Abstract
Initiation of translation in eukaryotes and in archaea involves eukaryotic/archaeal initiation factor (e/aIF)1 and the heterotrimeric initiation factor e/aIF2. In its GTP-bound form, e/aIF2 provides the initiation complex with Met–tRNAiMet. After recognition of the start codon by initiator tRNA, e/aIF1 leaves the complex. Finally, e/aIF2, now in a GDP-bound form, loses affinity for Met–tRNAiMet and dissociates from the ribosome. Here, we report a 3D structure of an aIF2 heterotrimer from the archeon Sulfolobus solfataricus obtained in the presence of GDP. Our report highlights how the two-switch regions involved in formation of the tRNA-binding site on subunit γ exchange conformational information with α and β. The zinc-binding domain of β lies close to the guanine nucleotide and directly contacts the switch 1 region. As a result, switch 1 adopts a not yet described conformation. Moreover, unexpectedly for a GDP-bound state, switch 2 has the “ON” conformation. The stability of these conformations is accounted for by a ligand, most probably a phosphate ion, bound near the nucleotide binding site. The structure suggests that this GDP–inorganic phosphate (Pi) bound state of aIF2 may be proficient for tRNA binding. Recently, it has been proposed that dissociation of eIF2 from the initiation complex is closely coupled to that of Pi from eIF2γ upon start codon recognition. The nucleotide state of aIF2 shown here is indicative of a similar mechanism in archaea. Finally, we consider the possibility that release of Pi takes place after e/aIF2γ has been informed of e/aIF1 dissociation by e/aIF2β.
Keywords: e/aIF2, G protein, initiation of translation, tRNA, start codon
Initiation of the translation of a mRNA into a protein involves a complex cascade of molecular events, leading to a translation-competent ribosome with a methionylated initiator tRNA in the P site, base-paired with the start codon on mRNA. A critical consequence of the entire process is the setting of the reading frame for mRNA decoding. In eukaryotic and archaeal cells, the initiator tRNA carrier is the eukaryotic/archaeal initiation factor (e/aIF)2 heterotrimer. In its GTP-bound form, this factor specifically binds Met–tRNAiMet and brings it to the initiation complex. After start codon recognition, the factor, in its GDP-bound form, loses affinity for Met–tRNAiMet and eventually dissociates from the initiation complex. This leaves Met–tRNAiMet in the P site of the small ribosomal subunit and allows the final steps of the initiation process to occur. Hence, selection of the start codon is achieved through control of the nucleotide state of the factor (1).
In eukaryotes, the nucleotide state of eIF2 is regulated with the help of several initiation factors. A 43S preinitiation complex comprising at least the small ribosomal subunit, the initiator tRNA, and the eIF1, eIF1A, eIF2, eIF3, and eIF5 factors binds the mRNA at its 5′ capped end to form a 48S complex (2–4). The preinitiation complex then scans the mRNA until an AUG start codon is found. eIF1 and eIF1A are important for promoting an open, scanning-competent 48S complex (5–8). By facilitating GTP hydrolysis on eIF2, the eIF5 factor plays the role of a GTPase-activating protein (GAP) (9–11). It was thought that the action of eIF5 was triggered by recognition of the start codon. However, recent results provide evidence that the activity of eIF5 is more directly related to the reaching of a competent state of the 48S ribosomal complex in mRNA scanning (12). Release of the inorganic phosphate (Pi) produced by GTP hydrolysis, rather than hydrolysis itself, would appear to be the key event triggered by recognition of a start codon (12, 13). In this view, the eIF1 factor would prevent translational starts on non-AUG codons by blocking Pi release from eIF2. In turn, upon start codon recognition, conformational changes in the preinitiation complex would promote the release of eIF1 and that of the Pi group (5, 8, 12, 13). After release of eIF2:GDP, a guanine nucleotide exchange factor, eIF2B, is required for the recycling of eIF2:GDP into eIF2:GTP.
In archaea, scanning does not occur, and the initiation codons are located either in the vicinity of a Shine–Dalgarno sequence or near the 5′ end of mRNA (14). Orthologs of eIF1, eIF1A, and eIF2 exist. This suggests that the molecular events that accompany start codon recognition are very similar in both archaea and eukaryotes. On the other hand, eIF3, eIF5, and eIF2B have no orthologs in archaea. Thus, GTP hydrolysis on archaeal aIF2 occurs without GAP assistance, and the recycling of aIF2:GDP into aIF2:GTP should be spontaneous.
e/aIF2 is composed of three subunits, α, β, and γ. The γ-subunit forms the core of the heterotrimer and contains the GTP-binding pocket. It binds methionylated initiator tRNA and closely resembles elongation factor EF1A (15, 16). As in the case of EF1A, productive binding of tRNA is GTP dependent and related to the “ON” conformations of two regions of the G domain, called switch 1 and switch 2 (17). In archaea, α strongly enhances the affinity of γ for tRNA (18, 19), probably by stabilizing the two switches in the ON conformation (20). The C-terminal domain of aIF2α (domain 3) is sufficient to confer on γ its full tRNA-binding activity (19, 21). The N-terminal part of eIF2β is specific to eukaryotes. It is involved in the binding of the C-terminal domains of eIF5 and eIF2B. The remaining C-terminal moiety of β is common to both eukaryotes and archaea (22–24). Three-dimensional structures of archaeal aIF2β have been obtained by NMR. They show that the conserved part of e/aIF2β is made of two subdomains: an α–β domain and a zinc-binding domain (ZBD). The N-terminal region is unstructured (25, 26).
Very recently, the crystal structure of the Pyrococcus furiosus aIF2βγ heterodimer (Pf-aIF2βγ) has been solved, and GDP binding has been studied by crystal soaking experiments (27). In this structure, the N-terminal region of β-folds as a helix that, together with the α–β domain of β, ensures binding to γ. The ZBD is oriented toward the solvent. Analysis of the structure has led the authors to propose that conformational change of the β-subunit facilitates GTP hydrolysis.
The present work describes two structures of aIF2 from S. solfataricus. The first structure (Ss-aIF2α3βγ, 3.2-Å resolution) contained a shortened form of the α-subunit and intact β- and γ-subunits. The second structure (Ss-aIF2α3β1γ, 2.15-Å resolution) also contained the shortened α-subunit and the intact γ-subunit, but only the N-terminal helix of β was visible. For both structures, crystallization was achieved in the presence of GDP and magnesium.
In Ss-aIF2α3βγ and Pf-aIF2βγ, the N-terminal helices of aIF2β were similarly anchored onto γ. Surprisingly, the rigid bodies formed by the α–β and ZBDs had diametrically opposed orientations onto γ. In the heterotrimer, the β-subunit sat astride the guanine nucleotide pocket, with its ZBD in contact with the γ-subunit and its α–β domain in the solvent. Being close to the bound guanine nucleotide, the β-subunit formed a direct contact with the switch 1 region of aIF2γ. In the γ-subunit, the GDP molecule was bound without an associated magnesium ion. Moreover, the switch regions of γ adopted an arrangement that was unexpected for a GDP-bound state. This arrangement appeared to be stabilized by a ligand near the nucleotide-binding site. This ligand, which fit within a cationic pocket of the protein, was most probably a phosphate ion. The data suggest that this GDP–Pi bound state of e/aIF2 is proficient for tRNA binding. These observations are consistent with the idea that release of Pi upon start codon recognition is at the origin of the release of e/aIF2 from the initiation complex.
Results
Overall Three-Dimensional Organization of the aIF2 Heterotrimer.
In the overall structure of the Ss-aIF2α3βγ heterotrimer (Fig. 1A), the γ-subunit formed the core on which the α3 domain and the β-subunit were anchored. α3 and β did not interact. As in the aIF2αγ complex (20), the βαββαβ domain 3 of the α-subunit was in contact with the long protruding loop 1 of the aIF2γ domain II. aIF2β was anchored on the opposite side of aIF2γ, near the nucleotide-binding pocket (Fig. 1A). A complete view of the aIF2 heterotrimer could be deduced by superimposing the γ-subunit of the Ss-aIF2α3βγ heterotrimer (this work) on that of the Ss-aIF2αγ heterodimer (ref. 20 and Fig. 1C). In the resulting model, α and β made no contact (Fig. 1B). This conclusion agrees with all available biochemical data (15, 18). The overall conformation of the γ-subunit in the heterotrimeric complex showed the G domain (domain I) packed onto the two β-barrel domains (domains II and III; Fig. 1). This organization closely resembled that observed in previously determined γ structures (15, 16, 20). The four-cysteine cluster that protruded from the γ-subunit (g-C59, g-C62, g-C74, and g-C77) did not include a zinc ion. (Note that residues and secondary structure elements belonging to the α-, β-, and γ-subunits are indicated throughout by a-, b- and g- prefixes, respectively.) Rather, two close interactions were formed between g-C59 and g-C74 and between g-C62 and g-C77. These interactions ensured folding of the knuckle structure. The γ-subunit in the C2 crystal was also devoid of zinc. Thus, the presence of citrate in the crystallization conditions of the Ss-aIF2α3βγ complex could not be the origin of a dissociation of the metal. Possibly, the absence of zinc reflected sequestration of the free metal by the 0.1 mM EDTA concentration used for purification and crystallization of the protein.
Fig. 1.
S. solfataricus aIF2αβγ. (A) Overall structure of aIF2α3βγ bound to GDP. aIF2γ is yellow, switch 1 is red (g-31 to g-49), and switch 2 is orange (g-93 to g-113). The aIF2α3 domain (a-174 to a-266) is cyan. The aIF2β-helix 1 (b-3 to b-17) is green. Residues b-23 to b-27 are not visible. The aIF2β central domain (b-28 to b-103) is dark green, and the aIF2β ZBD (b-104 to b-139) is light green. The same color coding is used in all four structures. (B) Putative structure of the full aIF2αβγ heterotrimer, as deduced from the available data. This structure results from superimposition of aIF2γ, as in the present aIF2α3βγ structure, to aIF2γ, as in the aIF2αγ:GDPNP-Mg2+ structure (20). The three domains of aIF2α are colored as follows: the N-terminal domain is dark blue (a-1 to a-84), the central domain is marine (a-85 to a-173), and the C-terminal domain is cyan (a-174 to a-266). (C) View of aIF2αγ:GDPNP-Mg2+ as obtained in ref. 20. Residues g-36 to g-43 in switch 1 are not visible. Switch 1 is ON, and the conserved g-T46 contacts the magnesium ion. Switch 2 is ON, and the NH group of g-G96 contacts the γ-phosphate. (D) Overall structure of Ss-aIF2α3β1γ bound to GDP–Mg2+. Residues g-35 to g-48 in switch 1 are not visible. Figs. 1–3 and SI Figs. 7–9 were drawn with PyMol (DeLano Scientific LLC).
A New Mode of Interaction Between the β- and γ-Subunits.
The β-subunit has three parts, as already observed in the solution structures of aIF2β alone (25, 26). An N-terminal α-helix (residues b-3 to b-17) was connected by a flexible linker to a central α–β domain (residues b-28 to b-103), followed by a C-terminal ZBD (residues b-104 to b-139; Fig. 2A).
Fig. 2.
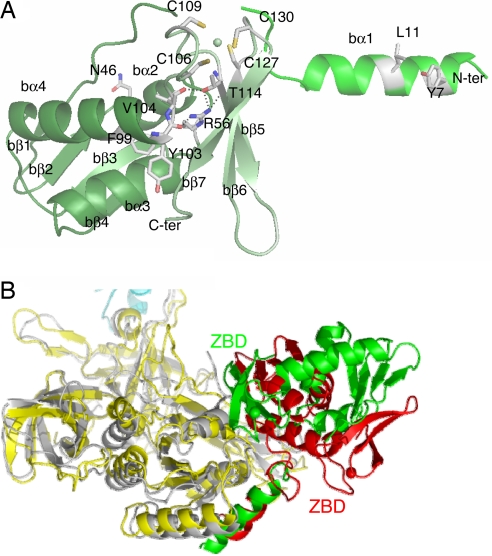
Structure of aIF2β within the Ss-aIF2α3βγ heterotrimer. (A) Overall structure of aIF2β. The color coding is the same as in Fig. 1A. The side chains of residues conserved in e/aIF2β sequences are drawn. C106, C109, C127, C130, V104, N46, and R56 are strictly conserved in all e/aIF2β (30 aIF2β and 43 eIF2β sequences) and eIF5 (60 sequences). Y7 is strictly conserved in all e/aIF2β sequences. L11 is strictly conserved in all aIF2β sequences and can be replaced by V (3 eIF2 sequences) or T (1 eIF2 sequence) in eIF2β. T114 is strictly conserved in the e/aIF2β sequences, but three exceptions exist in eIF5. Either F or Y are systematically found at positions 99 and 103 in e/aIF2β and eIF5. (B) Comparison of Ss-aIF2α3βγ and Pf-aIF2βγ (27) after superimposition of the two aIF2γ-subunits. Ss-aIF2β is green, Ss-aIF2γ is yellow, Ss-aIF2α3 is cyan, Pf-aIF2β is red, and Pf-aIF2γ is gray. This view shows the opposite orientations of the two aIF2β-subunits with respect to aIF2γ.
In the Ss-aIF2α3βγ electron density, the zinc ion of β was clearly visible. It was held by four cysteine residues (b-C106, b-C109, b-C127, and b-C130). The N-terminal α1-helix did not interact with the two other domains of the β-subunit. The ZBD was packed onto the central α–β domain, with which it formed a rigid body.
The overall organization of β in Ss-aIF2α3βγ can be compared with that in the recently solved structure of Pf-aIF2βγ (27). In the two complexes, the central domain and the ZBD of aIF2β had the same relative orientations (rmsd of 1.42 Å for 108 Cα atoms compared). The orientation of the two domains was maintained by hydrogen bonds that were conserved in the two structures. These bonds involved the side chain of b-R56 in the central domain, held in the ZBD by the main chain carbonyls of b-Y103 and b-T114 and by the side chain of b-T114 (Fig. 2A). Moreover, the main chain carbonyl of b-V104 was hydrogen bonded to the OH group of b-T114. Notably, b-R56, b-T114, and b-V104 were strictly conserved in e/aIF2β sequences [supporting information (SI) Fig. 5]. Y103 was highly conserved (of the 73 e/aIF2β sequences aligned, 67 had a Y and 6 an F at this position). Finally, anchoring of the ZBD to the α–β domain was also ensured by hydrophobic packing. Residues such as b-L30, b-I54, b-L100, and b-L112 were involved. The hydrophobic character of these residues was conserved in all e/aIF2β sequences. In Ss-aIF2α3βγ, the overall interaction surface of the central α–β domain with the ZBD was 1,388 Å2.
In the present structure, the N-terminal α1-helix and the C-terminal ZBD of aIF2β made contacts with aIF2γ, whereas the central α–β domain did not interact (Figs. 1A, 2B, and 3A). The N-terminal b-α1-helix was wedged between the g-α4- and g-α6-helices. Stacking interactions involved b-Y7 (b-α1) and g-Y163 (g-α4) on the one hand and b-Y15 (b-α1) and g-K156 (g-α4) on the other hand. Electrostatic interactions occurred between the side chains of b-R13 (b-α1), g-D192, and g-S193 (g-α6; Fig. 3A). Moreover, b-Y7 (b-α1) interacted with g-Q148 in the g-148-QNKVD-g-152 motif, and b-Y15 interacted with g-V154, which was just after the motif. The ZBD of aIF2β overhung the GDP-binding pocket of aIF2γ. The Nε group of b-K117 was hydrogen bonded to the 2′OH group of the GDP ribose, and the side chains of b-Q133 and b-E119 pointed toward the 2′ and 3′ group of the ribose. Finally, b-E119 was stacked on the side chain of g-W33 that belonged to switch 1 (Figs. 3B and 4A). The overall surface of interaction between aIF2β and aIF2γ was 2,529 Å2, with helix b-α1 contributing 1,691 Å2 and the ZBD contributing 838 Å2.
Fig. 3.
Communication between subunits. (A) Interaction of the b-α1-helix of Ss-aIF2β (green) with the G domain of Ss-aIF2γ (yellow). Relevant side chains are drawn. (B) Close-up view of the Ss-aIF2α3βγ heterotrimer highlighting the switch 1-mediated communication between the three subunits. The color coding is the same as that used in Fig. 1. Relevant side chains are drawn. Note the orientations of b-K117, b-E119, and b-Q133 toward the ribose hydroxyls of GDP and the stacking of b-E119 on g-W33.
Fig. 4.
Nucleotide binding site. (A) View of the Ss-aIF2α3βγ nucleotide-binding pocket showing the positions of relevant aIF2β and aIF2γ residues, as discussed in the text. The composite annealed 2Fo − Fc omit map was contoured at 1 σ. (B) View of the Ss-aIF2α3β1γ nucleotide-binding pocket, drawn in approximately the same orientation as in A. The composite annealed 2Fo − Fc omit map was contoured at 1 σ. Comparison of A and B shows the differences between the GDP–Pi and the GDP–Mg2+ states. (C) Close-up stereoview of the putative phosphate-binding pocket. The side chains of relevant residues are drawn. The water molecule linking the phosphate group to the NH of g-G96 is drawn as a red sphere. Hydrogen bonds surrounding the Pi group are shown. The 2Fo − Fc map contoured at 1 σ (firebrick) and the Fo − Fc map contoured at 4 σ (blue) were calculated before the addition of the Pi and water molecules in the model. B factors (in Å2) of the atoms coordinating the phosphate (72 Å2) and the water (29 Å2) are as follows: 34 (NH g-G96), 52 (Nδ g-H97), 55 (Nε g-K48), and 63 (Nδ g-H37).
Surprisingly, the docking of the β-subunit on the γ-subunit in the structure of Ss-aIF2α3βγ was different from that in Pf-aIF2βγ (27). Indeed, although the b-α1-helix ensured a similar anchoring between subunits in both structures, the rigid core formed by the central and C-terminal domains of β adopted opposite orientations (Fig. 2B). In the Pf-aIF2βγ heterodimer, the central domain of β was packed onto the γ-subunit, with the ZBD exposed to the solvent and making no contacts with γ (Fig. 2B). Here, in the Ss-aIF2α3βγ structure, the central domain of β had no contact with γ. Superimposing the two archaeal β-subunits showed that the two structures deviated from residue b-P31 (S. solfataricus numbering). In Ss-aIF2β, the main chain CO of b-N32 and NH of b-M33 interacted with the side chain of the strictly conserved b-N46 residue (Fig. 2A). In Pf-aIF2β, the side chain of the corresponding residue (b-N48) had a different conformation, leading to an interaction with the CO of b-G34 (equivalent to b-N32 in Ss-aIF2β) and with the side chain of b-D51.
Therefore, at least two conformations of the core domain of aIF2β with regard to the b-α1-helix are possible. The strictly conserved residue b-N46 (Ss numbering) may play a critical role in helping to switch between these two conformations.
A Conformational State for the Switch Regions in aIF2γ.
In Ss-aIF2α3βγ, a GDP molecule was clearly defined in the nucleotide-binding pocket of the γ-subunit (Fig. 4A). As in all G proteins, the nucleotide was bound, on the side of its guanine ring, by the g-184-SALH-g-187 and g-148-QNKVD-g-152 motifs and, on the side of the two phosphates, by the g-21-GKT-g-23 loop. However, even though the protein was in a GDP-bound state, the switch 2 region (g-93 to g-113) was in the typical ON conformation, already observed in Ss-aIF2αγ-GDPNP (ref. 20 and Fig. 1 A and C).
The conformation of switch 1 (g-31 to g-51) in the present structure was peculiar. It corresponded neither with the “OFF” conformation, seen in EF1A–GDP (28) and aIF2γ (15, 16, 27), nor with the ON conformation, seen in EF1A–GTP (17) and Ss-aIF2αγ-GDPNP (ref. 20 and Fig. 1C). Moreover, magnesium was absent from the electron density. Rather, the position expected for an Mg2+ ion was occupied by the intrusive side chains of g-K36 and g-H37 of switch 1. By interacting with the β-phosphate of GDP (Fig. 4A), these side chains ensured close contact between the N-terminal part of switch 1 and the core of the G domain. In particular, the switch 1 region interacted with both switch 2 and the P loop through contacts of g-K36 and g-H37 with g-T23, g-D93, and g-A94. Intriguingly, a large peak of electron density (8 σ on the Fo − Fc map) extended beyond the NH group of g-G96 (switch 2). This density would be nicely accounted for by a phosphate ion bound to this main chain NH via a water molecule (Fig. 4C). Ss-aIF2α3βγ triammonium citrate crystals were also obtained, even when the last addition of GDP–Mg2+ after the superdex was omitted. The electron density corresponding to GDP and Pi could still be observed. Washing the crystals with either crystallization liquor without GDP or crystallization liquor containing 1 mM GTP–Mg2+ failed to remove this electron density.
Given that no phosphate had been introduced into the crystallization medium, we searched for and eventually evidenced the presence of Pi as a contaminant [2.5% (mol/mol)] in the GDP solutions used in this study (see Materials and Methods). Thus, the presence of a phosphate ion in Ss-aIF2α3βγ would be plausible. Under this idea, the phosphate group would interact with the Nδ of g-H37 (switch 1) and g-H97 (switch 2), as well as with the Nε of g-K48 (switch 2; Fig. 4C). Interaction of the NH group of g-G96 with the phosphate ion would account for the ON position of switch 2. Indeed, in the case of the GDPNP-bound state, in which the ON position also is obtained, this NH would interact with the γ-phosphate of the nucleotide (20). In support of the above interpretation of the data, (i) g-H37 was strictly conserved in archaeal aIF2γ, (ii) a basic residue was strictly conserved at the position of g-K48 in all e/aIF2γ, and (iii) g-G96 and g-H97 were strictly conserved in all e/aIF2γ (SI Fig. 6).
In contrast with all previous structures of aIF2γ, the central part of switch 1 (g-E39 to g-I47) appeared fully defined in Ss-aIF2α3βγ. It interacted with the conserved g-219-RSFD-g-222 region in the g-β7 strand of domain II and with the conserved g-D309 residue of domain II (Fig. 3B). Moreover, the two γ regions ensuring contacts between switch 1 and domain II were immediately adjacent to the two regions of γ that contacted the α-subunit, i.e., the long g-L1 loop and the g-300 region (Fig. 3B). Finally, on the N-terminal side of switch 1, the side chain of g-W33 was stacked on that of b-E119. This network of interactions illustrates how the switch 1 region can exchange information with the α- and β-subunits (Fig. 3B). Contacts of the β-subunit with both the nucleotide and the switch 1 region in γ suggest that binding of β participates in fixing the peculiar conformation of the GDP binding site, as observed here.
An involvement of the core domain of aIF2β in such an unusual conformation of aIF2γ was indirectly supported by the structure of Ss-aIF2α3β1γ in the C2 crystals (Table 1). These crystals and the hexagonal crystals were obtained from the same batch of purified heterotrimer (α3βγ). However, probably because of proteolysis of the core domain of β, only the b-α1-helix was visible in the C2 crystals. Accompanying this modification, the GDP and the magnesium ion became clearly visible in the electron density map (Fig. 4B). The Pi density no longer occurred, and the two switch regions displayed canonical OFF conformations (Fig. 1D and SI Fig. 7). The magnesium ion was hexacoordinated by four water molecules, one oxygen of the β-phosphate, and the side chain of g-T23. Residue g-T46 did not interact with the magnesium ion. As a result, switch 1 was OFF and partly disordered from g-36 to g-48. Moreover, because the NH group of g-G96 was released from the nucleotide-binding pocket, switch 2 could adopt the OFF conformation.
Table 1.
Crystallographic and refinement data
| Parameter | Crystal |
|
|---|---|---|
| α3β1γ:GDP-Mg2+ | α3βγ:GDP-Pi | |
| Data collection wavelength, Å | 0.9795 | 1.007 |
| X-ray source | id14eh4 | id23eh1 |
| Space group | C2 | P3221 |
| Cell parameters | ||
| a/b/c, Å | 122.1/51.8/98.0 | 118.6/118.6/160.9 |
| β, ° | 94.4 | — |
| Unique reflections | 31,835 | 21,823 |
| Resolution, Å | 2.15 | 3.2 |
| Completeness, % | 95.3 | 98.7 |
| Redundancy | 2.3 | 4.3 |
| Rsym (I),* % | 5.2 (32.0) | 11.1 (33.8) |
| R/RFree,† % | 21.7/26.4 | 25.4/31.4 |
| rmsd bonds, Å | 0.0067 | 0.0089 |
| rmsd angles, ° | 1.337 | 1.708 |
| Residues in model | ||
| γ | 2–34; 49–340; 348–415; GDP-Mg2+ | 2–415; GDP, Pi |
| β | 2–19 | 3–22; 28–139; Zn |
| α3 | 176–265 | 174–266 |
| Mean B values | ||
| γ | 47 | 57 |
| β | 62 | 64 |
| α3 | 38 | 113 |
| GDP | 59 | 67 |
| Pi | — | 72 |
| Mg2+ | 51 | — |
| Zn (β subunit) | — | 58 |
| Water | 48‡ | 29§ |
Ramachandran plots are shown in SI Fig. 10.
*Values in parentheses are Rsym (I) in the highest shell of resolution.
†Rfree was calculated with 6% of the reflections.
‡Included in the model were 253 water molecules.
§This value corresponds to the B value of the Pi bound water molecule.
Discussion
Control of the nucleotide cycle of e/aIF2 is crucial for the accuracy of the initiation of translation. In eukaryotes, eIF5 and eIF2Bε, the catalytic subunit of eIF2B, modulate the nucleotide state of the heterotrimeric protein eIF2. On the one hand, the AA boxes in the C-terminal regions of eIF5 and eIF2Bε bind the eukaryotic specific N-terminal K boxes in eIF2β. Such contacts might be responsible for some allosteric regulation by eIF5 and eIF2Bε of the eIF2γ G domain function, relayed by the eIF2β-subunit (22, 23, 29, 30). On the other hand, recent reports show that eIF5 and eIF2Bε can directly bind the G domain of eIF2γ (31). These interactions involve the N-terminal domain of eIF5 (residues 50–100) and the C-terminal part of eIF2Bε (residues 524–712) (31). Thus, eIF5 and eIF2Bε may directly interfere with eIF2 function at the level of the eIF2γ G domain. Archaea do not contain proteins equivalent to eIF5 or eIF2Bε. Hence, GTP hydrolysis by aIF2 is likely not to be transactivated. Moreover, recycling of the aIF2-GDP factor is thought to be spontaneous.
Comparison of the structure of Ss-aIF2α3βγ bound to GDP (present work) with the recently published structure of Pf-aIF2βγ (27) indicates two modes of binding of the β-subunit onto the γ-subunit (Fig. 2B and SI Fig. 8). It cannot be excluded that these two modes of binding are induced by the crystallization conditions and the crystal contacts. However, it is worth considering the possibility that the two resulting structures of aIF2β reflect snapshots of the aIF2β conformations in aIF2 at different steps of the nucleotide cycle. An important contribution of β to the function of yeast eIF2 is indicated by analysis of mutations that allow translation initiation at non AUG codons (32–35). Isolated mutations in β map to the vicinity of the zinc-binding region, within the C terminus of the protein, or in the α1-helix (1, 36). Among these, two mutations increase the intrinsic GTPase activity of eIF2, independently of the presence of eIF5 or of that of the ribosome (1). These mutations, L254P and S264Y, affect residues that correspond to b-S122 and b-A132. In the structure of Ss-aIF2α3βγ, these two residues were at the surface of β, close to the nucleotide-binding site of γ (SI Fig. 8). This argues in favor of the idea that the observed conformation of β has functional significance.
In the ZBD of β, the side chains of b-K117, b-E119, and b-D133 pointed toward the ribose group of GDP, and stacking interactions occurred between b-E119 and g-W33 of switch 1. Switch 1 adopted a conformation with the two intrusive g-K36 and g-H37 side chains occupying the position of the normally GDP-bound magnesium ion. We propose that such peculiar positions of g-K36 and g-H37 create a cationic binding site for free phosphate. Therefore, the present structure indicates a state of aIF2 one step after GTP hydrolysis into GDP and Pi and one step before the pairing of the tRNA with the start codon.
Transient occurrence of a distinct phosphate-binding site on eIF2γ has already been predicted in the case of eukaryotes. The prediction has been made based on a biochemical analysis showing that Pi release from eIF2, and not GTP hydrolysis itself, is the actual step triggered by recognition of an AUG start codon (12). This view implies that initiator tRNA can still associate with e/aIF2 when the factor is in its GDP–Pi nucleotide state. It was shown that, when in the GDPNP state, the two switch regions open a channel between domain II and the G domain of aIF2γ. The switches are said to be in the ON conformation. This channel allows the binding of tRNA (20). In the GDP–Pi nucleotide state, as shown here, the ON position of switch 2 and the novel conformation of switch 1 kept the channel in a nearly open conformation. Then, complex formation with initiator tRNA easily could be envisaged, provided that a simple flip was imposed on the central loop of switch 1 (g-E39 to g-I47; Figs. 1A and 3B).
The absence of magnesium in the GDP–Pi state discussed above deserves further comment. Removal of the magnesium ion may have been facilitated by the presence of citrate ions in the crystallization conditions. Even if this were the case, the present structure showed a stable state that was obtained with the help of conserved residues. This favors the idea that this state indeed occurs in vivo. In the case of eukaryotes, it has been proposed that dissociation of magnesium from factor-bound GDP is the rate-limiting step in the GDP–GTP exchange reaction catalyzed by the guanine exchange factor (37). In archaea, the GDP–GTP exchange is thought to be spontaneous. Thus, to obtain the exchange, it would be logical that archaeal aIF2 would dissociate from the ribosome in a magnesium-free GDP state. In comparison with the GDP–Pi state that we propose here, the metal should be lost as soon as the γ-phosphate bond has been cut.
Can the structure of the above GDP–Pi state be transposed to the case of eukaryotes? In eukaryotic eIF2γ, the residues corresponding to archaeal residues g-G96 and g-H97 were strictly conserved (72 sequences), and g-K48 was almost strictly conserved (replaced by R in two cases, and by H in three cases). In contrast, g-H37 of switch 1 was systematically replaced by an F (SI Fig. 6). This implies that the role of this histidine in phosphate binding would be played in eukaryotes by another residue. One possible candidate is a K systematically present after the F. Another idea is that the function of g-H37 is contributed by eIF5. Finally, we must consider that, in the case of eukaryotes, GDP–GTP exchange does not occur spontaneously and requires the action of eIF2Bε. As noted above, removal of the GDP-bound magnesium ion, necessary for nucleotide exchange, is most probably catalyzed by eIF2Bε. This suggests that, in the case of eukaryotes, the binding of a magnesium ion in the GDP–Pi state is more stable than in the case of archaea. In this view, it should be noted that the position of Pi, such as it is described here, is compatible with the simultaneous presence of a divalent metal.
As mentioned above, the N-terminal part of eIF5, comprising an α–β domain and a ZBD, binds the G domain of eIF2γ (31). The NMR structure of the N-terminal part of yeast eIF5 showed that the α–β and ZBD domains were homologous to those in eIF2β (SI Fig. 9) (38). Although the NMR data did not enable to unambiguously determine the relative orientations of the eIF5 subdomains, the preferred orientation was close to that observed in the present Ss-aIF2α3βγ structure and to that in Pf-aIF2βγ (27). Sequence alignment of the eIF5 and e/aIF2β sequences indicated that the residues of the two proteins involved in the bridging of the α–β domain to the ZBD are strictly or highly conserved (see legend of Fig. 2A). Therefore, we propose that the two subdomains composing the N-terminal part of eIF5 adopt orientations similar to those in aIF2β. In addition, eIF1, the other key factor involved in the nucleotide cycle of eIF2, also displayed a structure similar to that of an α–β domain in e/aIF2β (SI Fig. 9) (38, 39). All of these observations give credit to the previous idea (38) that the three factors eIF2β, eIF5, and eIF1 play concerted roles during the nucleotide cycle of eIF2. Indeed, because of their structural similarities, the factors may target the same region on the G domain of eIF2γ. Archaea possess an equivalent of eIF1. Thus, in the context of the above idea, one can imagine that, either in eukaryotes or in archaea, e/aIF1 release would be sufficient to trigger rearrangement of the full eIF2 structure.
Materials and Methods
Expression and Purification of aIF2 Subunits from Sulfolobus solfataricus.
Expression in Escherichia coli of the S. solfataricus genes encoding either domain 3 of aIF2α (α3), or intact aIF2β- or aIF2γ-subunits, were performed as described (20). Purification of Ss-aIF2α3βγ was performed as the purification of Ss-aIF2 described previously (20). Before the final Superdex 200 chromatographic step, 1 mM GDP and 1 mM MgCl2 were added to the sample. The recovered protein was stored in the presence of 1 mM GDP–Mg2+. tRNA-binding activity (20) of the Ss-aIF2α3βγ form was identical to that of the intact Ss-aIF2αβγ form.
Crystallization of aIF2α3βγ.
Crystallization trials with a same preparation of the Ss-aIF2α3βγ heterotrimer in the presence of GDP–Mg2+ gave two crystal forms at 24°C. One form was obtained over 2 days in 1.6 M triammonium citrate at pH 7.0 (Index screen, Hampton Research). It belongs to space group P3221 and diffracts to 3.2-Å resolution. The other form was obtained over nearly 1 month in the presence of 15% PEG3350 as precipitating agent and 0.1 M magnesium formate (Index screen, Hampton Research). This second type of crystal belongs to space group C2 and diffracts to 2.15-Å resolution.
In the P3221 crystalline form, all residues of α3, β (except residues 23–27), and γ, were visible. The corresponding model was denoted Ss-aIF2α3βγ.
In the C2 form, α3 and γ (with the exception of residues 35 to 48 in the switch 1 region and the loop 341 to 347 in the domain III of aIF2γ) were visible. Only the α1-helix of the β-subunit was observed (Table 1). SDS/PAGE analysis of dissolved C2 crystals indicated proteolysis of the β-subunit. Thus, in the crystallization conditions, the β-subunit was subject to degradation. Very likely, in solution, the α1-helix resisted proteolysis through its strong binding to the γ-subunit. The corresponding model was denoted Ss-aIF2α3β1γ.
Pi Quantification.
The 200 mM GDP (Sigma–Aldrich) solution used in the crystallization assays was examined for the presence of contaminating Pi by using the EnzChek Phosphate Assay Kit (Molecular Probes). The presence of 2.5% (mol/mol) of Pi in the GDP sample was detected. Hence, in our crystallization conditions, the final concentrations of GDP and Pi were of 1 mM and at least 25 μM, respectively. Because Pi can also contaminate the protein preparations and the crystallization buffers, the value of 25 μM was an underestimate.
Structure Determination and Refinement.
Data corresponding to Ss-aIF2α3βγ crystals (P3221 form) were collected at 100 K by using the synchrotron source at the ESRF (Grenoble, France; ID14eh4 beamline, Table 1). Diffraction images were analyzed by using XDS (40), and the data were processed with programs of the CCP4 package (ref. 41 and Table 1). The structure of Ss-aIF2α3βγ was solved by molecular replacement by using PHASER (42) and Ss-aIF2γ as a model (20). The structure of the γ-subunit was refined by cycles of manual model building and energy minimization with CNS (43) by using data between 12 and 3.2 Å resolution (2-σ cutoff). After a few steps of refinement, the density around the γ-subunit became more organized. This allowed the positioning of domain 3 of aIF2α from P. abyssi in the density map and the building of the aIF2β-subunit. The structure was refined at 3.2 Å resolution with final R and Rfree factors of 25.4% and 31.4%, respectively. Note that the quality of the electron density of the α3 domain was relatively low, in agreement with the high B values associated with the atoms in this domain (117 Å2; Table 1). However, the α3 domain from Ss-aIF2αγ structure (Protein Data Bank ID code 2AHO) could be fitted unambiguously in the density. The mean B value of the structure (66 Å2) matches that deduced from the Wilson plot (66 Å2).
Data corresponding to Ss-aIF2α3β1γ crystals (C2 form) were collected at 100 K by using the synchrotron source at the ESRF (ID23eh1 beamline; Table 1). The same protocol as above was used to solve the Ss-aIF2α3β1γ structure. The model was refined at 2.15-Å resolution with final R and Rfree factors of 21.7% and 26.4%, respectively.
Supplementary Material
Acknowledgments
We thank Thomas Simonson for critical reading of the manuscript. This work was supported in part by Les Actions Concertées Incitatives (Biologie Cellulaire, Moléculaire et Structurale) from the Ministère de la Recherche and by L'Agence Nationale de la Recherche (Physique et Chimie du Vivant).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2QMU and 2QN6).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706784104/DC1.
References
- 1.Huang HK, Yoon H, Hannig EM, Donahue TF. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. EMBO J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. Mol Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 6.Pestova TV, Borukhov SI, Hellen CU. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 7.Pestova TV, Kolupaeva VG. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Das S, Ghosh R, Maitra U. J Biol Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- 10.Paulin FE, Campbell LE, O'Brien K, Loughlin J, Proud CG. Curr Biol. 2001;11:55–59. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 11.Majumdar R, Maitra U. EMBO J. 2005;24:3737–3746. doi: 10.1038/sj.emboj.7600844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Algire MA, Maag D, Lorsch JR. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolstrup N, Sensen CW, Garrett RA, Clausen IG. Extremophiles. 2000;4:175–179. doi: 10.1007/s007920070032. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E, Blanquet S, Mechulam Y. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roll-Mecak A, Alone P, Cao C, Dever TE, Burley SK. J Biol Chem. 2004;279:10634–10642. doi: 10.1074/jbc.M310418200. [DOI] [PubMed] [Google Scholar]
- 17.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 18.Pedulla N, Palermo R, Hasenohrl D, Blasi U, Cammarano P, Londei P. Nucleic Acids Res. 2005;33:1804–1812. doi: 10.1093/nar/gki321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatime L, Schmitt E, Blanquet S, Mechulam Y. J Biol Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- 20.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure (London) 2006;14:119–128. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Yatime L, Schmitt E, Blanquet S, Mechulam Y. Biochemistry. 2005;44:8749–8756. doi: 10.1021/bi050373i. [DOI] [PubMed] [Google Scholar]
- 22.Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Maitra U. Mol Cell Biol. 2000;20:3942–3950. doi: 10.1128/mcb.20.11.3942-3950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyrpides NC, Woese CR. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho S, Hoffman DW. Biochemistry. 2002;41:5730–5742. doi: 10.1021/bi011984n. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez P, Osborne MJ, Siddiqui N, Trempe JF, Arrowsmith C, Gehring K. Protein Sci. 2004;13:659–667. doi: 10.1110/ps.03506604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokabe M, Yao M, Sakai N, Toya S, Tanaka I. Proc Natl Acad Sci USA. 2006;103:13016–13021. doi: 10.1073/pnas.0604165103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berchtold H, Reshtnikova L, Reiser COA, Schirmer NK, Sprinzl M, Hilgenfeld R. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 29.Das S, Maiti T, Das K, Maitra U. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- 30.Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alone PV, Dever TE. J Biol Chem. 2006;281:12636–12644. doi: 10.1074/jbc.M511700200. [DOI] [PubMed] [Google Scholar]
- 32.Castilho-Valavicius B, Thompson GM, Donahue TF. Gene Expr. 1992;2:297–309. [PMC free article] [PubMed] [Google Scholar]
- 33.Donahue TF, Cigan AM, Pabich EK, Valavicius BC. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- 34.Laurino JP, Thompson GM, Pacheco E, Castilho BA. Mol Cell Biol. 1999;19:173–181. doi: 10.1128/mcb.19.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto NN, Carnevalli LS, Castilho BA. Biochem J. 2002;367:359–368. doi: 10.1042/BJ20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson GM, Pacheco E, Melo EO, Castilho BA. Biochem J. 2000;347:703–709. [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen GR, Pedersen L, Valente L, Chatterjee II, Kinzy TG, Kjeldgaard M, Nyborg J. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 38.Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, Smerdon SJ, Proud CG. Biochemistry. 2006;45:4550–4558. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- 39.Fletcher CM, Pestova TV, Hellen CU, Wagner G. EMBO J. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch WJ. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 41.Collaborative Computational Project. Acta Crystallogr D. 1994;50(5):760–763. Number 4. [Google Scholar]
- 42.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D. 2004;60(3):432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54(5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.