nm, or close to near 0°, bringing the terminal globular domains together.
A surprising observation is that the ∼300–amino acid–long coiled coils are in an antiparallel arrangement. Known coiled coils are almost all parallel, and the longest antiparallel coiled coils known previously are 35–45 amino acids long. This antiparallel arrangement produces a symmetrical molecule with both an NH2- and a COOH-terminal domain at each end. The SMC molecule therefore has two complete and identical functional domains at the ends of the long arms. The bifunctional symmetry and a possible scissoring action at the hinge should provide unique biomechanical properties to the SMC proteins.
Keywords: SMC, MukB, coiled coil, electron microscopy, chromosome
The structural maintenance of chromosomes (SMC)1 proteins are a family of DNA processing proteins almost ubiquitous in bacteria, archaea, and eukaryotes (for reviews see Hirano et al., 1995; Saitoh et al., 1995; Koshland and Strunnikov, 1996; Heck, 1997; Jessberger et al., 1998). Eukaryotic SMC proteins function in chromosome condensation and segregation from fungi to vertebrates (Strunnikov et al., 1993; Hirano and Mitchison, 1994; Saka et al., 1994; Koshland and Strunnikov, 1996), X chromosome dosage compensation in Caenorhabditis elegans (Lieb et al., 1998), sister chromatid cohesion (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998), and other aspects of DNA processing and repair (Jessberger et al., 1996, 1998). The names “condensin” and “cohesin” have been applied to the SMC proteins and their complexes that function in DNA condensation and sister chromatid cohesion (Heck, 1997). SMC proteins are also found in bacteria and archaea. In contrast to eukaryotic SMC proteins, which are generally heterodimers, the prokaryotic SMCs are predicted to be homodimers since there is only one SMC sequence in each genome. In addition to the two SMC peptides that form the dimer, eukaryotic SMC proteins (Guacci et al., 1997; Hirano et al., 1997; Michaelis et al., 1997; Jessberger et al., 1998; Lieb et al., 1998) and Escherichia coli MukB (Yamanaka et al., 1996) have associated proteins that are essential for function. Probably all SMC proteins have accessory proteins. For the present study, however, we will focus on the Bacillus subtilis SMC and E. coli MukB proteins alone, which can form dimeric structures in the absence of the other proteins.
E. coli MukB has been studied extensively both for phenotype of mutants and by in vitro biochemistry (Niki et al., 1991, 1992; Yamanaka et al., 1994; Saleh et al., 1996). Although it has a similar domain structure to the SMCs, its NH2- and COOH-terminal domains are much more distant in sequence than any of the other SMCs, and it has not been considered as a member of the SMC family until recently. In addition to MukB's structural similarity to SMCs, however, mutant phenotypes suggest that the proteins may be functionally analogous. Thus, MukB mutants cause the production of anucleate cells, cells with two nucleoids, and cells with small amounts of DNA caused by a “guillotine effect” (Niki et al., 1991), similar to the phenotype of Cut3/Cut14 mutants in fission yeast (Saka et al., 1994). Britton et al. (1998) recently achieved a gene knockout of the SMC protein of B. subtilis and commented that “the smc phenotypes are remarkably similar to those caused by mukB mutations in E. coli.” The structural analysis we present here adds additional evidence that MukB and Bacillus subtilis SMC (BsSMC) are closely related proteins.
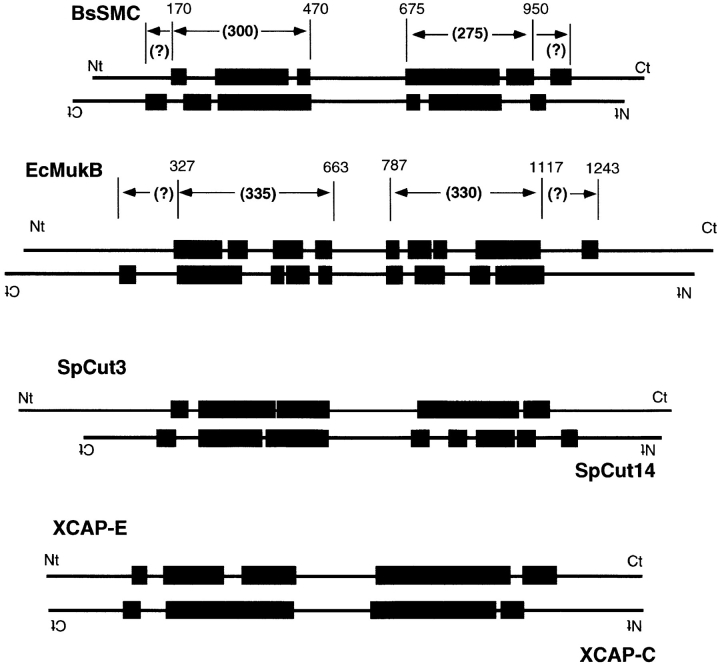
One of the most intriguing features of SMC proteins is the presence of the conserved NTP-binding domain, with potential motor function. Fig. 1 illustrates the domain structure of SMC proteins. For the moment consider only the top line of BsSMC and MukB. The NH2-terminal domain contains a conserved NTP-binding motif (Walker A), and the COOH-terminal domain has been suggested to have a Walker B motif, which is defined as an aspartic acid preceded by four hydrophobic amino acids (aa) (Saitoh et al., 1994, 1995). The NH2- and COOH-terminal domains are separated by a very long coiled coil, which is broken near the middle by a noncoil domain of ∼200 aa. The predicted coils also show minor discontinuities, but as shown below, the coils appear continuous in the electron micrographs. In order for the COOH-terminal domain to contribute to nucleotide binding or hydrolysis, it would have to be physically adjacent to the NH2-terminal domain in the dimeric structure. This could be achieved if the heterodimer were an antiparallel coiled coil, bringing the NH2-terminal domain of one subunit next to the COOH-terminal domain of the other, or if the molecule were bent at the hinge, bringing all the terminal domains together. These two possibilities were suggested by Saitoh et al. (1994) for the chick SCII protein. Remarkably, we find that both structural features are realized by SMC proteins.
Figure 1.
Coiled-coil segments predicted by the program Protean (DNAstar) are shown as black rectangles. Numbers above the vertical lines indicate the aa number, and numbers in parentheses between arrows indicates the total number of aa between the lines. The 275–300-aa segment for BsSMC and the 330–335-aa segment of EcMukB were initially selected as the coiled coil because they matched the 41- and 51-nm lengths measured by EM. However, measurements of the truncated construct MukBcoil (see Discussion) indicate that the coiled coil of MukB probably includes the short segment 1205–1243. The boundaries and alignment of the coiled-coil segments are therefore still ambiguous. Heterodimeric SMCs may also pair by the antiparallel coiled-coil arrangement, as illustrated for Cut3/Cut14 and XCAP-E/XCAP-C.
The most detailed structural studies to date are of E. coli MukB, which has been visualized by EM (Niki et al., 1992). The molecules showed a large and a small globular domain separated by a 48-nm-thin rod. The authors recognized that a single ∼300-aa coiled coil segment would fit this length, so they identified this as the NH2-terminal coiled coil. They assigned the entire COOH-terminal half of the molecule to the large globular domain, so the structure was interpreted simply as two globular domains separated by a coiled coil. We have now obtained higher-resolution images of both E. coli MukB and B. subtilis SMC that show a different and much more elaborate molecular architecture.
Materials and Methods
The BsSMC cDNA was cloned from genomic DNA by PCR, using pfu polymerase (Lu and Erickson, 1997) and adding NdeI and BamHI sites to allow subcloning into the expression vector pET11 (Studier et al., 1990). When induced in BL21 at 37°C, the protein was abundantly expressed but totally insoluble, and attempts to solubilize and renature it were unsuccessful. However, when the bacteria were maintained at 22°C during induction, ∼1/3 of the protein remained soluble. The bacterial supernatant was partially purified by passing over a Sephacryl HR-500 (Pharmacia Biotech, Piscataway, NJ) column in 20 mM Tris, 100 mM NaCl, pH 8, where the BsSMC eluted ahead of most bacterial proteins. The BsSMC from the leading and trailing Sephacryl fractions sedimented identically at 6.3 S on glycerol gradients, indicating a homogeneity of the expressed molecules. The Sephacryl fractions showed some contamination, and in two cases were further purified by chromatography on mono Q (Pharmacia Biotech), where it eluted at 0.2–0.25 M NaCl. However, the best preparations for EM were obtained by running the Sephacryl-purified BsSMC directly on glycerol gradients.
We prepared a truncated form of MukB and an additional chimeric construct. MukBcoil is missing both the NH2- and COOH-terminal domains and includes only the coiled coils and hinge (aa 319–1125; our analysis of heptad repeats was slightly different from the coil prediction of DNAstar [Madison, WI], Fig. 1). MukBcoil tended to aggregate by sticking to small bits of bacterial debris; however, some largely monomeric fractions could be obtained from glycerol gradient sedimentation. (We also made a slightly larger coil construct, keeping the NH2 terminus at 319 and extending the COOH terminus to aa 1256, which includes the last predicted segment of coiled coil [Fig. 1]. This longer coil segment was highly aggregated into rosettes, and there were no monomers. We concluded from this that the extra segment of predicted coiled coil is probably missing a partner to pair with, and therefore denatured, leading to aggregation.) We then used the shorter MukBcoil as the basis for a chimera, FN-MukBcoil, in which the 40-kD cell adhesion domain of fibronectin, FN7-10 (Leahy et al., 1996), was added to the NH2 terminus.
We purified MukB as previously described using the expression plasmid pAX814 kindly provided by Dr. Sota Hiraga, Kumamoto University, Kumamoto, Japan (Niki et al., 1992). We substituted a mono Q column for the DEAE Sephacel and MukB eluted at 0.4 M NaCl. The best EM results were obtained with material from the mono Q column, subsequently sedimented over glycerol gradients. MukBcoil and FN-MukBcoil were purified by Sephacryl chromatography followed by glycerol gradient sedimentation, omitting the mono Q step.
Sedimentation coefficients were estimated by zone sedimentation through 15–40% glycerol gradients in 0.2 M ammonium bicarbonate (using the model SW50.1 or SW55.1 rotor; Beckman Instruments, Fullerton, CA) (Erickson and Briscoe, 1995). Standard proteins catalase (11.3 S) and BSA (4.6 S) were run in the same gradient as the samples, and the S value was estimated by linear interpolation between these standards (sedimentation of other proteins has shown our gradients to be linear in this region). The Stokes' radius, R s in nm, was estimated by gel filtration on a Superose 6 column using the following standards: thyroglobulin, R s = 8.5 nm; catalase, R s = 5.2 nm; and aldolase, R s = 4.8 nm. S and R s were used to calculate an experimental molecular weight as described by Siegel and Monte (1966) (see Ohashi and Erickson, 1997, for an example and our assumed parameters). Samples from the glycerol gradients were rotary shadowed (Fowler and Erickson, 1979) and photographed at 50,000 magnification in an electron microscope (model 301; Philips Electron Optics, Mahwah, NJ).
Results
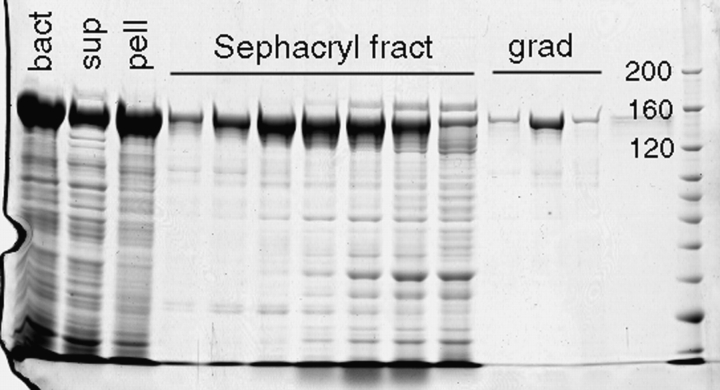
Fig. 2 shows the purification of BsSMC after overexpression in E. coli using the pET system. Because the protein is so large and elongated, the gel filtration chromatography results in a substantial purification from the smaller bacterial proteins. A final step of glycerol gradient sedimentation resulted in fractions that showed only small contamination with other protein bands by SDS-PAGE and little obvious contamination by EM.
Figure 2.
Expression and purification of BsSMC. The first three lanes show bacterial lysate, supernatant, and pellet. About one-third of the BsSMC is in the supernatant in these expressing bacteria grown at 22°C. The next seven lanes show fractions from the Sephacryl column, and the final three lanes show fractions from the glycerol gradient (of the peak Sephacryl fraction). The molecular mass markers at 200, 160, and 120 kD are indicated.
The sedimentation coefficient of BsSMC was 6.3 S, based on four determinations ranging from 6.2 to 6.4 S, and excluding three earlier results from 7 to 8 S. The sedimentation coefficient of MukB was 9.6 S, based on six measurements ranging from 9.2 to 10.3 S, and excluding two at 6.7 and 7.5 S. In several MukB preparations, some of the protein was aggregated into rosettes, which sedimented ahead of the monomers. The value of 14.3 S reported by Niki et al. (1992) may have been due to this aggregation. We confirmed by EM that the 9.6-S fraction was composed exclusively of monomers. The Stokes' radius was measured for BsSMC, MukB, and FN-MukBcoils by chromatography on a calibrated gel filtration column. The molecular masses calculated from these experimental values of S and R s are close to those predicted for a homodimer of each protein (Table I). The hydrodynamic data for these full-length proteins and other constructs are collected in Table I and addressed further in the Discussion.
Table I.
Hydrodynamic Properties of SMC Proteins
| Protein | M kD aa seq | S Sved. | R s nm | M kD experim | f/f min | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| BsSMC | 2 × 135 | 6.3 | 10.3 | 269 | 2.5 | |||||
| BsSMCΔN* | 2 × 117 | 6.1 | 2.3 | |||||||
| XCAP-C/E | 147 + 136 | 8 | 2.0 | |||||||
| MukB | 2 × 170 | 9.6 | 7.6 | 328 | 1.9 | |||||
| FN-MukBcoil | 2 × 133 | 7.0 | 9.6 | 272 | 2.3 |
“M kD aa seq” is the mass calculated for the presumed dimer from the aa sequence. S and R s were experimentally determined as described, and “M experim” was calculated from these values (Siegel and Monte, 1966). f/f min is the ratio of the experimental frictional coefficient (determined from S) to that of an unhydrated sphere of the same mass.
BsSMC with the NH2-terminal domain truncated.
Fig. 3 shows electron micrographs of rotary shadowed MukB and BsSMC. The fields in Fig. 3, A and B, were selected to show the variety of conformations. The fraction of molecules in the different conformations varied considerably from one field to another and is probably affected by the local conditions as the molecules are deposited on the mica and dried. Both MukB and BsSMC show the same three or four characteristic conformations, which is strong evidence that their basic structure is the same.
Figure 3.
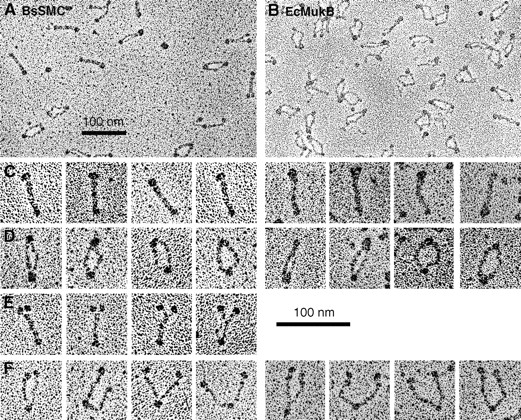
Electron micrographs of BsSMC and MukB. Selected fields are presented in A and B. (C) The most common conformation, “folded-rod.” (D) The “coils-spread” conformation. (E) Molecules with the coils together but with the terminal domains split, which was seen reproducibly for BsSMC. (F) The most informative “open-V” conformation. Note the symmetry of the molecules in the open-V conformation: whenever one arm shows two small globular domains, the other arm does also. Bars, 100 nm.
The first conformation, which we call “folded-rod,” is a rod-shaped molecule with a large globular domain at one end and a smaller one at the other (Fig. 3 C). This is similar to some of the images of Niki et al. (1992). The second conformation, which we call “coils-spread,” has similar large and small globular domains, but the rod connecting them is now split into two thinner rods (Fig. 3 D). These thinner rods are sometimes quite close together and sometimes bowed apart. A third conformation was seen frequently for BsSMC but was rare for MukB (Fig. 3 E). These molecules have the large globular domain split into two smaller domains. The rods are together over most of the molecule but veer apart near the pair of globular domains. The fourth conformation, which we call “open-V,” has both the rod and the large globular domain split into two and splayed apart (Fig. 3 F).
The open-V conformation in Fig. 3 F shows the structure of the molecule most clearly and explains the other forms. The globular domain in the middle is identified as the hinge, and the thin rods extending on each side are each a coiled coil. At the end of the coiled coils are the globular domains, discussed below. The coils-spread conformation is now interpreted as having the globular domains attached to each other but the two coiled coils separated and bowed out. The folded-rod conformation, giving the simple rod shape, is produced when the globular domains are attached to each other and the two coiled coils lie close to each other. Measurements of several aspects of the molecules are tabulated and explained in Table II and will be addressed in the Discussion.
Table II.
Measurements of MukB and BsSMC Lengths and Calculations
| Dimensions measured or calculated | Measured length | Calculated length of coil | ||
|---|---|---|---|---|
| nm ± SD | nm | |||
| 1. Whole MukB, length | 65.3 ± 2.6 | |||
| n = 30 | ||||
| 2. MukB hinge, diam | 6.7 ± 1.0 n = 52 | |||
| 3. Outer globular domain, diam | 7.3 ± 1.0 n = 17 | |||
| 4. Coil length: hinge to outer globular domain | 51.3 | |||
| 5. Whole MukBcoil, length | 49.3 ± 2.6 n = 21 | |||
| 6. Coil length: MukBcoil minus hinge | 42.6 | |||
| 7. Whole BsSMC, length | 58.0 ± 1.1 n = 24 | |||
| 8. Outer globular domain, diam | 9.5 ± 0.9 n = 22 | |||
| 9. BsSMC hinge, diam | 7.1 ± 1.1 n = 23 | |||
| 10. Coil length: hinge to globular domain | 41.4 |
All dimensions listed here have been corrected for the presumed 1-nm shell of metal. 1. The most definitive measurement was the length of the whole molecule in the folded-rod conformation and the lengths of single arms in the open-V; these were the same, and the measurements given here include both forms. 2. The smaller globular domain was identified as the hinge and had the same diameter in all three conformations. 3. Molecules in the open-V conformation usually showed the terminal domain resolved into two globular domains. The diameter of the terminal globular domain is given here. 4. The length of the coiled-coil was estimated by subtracting the diameters of the hinge and outer globular domains from the length of the whole molecule. 5. The length of the MukBcoil was measured from the outside of the globular hinge domain to the termination of the thin coil. This termination was sometimes ambiguous, hence the larger standard deviation of these measurements. 6. The length of the coil was estimated by subtracting the diameter of the hinge (2) from the length of MukBcoil (5). 7. The length of whole BsSMC molecules was measured, as in 1. 8. The larger (nonhinge) globular domain was approximately spherical in all conformations of BsSMC, and its diameter is measure here. 9. The diameter of the hinge domain, measured as in 2. 10. The coil of BsSMC was estimated by subtracting the diameters of the hinge and outer globular domain from the length of the whole molecule.
We initially anticipated that the coiled coils would be parallel since all known long coiled coils are parallel. This arrangement would place the two NH2-terminal domains at one end and the two COOH-terminal domains at the other end of an open-V molecule. We therefore expected to see some difference in size or shape in the terminal globular domains that would indicate this polarity. We also expected to see some difference in the lengths of the two arms since they would be formed from different segments of coiled coils. Instead, we were surprised to find that both BsSMC and MukB showed a striking symmetry. The arms were indistinguishable in length, and most importantly the domains at the two ends always appeared identical (Fig. 3 F). Some molecules showed a simple globular domain on each arm, but the best images of both BsSMC and MukB showed division of the terminal domain into a larger terminal globe and a smaller one somewhat closer to the hinge. Remarkably, whenever the two-part domain structure could be resolved on one arm, an identical structure was seen on the other (Fig. 3 F). The apparent twofold symmetry would only be possible with an antiparallel arrangement of the coiled coils.
To address the question of polarity, we prepared constructs of MukB with markers at the NH2 terminus. We had already prepared a truncated MukB, MukBcoil, in which we had deleted both the NH2- and COOH-terminal domains, leaving only the two coiled-coil segments and the hinge. This molecule frequently appeared as a rod where the hinge was obvious as a single globular domain; sometimes the two coiled-coil segments splayed apart in an open-V or coils-spread conformation (Fig. 4 A). The MukBcoil does not indicate the polarity because the NH2 and COOH termini are both truncated α helices. To visualize the polarity, we made a new chimeric protein, FN-MukBcoil, in which we fused FN7-10 onto the NH2 terminus of MukBcoil. FN7-10, a 40-kD fragment of fibronectin, is well characterized by x-ray crystallography (Leahy et al., 1996) as a rod-shaped molecule about 3 nm in diameter and 14 nm long, which should easily stand out from the thinner coiled coil. If the molecules were a parallel coiled coil, we expected to see two fat FN rods projecting from one end. If they were antiparallel, we expected to see one fat rod at each end.
Figure 4.
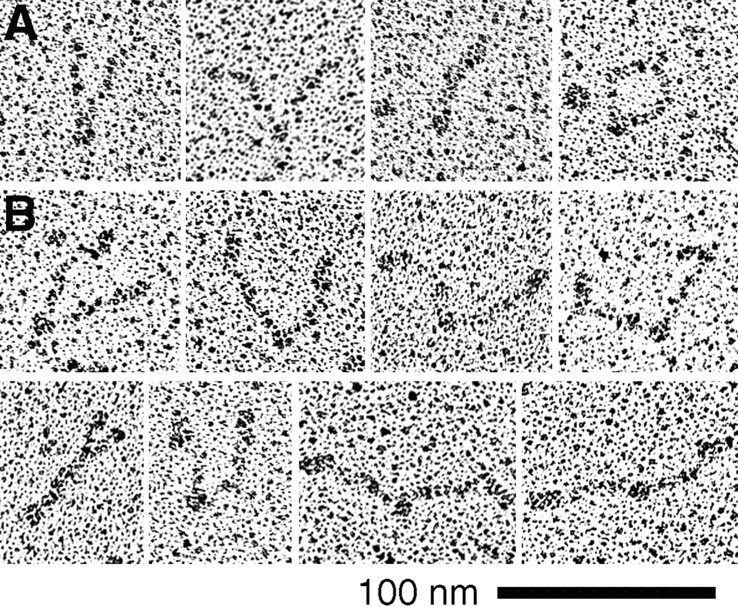
Electron micrographs of truncated and chimeric constructs. (A) MukBcoil, in which both the NH2- and COOH-terminal domains were deleted. The globular domain is the hinge. The ends seem to be sticky and are frequently together (right-hand images), although some open-V molecules could be found. (B) FN-MukBcoil, in which a thick segment of fibronectin was attached to the NH2 terminus of the coiled coil. This segment completely eliminated the stickiness of the ends, and all molecules are in the open-V configuration. The thick FN segment is seen projecting from each end, confirming the antiparallel coiled-coil arrangement.
The FN-MukBcoil molecules visualized in the EM were nearly all in the open-V conformation and showed the FN segment at each end (Fig. 4 B). This provides the most compelling evidence for the antiparallel arrangement of the coiled coils. There were several interesting contrasts between the structures of MukBcoil and FN-MukBcoil. MukBcoil tended to aggregate, sedimenting as a smear from 8 to 15 S, and the heavier fractions appeared as rosettes with the terminal segments of the coil aggregated at the center and the hinge projecting out. We believe that the ends of these coils are sticky, probably because we misjudged the termini of the coils. The sticky ends were also evident in single molecules, since the two ends usually remained in contact giving the folded-rod or coils-spread conformations (Fig. 4 A). In contrast, FN-MukBcoil sedimented as a sharp peak at 7 S, and the molecules were unaggregated and primarily in the open-V conformation in the EM. Thus, the FN segment at the terminus of the rods seems to block the sticky sites and prevent association either within or between molecules.
In addition to demonstrating the antiparallel arrangement of the coiled coils, FN-MukBcoil provides definitive proof that the hinge is flexible. Fig. 5 shows a histogram of the angles of the two arms measured for different molecules. We do not believe the apparent peaks and dips are significant, and we conclude that the angles are probably randomly distributed. It is perhaps of interest to ask whether the hinge can open to more than 180°, but these measurements cannot distinguish 270° from 90°. We conclude that the hinge is quite flexible between 1 and 180° and perhaps can bend even further.
Figure 5.
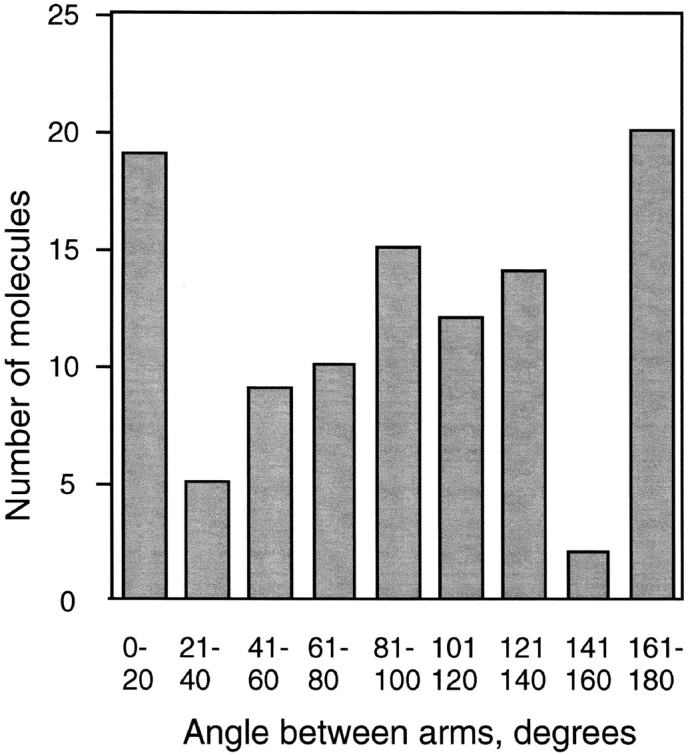
Molecules of FN-MukBcoil in the open-V conformation were measured for the angle of the two arms. The number of molecules found in 20° increments is shown as a histogram. Note that angles larger than 180° are possible but could not be distinguished, so they would be grouped with the smaller angle.
Discussion
Conformations and Dimensions
The frequency of the different conformations seen by EM varied from field to field, but in most cases the folded-rod was the predominant conformation. The open-V conformation was the most important for demonstrating the fundamental structure of the molecule. These open-V molecules also demonstrate the potential of the terminal domains to separate and undergo a scissoring motion, which is likely to be important for biochemical functions.
The hydrodynamic data are consistent with the elongated shape seen by EM and provide insight into the question of whether the molecule is in the folded-rod or open-V conformation in solution. The shape of a molecule is best indicated by the frictional ratio f/f min, where f min is the frictional coefficient of an unhydrated sphere of the same mass and density as the protein in question (Tanford, 1961). f/f min was calculated from the measured sedimentation coefficient, and the mass was predicted from the subunit aa sequence. Values are presented in Table I for the four molecules we purified and for the 8-S XCAP-C/ XCAP-E heterodimer (Hirano et al., 1997). The FN-MukBcoil provides an important benchmark for comparison because EM indicated that this construct was entirely in the open-V conformation. We believe that its f/f min = 2.3 is characteristic of the open-V conformation. The lower values of 1.9–2.0 for MukB and XCAP are probably characteristic of the folded-rod conformation.
The f/f min = 2.5 for BsSMC is the largest of the group and strongly suggests that this molecule is open-V in solution. Also, both BsSMC and FN-MukBcoil have a larger Stokes' radius than MukB, which is consistent with the interpretation that they are open-V, and MukB is folded-rod in solution. The only contradiction is that most of the BsSMC molecules appeared to be folded-rods in the EM. It is possible that the folded-rod conformation of BsSMC was generated by the high glycerol and salt concentrations when the specimens were dried on the mica.
There were two possibilities for the mechanics of the hinge: it could be quite flexible, permitting free scissoring motion of the two coiled coils, or it could be relatively rigid, locking the coils into the parallel configuration seen in the folded-rod. A flexible hinge was implied by the coils-spread conformation, which frequently showed an angle of 60° or more for the coils at the hinge. The flexibility of the hinge was demonstrated most convincingly by the FN-MukBcoils construct, which appears by EM to be entirely in the open-V conformation. Measurements of angles (Fig. 5) suggest complete flexibility of the hinge. Since the hinge is flexible, the folded-rod conformation must be stabilized by an interaction of the terminal domains with each other.
The measurements of the molecules in Table II show that MukB is longer than BsSMC (65 vs. 58 nm), which is consistent with its larger mass and longer estimated coiled coil (Fig. 1). We particularly wanted to determine the length of the coiled coil, as a guide to aligning the sequences. As shown in Table II, the measurement of the thin rod from the hinge to the outer globular domain (subtracting the diameters of these domains from the length of the whole molecule) gives 51 and 41 nm for MukB and BsSMC. These would actually estimate the minimum length of the coiled coil since the coils could extend into the hinge and globular domain. Using the spacing of 0.15 nm per aa in an α helix, these lengths would predict coiled coils of 340 aa for MukB and 273 aa for BsSMC. The assignments indicated in Fig. 1 fit these predictions reasonably well.
There is, however, a discrepancy indicated by the measurements of MukBcoil. This construct included aa 319– 1125, extending in both directions slightly beyond the entire 330–334-aa coiled-coil segments indicated in Fig. 1. However, the length of the coil measured from this construct was only 41 nm, corresponding to 273 aa. It therefore seems likely that the short predicted coil from 1205– 1243, which is missing from MukBcoil, is a part of the 51-nm rod of the whole MukB. It is not clear what this segment would be pairing with to make a coiled coil. It is interesting that a mutation D1201N, just before this predicted coil segment, completely disrupts MukB function (Yamanaka et al., 1994).
The precise alignment of the coiled coils is complicated by the appearance of gaps in the predicted coils. Some of these gaps may actually form coils and continue the coiled coil, but others may bud out, shortening the length of the rod. Most of these interruptions are small and might not be visible even if they did fold into a globular domain. We note that the thin rods of the open-V molecules appear as a uniform, thin diameter between the hinge and the two globular domains at the ends.
The heterodimeric SMCs of eukaryotes pose additional ambiguity in alignment of the coils since there are now four separate sequences that have to form the two coiled coils. As shown in Fig. 1, reasonable alignments can be made for Cut3/Cut14 of Schizosaccharomyces pombe and for XCAP-C/XCAP-E of Xenopus laevis. The antiparallel arrangement provides as good a match as a parallel arrangement.
Phylogenetic Tree of the SMC Family and Consideration of Heterodimers
The following two sections diverge from the structural analysis to discuss general features of the SMC family of proteins.
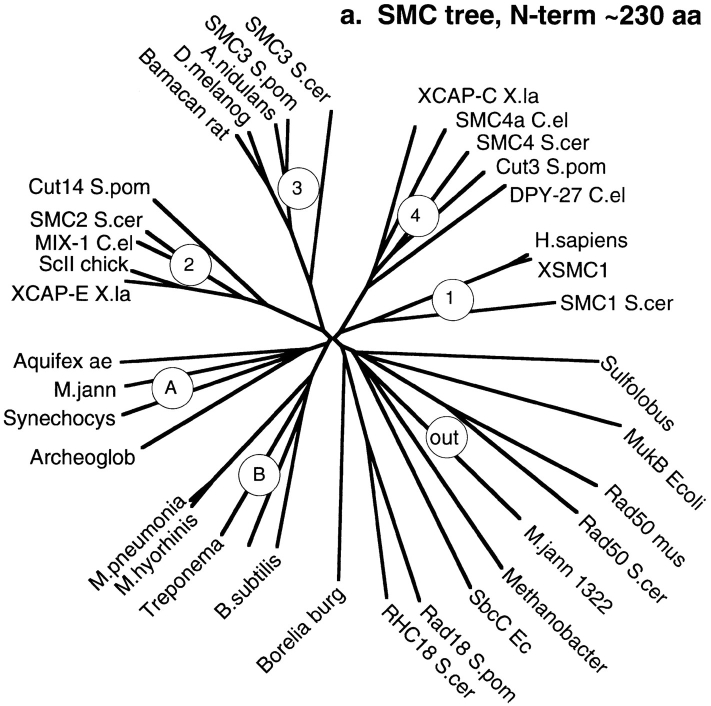
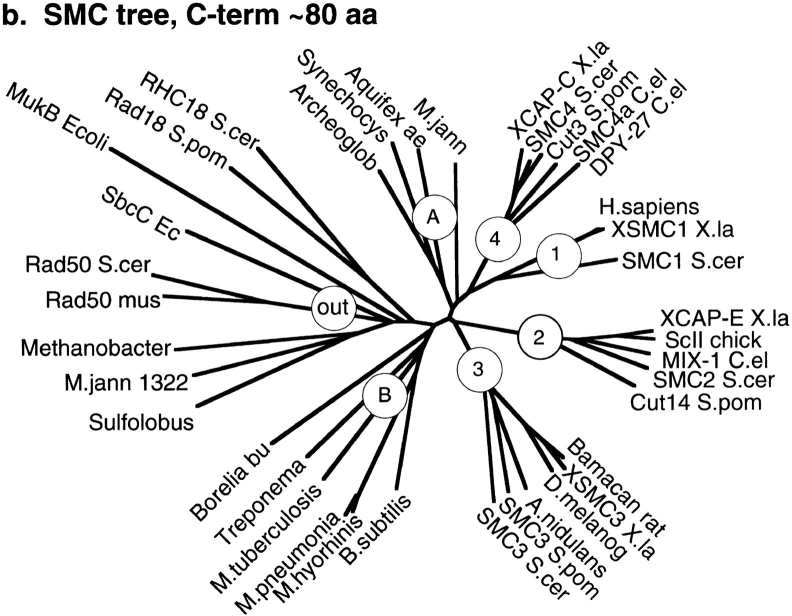
From the databases, we have collected 18 eukaryotic SMC sequences, 6 bacterial sequences, and 4 archaeal sequences that appear to be bona fide SMCs: MukB from E. coli, which has the SMC structure but limited sequence identity; and 8 sequences that show limited sequence identity at the NH2- and COOH-terminal domains and have long coiled-coil segments between them. We did a separate sequence alignment of the NH2-terminal domain of ∼230 aa and the COOH-terminal domain of 80 aa from each of the 34 sequences. The sequences were aligned by the Clustal algorithm of DNAstar and then adjusted by hand. Fig. 6 shows the two independently derived phylogenetic trees from the NH2- and COOH-terminal sequence alignments (trees were drawn with the DrawTree algorithm of phylip, http://evolution.genetics.washington.edu/phylip.html).
Figure 6.
Phylogenetic trees of the SMC proteins and some outliers more distantly related. Separate trees are shown for the ∼220–240-aa NH2-terminal domain (FLKRL...LEHVE in BsSMC), and the ∼80-aa COOH-terminal domain (LSGGE...YSSDT in BsSMC). The circles indicate the six groups of SMCs: 1–4, eukaryal SMCs; B, bacterial; A, archaeal. The group under the circle “out” are outliers, distantly related to SMCs. Each of the outlier sequences has an NH2-terminal domain with a related ATP-binding motif, some limited sequence identity in the COOH-terminal domain, and long coiled coils separating the two, but usually no hinge. Accession numbers for SMC sequences (mostly Swiss Prot). Group 1 SMC: SMC1 (S. cer), P41003; H. sapiens, S78271; XSMC1 (X. la), AF051784. Group 2 SMC: SMC2 (S. cer), P38989; Cut14 (S. pom), P41003; MIX-1 (C. el), U96387; XCAP-E (X. la), P50533; ScII (chick), Q90988. Group 3 SMC: SMC3 (S. cer), P47037; SMC3 (S. pom), AL009197; A.(E.) nidulans, S65799; D. melanog., U30492; Bamacan (rat), U82626; XSMC3 (X. la), AF051785. Group 4 SMC: SMC4 (S. cer), U53880; Cut3 (S. pom), P41004; DPY-27 (C. el), P48996; SMC4a (C. el), Z46242; XCAP-C (X. la), P50532. Group B Bacterial SMCs: Bacillus subtilis, P51834; Mycobacterium tuberculosis, Q10970; Treponema palidum, ORF00437; Mycoplasma hyorhinis, P41508; Mycoplasma pneumoniae, P75361. Group A (mostly) archaeal SMCs: Methanococcus jannaschii, U67604; Aquifex aeolicus, AE000699; Archeoglobus fulgidus, AE000995; Synechocystis, D90905; Pyrococcus horikoshii, D90905. Outliers—proteins distantly related to SMC: MukB (E. coli), P22523; M. jannaschii 1322, A64465; Methanobacterium thermoautotrophican, AE000837; Sulfolobus acidophilum, Y10687; Rad50 (mouse), U66887; Rad50 (S. cer), P12753; Rad18 (S. pom), P53692; RHC18 (S. cer), Q12749; SbcC (E. coli), P13458.
The trees from the NH2- and COOH-terminal domains are remarkably similar, and a tree calculated from the hinge region (not shown) also shows the same grouping. All three trees cluster the SMC sequences into six groups of SMC proteins and a group of outliers. The six-SMC groups comprise four eukaryotic SMC groups, each containing one of the four S. cerevisiae SMCs; a group of bacterial SMCs; and a group of mostly archaeal SMCs (which, however, also includes the cyanobacterium Synechocystis). We have labeled the eukaryotic groups 1–4, corresponding to the S. cerevisiae SMC in the group. (This is consistent with the previously proposed nomenclature of Koshland and Strunnikov [1996] and a recent review by Jessberger et al. [1998], which did not include the bacterial and archaeal SMCs.) B is for the bacterial group, and A is for the mostly archaeal group.
Jessberger et al. (1998) have proposed that there are two classes of heterodimers: SMC2/4 and SMC1/3. There are three well-established examples of pairing a member of the SMC2 group with a member of the SMC4 group: XCAP-E + XCAP-C (Hirano and Mitchison, 1994), Cut14 + Cut3 (Saka et al., 1994), and MIX-1 + DPY-27 (Lieb et al., 1998). It is attractive to speculate that an SMC of group 2 always pairs with a group 4 partner. MIX-1 provides an interesting variation, since it is known to pair with DPY-27 for its role in dosage compensation, but it probably has a different SMC partner for its role in mitosis (Lieb et al., 1998). C. elegans has a second SMC in group 4, so far identified only from genomic sequencing. This CeSMC4a may be the second partner of MIX-1.
Heterodimers of a group 3 and group 1 SMC have been identified for bovine SMC (Jessberger et al., 1998) and Xenopus (Losada et al., 1998). In S. cerevisiae, SMC3 and SMC1 were each identified in a screen for mutants affecting sister chromatid cohesion (Michaelis et al., 1997), consistent with their forming a pair. A possible exception to the generalization is that SMC2 from S. cerevisiae has been shown to associate both with itself and with SMC1 (Strunnikov et al., 1995). It was not determined that either of these was a specifically heterodimer association, but this observation raises the possibility that heterodimer associations may be more complex than simply two classes.
Of 18 bacterial or archaeal genomes completely or almost completely sequenced, 13 have a clear SMC, and 3 have no SMC but a MukB (which we now consider a divergent SMC; see also Britton et al., 1998). Methanococcus thermoautotrophican has only a distantly related protein but no SMC or MukB, and Borelia burgdorferi has an SMC-like protein that is shorter than most and branches ambiguously between bacterial SMCs and outliers. Helicobacter pylori has nothing matching the conserved NH2 or COOH termini of SMC, MukB, or the distantly related proteins (Erickson, H.P., unpublished sequence analysis; sequence data for partially completed genomes were obtained from The Institute for Genomic Research website at http://www.tigr.org).
Conserved Sequences and the Defining Characteristics of SMC Proteins
The most distinctive structural feature of the SMC proteins is the presence of five domains: NH2-terminal, long coiled coil, hinge, long coiled coil, and COOH-terminal. The coiled-coil segments of SMC proteins are not highly conserved; however, the NH2-terminal, COOH-terminal, and hinge domains show a high level sequence match over several motifs that seem characteristic of the SMC proteins. For the present discussion, we will not show the full alignment but will designate the motifs according to aa numbers of the BsSMC sequence (Swiss Prot P51834).
In the NH2-terminal domain, the most striking motifs are FKS (11–13), GxNGSGKSN (31–39), which is the Walker A motif for NTP binding, and the dipeptide QG (143–144). The FKS and GxNGSGKSN are almost totally conserved in the bona fide SMC proteins, and even the outliers show mostly conservative substitutions. MukB conserves these sequences no better than the outliers. A most intriguing sequence motif, not previously noted, is the QG, which is conserved in all 26 SMC sequences examined. It is also conserved in all three known MukB proteins (from E. coli, H. influenzae, and Vibrio cholerae [sequence data for V. cholerae were obtained from The Institute for Genomic Research website at http://www. tigr.org]). QG is conserved in Methanococcus jannaschii 1322 and the Sulfolobus protein, but not in Methanobacter thermoautotrophican nor in any of the Rad proteins. We suggest that this QG is a diagnostic signature of the SMC proteins and is another argument for including MukB in this group.
The hinge region shows no extended conserved motif, but the segment from aa 517 to 666 contains several isolated aa that are highly conserved. It is interesting that Rad18 and RHC18 show a hinge region that matches that of the SMC proteins. MukB has a 125-aa noncoil segment that obviously functions as a hinge, but it shows no sequence similarity to the SMC hinge sequence.
The COOH-terminal domain as indicated in Fig. 1 includes 200–300 aa from the end of the coiled coil, but only a short terminal segment of about 80 aa shows significant sequence identity. This segment begins with the highly conserved LSGG (1091–1094) and then the PxPhhhhDEhDAALD (1112–1126), where “h” is a hydrophobic aa. The hhhhD may correspond to the Walker B site, which is very loosely defined as an aspartic acid preceded by four hydrophobic residues. See Saitoh et al. (1995) for a more detailed analysis of this motif and its use in placing the SMC proteins in a larger family of transporters, helicases, and repair enzymes.
A recent study of the E. coli DNA-processing protein SbcC has proposed including it as a member of the SMC family (Connelly et al., 1998). This protein falls among the outliers in our alignments of NH2- and COOH-terminal domains (Fig. 6), and moreover has no functional hinge (see below). Although one could expand the definition of the SMC family to include the Rad proteins and other outliers, we suggest for the present to limit the SMC designation to the family members defined by the above characteristics, including specifically a functional hinge.
The Uniqueness of the Long, Antiparallel Coiled Coil
The antiparallel arrangement of the SMC coiled coils is a completely novel discovery. The antiparallel orientation seems preferred for the short, interacting α helices found in globular proteins, but coiled-coil proteins are predominantly parallel. Two exceptions are the antiparallel, 35-aa coiled coil projecting as a helical arm of the bacterial seryl tRNA synthetases (Oakley and Kim, 1997) and a 45-aa coiled coil of F1-ATPase (Abrahams, 1994). The ∼300-aa coiled coils of the SMC proteins demonstrate for the first time that the antiparallel arrangement can be used to form very long coiled coils.
We predict that the eukaryotic SMC heterodimers will have the same antiparallel coiled coil structure (Fig. 1), and this may also be true for the several proteins (Rad50, Rad18, E. coli SbcC, M. jannaschii 1322) indicated as outliers in the phylogenetic tree. A recent study of one of these outliers, SbcCD, included an electron micrograph that provides some important insights (Connelly et al., 1998). The SbcCD showed two large globular domains separated by a single coiled coil 80 nm long. Although the sequence indicates a short break in the middle of this coiled coil, the images show no indication of a hinge in the structure. The coiled coil shows a gentle curvature that can bring the terminal domains within ∼40 nm of each other, but this molecule seems incapable of achieving a folded-rod conformation. The question of parallel or antiparallel arrangement was not addressed in that study, and indeed the structure is described as “head-rod-tail.” However, there is no evidence of asymmetry in the structure; the globular domains at the two ends actually appear to be the same size, consistent with an antiparallel arrangement of the coiled coils in SbcCD.
It is also interesting to note that the antiparallel structure provides a mechanism for bringing the proposed Walker A and B motifs together because the NH2- and COOH-terminal domains are paired at each end of the molecule. Alternatively, in the SMC proteins they could be brought together in the folded-rod conformation. However, since SbcCD seems incapable of forming the folded-rod conformation, the antiparallel structure seems the only possibility for bringing its NH2- and COOH-terminal domains together. We should caution that the resolution of the analysis is not sufficient to actually demonstrate contact of the domains, much less a functional association of the Walker A and B motifs. However, the antiparallel structure provides strong evidence of this possibility.
The antiparallel coiled coil arrangement also suggests the possibility of a twofold axis of symmetry passing through the hinge. The twofold symmetry could be exact for the homodimers and could be approximate for the heterodimers. This would relate the NH2- and COOH-terminal domains on opposite arms by a 180° rotation, bringing identical faces into contact in the folded-rod conformation. This is illustrated in the model in Fig. 7.
Figure 7.
The model of the SMC protein structure. Arrows indicate the N→ C direction of the polypeptide. The NH2- and COOH-terminal domains are shown schematically, without attempting to identify them with the two globular domains seen by EM in some open molecules. The coiled-coil rods are rather rigid, but the hinge is quite flexible, permitting a scissoring movement with the coils separated at angles from 0 to 180° or more. The terminal domains associate with each other to lock the molecule into the “folded-rod” conformation, but this association is reversible, permitting transition to the open-V conformation.
Perhaps the most important feature of the new model is that the molecule is not a polar structure, with an ATP-binding domain at one end and a DNA-binding domain at the other, but each terminus of the molecule contains a complete and identical functional unit. This means that the two ends of the molecule can operate identically on two strands of DNA, separated by 100 nm for the fully open molecule, or brought into contact in the folded-rod. One function well established for 13-S XCAP condensin is the generation of DNA supercoils in vitro (Kimura and Hirano, 1997), and this may be the basis for DNA condensation. The symmetrical molecule with two complete functional units should suggest novel mechanisms for how the SMC complex might operate on DNA to generate supercoils, and for how related complexes might manipulate DNA for cohesion or repair.
Acknowledgments
We thank Ms. Carmen Lucheveche for preparation of the rotary shadowed specimens. We thank The Institute for Genomic Research website at http://www.tigr.org, for providing unpublished sequence data from several genomes.
Supported by National Institutes of Health grant GM28553.
Abbreviations used in this paper
- aa
amino acid(s)
- BsSMC
Bacillus subtilis SMC protein
- FN-MukBcoil
MukBcoil with the fibronectin cell adhesion segment fused to the NH2 terminus
- MukBcoil
MukB with both NH2- and COOH-terminal domains truncated
- NTP
nucleotide triphosphate
- SMC
structural maintenance of chromosomes
Footnotes
Address all correspondence to Harold P. Erickson, Department of Cell Biology, Duke University Medical Center, Durham, NC 27710-3011. Tel.: (919) 684-6385. Fax: (919) 684-3687. E-mail: H.Erickson@cellbio.duke.edu
References
- Abrahams JP. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DRF. The SbcCD nuclease of Escherichia coliis a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, H.P., and G. Briscoe. 1995. Tenascin, laminin and fibronectin produced by cultured cells. In Extracellular Matrix: A Practical Approach. M.A. Haralson and J.R. Hassell, editors. Oxford University Press, Oxford, UK. 187–198.
- Fowler WE, Erickson HP. Trinodular structure of fibrinogen. Confirmation by both shadowing and negative stain electron microscopy. J Mol Biol. 1979;134:241–249. doi: 10.1016/0022-2836(79)90034-2. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. . Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MMS. Condensins, cohesins, and chromosome architecture: how to make and break a mitotic chromosome. Cell. 1997;91:5–8. doi: 10.1016/s0092-8674(01)80002-7. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ, Swedlow JR. The SMC family: from chromosome condensation to dosage compensation. Curr Opin Cell Biol. 1995;7:329–336. doi: 10.1016/0955-0674(95)80087-5. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophilabarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Baechtold H, Akhmedov AT. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO (Eur Mol Biol Organ) J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Jessberger R, Frei C, Gasser SM. Chromosome dynamics—the SMC protein family. Curr Opin Genet Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: an essential component of the C. elegansmitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of XenopusSMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Erickson HP. Expression in Escherichia coli of the thermostable DNA polymerase from Pyrococcus furiosus. . Protein Expr Purif. 1997;11:179–184. doi: 10.1006/prep.1997.0775. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. . EMBO (Eur Mol Biol Organ) J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. E. coliMukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/ GTP binding activities. EMBO (Eur Mol Biol Organ) J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley MG, Kim PS. Protein dissection of the antiparallel coiled coil from Escherichia coliseryl tRNA synthetase. Biochemistry. 1997;36:2544–2549. doi: 10.1021/bi962391t. [DOI] [PubMed] [Google Scholar]
- Ohashi T, Erickson HP. Two oligomeric forms of plasma ficolin have differential lectin activity. J Biol Chem. 1997;272:14220–14226. doi: 10.1074/jbc.272.22.14220. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. BioEssays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO (Eur Mol Biol Organ) J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh AZM, Yamanaka K, Niki H, Ogura T, Yamazoe M, Hiraga S. Carboxyl terminal region of the MukB protein in Escherichia coliis essential for DNA binding activity. FEMS Microbiol Lett. 1996;143:211–216. doi: 10.1111/j.1574-6968.1996.tb08482.x. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monte KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiaegene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tanford, C. 1961. Physical Chemistry of Macromolecules. John Wiley, New York. 356–364.
- Yamanaka K, Mitani T, Feng J, Ogura T, Niki H, Hiraga S. Two mutant alleles of mukB, a gene essential for chromosome partition in Escherichia coli. . FEMS Microbiol Lett. 1994;123:27–31. doi: 10.1111/j.1574-6968.1994.tb07196.x. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241–251. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]