Abstract
From a patient with acute myeloid leukemia (AML), we have identified IL-27Ra (also known as TCCR and WSX1) as a gene whose expression can induce the transformation of hematopoietic cells. IL-27Ra (IL-27R) is a type I cytokine receptor that functions as the ligand binding component of the receptor for IL-27 and functions with the glycoprotein 130 (gp130) coreceptor to induce signal transduction in response to IL-27. We show that IL-27R is expressed on the cell surface of the leukemic cells of AML patients. 32D myeloid cells transformed by IL-27R contain elevated levels of activated forms of various signaling proteins, including JAK1, JAK2, STAT1, STAT3, STAT5, and ERK1/2. Inhibition of JAK family proteins induces cell cycle arrest and apoptosis in these cells, suggesting the transforming properties of IL-27R depend on the activity of JAK family members. IL-27R also transforms BaF3 cells to cytokine independence. Because BaF3 cells lack expression of gp130, this finding suggests that IL-27R-mediated transformation of hematopoietic cells is gp130-independent. Finally, we show that IL-27R can functionally replace a homodimeric type I cytokine receptor in the activation of JAK2-V617F, a critical JAK2 mutation in various myeloproliferative disorders (MPDs). Our data demonstrate that IL-27R possesses hematopoietic cell-transforming properties and suggest that, analogous to homodimeric type I cytokine receptors, single-chain components of heterodimeric receptors can also enhance the activation of JAK2-V617F. Therefore, such receptors may play unappreciated roles in MPDs.
Keywords: acute myeloid leukemia, myeloproliferative disease, oncogene, signaling
Acute myeloid leukemia (AML) is a disease of the myeloid compartment of the hematopoietic system characterized by an accumulation of undifferentiated blast cells in the peripheral blood and bone marrow. AML is caused by multiple genetic and epigenetic changes that result in stimulation of mitogenic signals and deregulation of apoptosis and differentiation. It has been proposed that mutations in two different classes of oncogenes are required to induce AML. Mutations in class I oncogenes (e.g., Ras, Flt3) result in stimulation of proliferative and cell survival signals, whereas mutations in class II oncogenes (e.g., AML1-Eto, PML-RARα) lead to inhibition of differentiation and subsequent cell death by apoptosis (1–3). Although many mutations are recurrently found in AML patients, it is believed that additional mutations in AML exist and have yet to be identified (1, 2, 4–6). Even with a large knowledge base about the causative genetics of AML, a more complete understanding of the molecular players is needed to identify targets for future therapeutic treatment for this disease.
We have previously performed functional genetic screens to identify oncogenes in AML by using an efficient retroviral delivery, expression, and cDNA recovery system (7, 8). Using this approach, we identified an activating deletion mutation in the TrkA tyrosine kinase in a patient with AML (7). This discovery provided evidence that TrkA may play a role in leukemogenesis. The deletion mutation we identified has been shown to be leukemogenic in mice (9), further validating our approach to identify genes that contribute to leukemia formation.
In this study, we have used a functional genetic screen to identify genes that contribute to myeloid cell transformation and perhaps AML formation. Here, we describe our identification of the ligand binding component of the receptor for IL-27 (IL-27R), as a unique transforming gene product. We show that IL-27R is expressed on the cell surface of leukemic cells of AML patients and demonstrate that IL-27R can transform hematopoietic cells in a JAK-dependent manner. We also show that IL-27R activates JAK2-V617F. Previously, only homodimeric type I cytokine receptors have been reported to activate this important JAK2 mutant. Our data indicate that a single component of a heterodimeric cytokine receptor has the ability to aberrantly activate signaling pathways in hematopoietic cells. Thus, it is possible that components of heterodimeric cytokine receptors may play a role in myeloid diseases either through aberrant expression or activating mutations.
Results
Functional Genetic Screens to Identify Myeloid Cell Oncogenes Uncover Unique Transforming Properties of IL-27R.
To identify oncogenes involved in AML, or perhaps other myeloid disorders, we developed functional genetic screens in myeloid cells. We used 32D myeloid progenitor cells, which depend on IL-3 for growth and viability (10). When 32D cells are cultured in the absence of IL-3, they undergo cell cycle arrest and apoptosis (11). These cells have served as a model cell system for studying the transforming properties of various leukemia-associated oncogenes, such as Bcr/Abl and Flt3, among others (12, 13). For our approach, we used cDNA representing the genes expressed in the leukemic cells of patients with AML. This cDNA was cloned into a retroviral vector (pEYK3.1; a gift of George Daley, Harvard Medical School, Children's Hospital, Boston) designed for efficient delivery and proviral recovery (14). We generated retrovirus containing cDNA libraries from AML patients and used this virus to infect 32D cells that exogenously express B-cell CLL/lymphoma 2 (Bcl2).
We used 32D/Bcl2 cells for multiple reasons. First, removal of IL-3 leads to a very rapid apoptotic cell death of 32D cells, and our screens of parental 32D cells resulted in no cytokine-independent isolates. Second, oncogenic expression in 32D cells needs to elicit an antiapoptotic and a mitogenic signal to transform these cells to cytokine independence. Third, it is believed that transformation of myeloid cells in AML is caused by mutations that activate cooperating signaling pathways (1–3). Culturing 32D cells without IL-3 resulted in cell death of the entire culture in a little over 2 days. However, expression of Bcl2 led to enhanced cell survival in the absence of IL-3 without inducing cell growth (data not shown), which is in agreement with previous studies (15, 16). Two days after infection, 32D/Bcl2 cells were plated in medium lacking IL-3 to identify IL-3-independent transformants. Using a cDNA library from an AML patient who exhibited AML of FAB subtype M5b, a monocytic leukemia with 93% blast cells and normal cytogenetics, we isolated a cDNA representing a WT IL-27Ra (TCCR, WSX-1) (17) gene from multiple independent isolates of IL-3-independent cells. For simplicity, we will refer to this gene and protein as IL-27R.
Transformation of 32D cells by IL-27R.
Expression of IL-27R in 32D/Bcl2 cells is sufficient to transform these cells to IL-3 independence (data not shown). Interestingly, IL-27R also transforms parental 32D cells to cytokine independence (Fig. 1A). These results suggest that in the context of the expression of an entire library of cDNAs, expression of Bcl2 in 32D cells during our initial screen likely sensitized these cells to transformation to IL-3 independence. Immunoblot analysis confirmed expression of IL-27R in these cells (Fig. 1B). The two protein bands of IL-27R are the result of glycosylation (data not shown). This finding is consistent with the presence of multiple glycosylation sites in the extracellular region of the receptor (17).
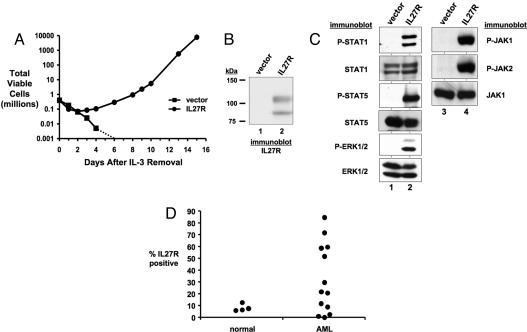
Fig. 1.
IL-27R transforms 32D cells to IL-3 independence. (A) 32D cells stably expressing IL-27R (●) or a control vector (■) were cultured in the absence of IL-3 starting on day 0. The total number of viable cells was determined at each time point by trypan blue exclusion. The dotted line represents the total number of viable cells going below the limit of detection of a hemacytometer and to zero. (B) Cell lysates of 32D cells stably expressing a control vector (lane 1) or IL-27R (lane 2) were immunoblotted with anti-IL-27R antibodies. (C) Antibodies that recognize the indicated proteins were used to immunoblot lysates from control 32D cells expressing an empty vector (lanes 1 and 3) or IL-27R (lanes 2 and 4) after 1 h of growth factor starvation. (D) Bone marrow mononuclear cells from normal and AML patients were analyzed by flow cytometry using an Alexa Fluor-647-conjugated anti-IL-27R antibody. The percentage of IL-27R-positive cells of each patient sample (represented by ●) is plotted.
IL-27R is a type I cytokine receptor that functions as the ligand binding component of the receptor for IL-27 (18, 19). Signaling induced by IL-27 activates JAK and STAT proteins, including JAK1, JAK2, Tyk2, STAT1, STAT2, STAT3, STAT4, and STAT5 in various cell types (20, 21). There is clear evidence that JAK/STAT proteins are activated in myeloid disorders. STAT1, STAT3, and STAT5 are frequently activated by leukemogenic oncogenes and are activated in AML (22, 23). JAK proteins, in particular JAK2, are mutationally activated in myeloproliferative disorders (MPDs) (24–28) and a small fraction of AML (29, 30). Therefore, we analyzed the activation state of these proteins in 32D cells transformed by IL-27R and determined that these cells have increased phosphorylated STAT1, STAT5, JAK1, and JAK2 compared with vector control cells (Fig. 1C). STAT3 is also activated in 32D cells transformed by IL-27R (see Fig. 2E). Because transformation of 32D cells involves mitogenic signaling and inhibition of apoptosis, we also determined that ERK1/2 is activated in these cells (Fig. 1C). These data indicate that IL-27R-mediated transformation of 32D cells is associated with activation of JAK/STAT and ERK pathways. In addition to STATs, ERKs are commonly activated in AML cells (31). Thus, transformation by IL-27R results in activation of pathways that are frequently activated in AML. Activation of transforming signals by IL-27R is not associated with the autocrine production of factors that stimulate cell growth or survival (data not shown). In addition, a neutralizing antibody to IL-27 did not affect the growth or viability of cells transformed by IL-27R [supporting information (SI) Fig. 6].
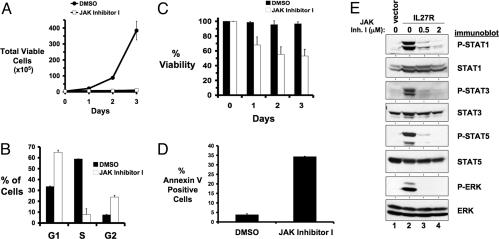
Fig. 2.
Transformation of 32D cells by IL-27R requires JAK family kinase activity. 32D cells transformed to cytokine independence by IL-27R were cultured in the presence of 0.1% DMSO [● (A) and filled bars (C)] or 0.5 μM JAK inhibitor I [○ (A) and empty bars (C)] on day 0. (A) The total number of viable cells for each treatment was determined daily by trypan blue exclusion. Error bars indicate standard deviation within a representative experiment. (B) After 24 h of JAK inhibitor I treatment, the DNA content present in each phase of the cell cycle was determined by propidium iodide staining and flow cytometry. The percentage of cells that were in G1, S, and G2 are plotted. Error bars indicate standard deviation of triplicate samples of a representative experiment. (C) Cell viability was determined by trypan blue exclusion. Error bars indicate standard deviation within a representative experiment. (D) After 24 h of JAK inhibitor I treatment, apoptosis was determined by annexin V binding. Error bars indicate standard deviation of triplicates within a representative experiment. (E) 32D cells transformed by IL-27R were starved of serum/IL-3 for 3 h in the presence of 0.1% DMSO (lane 2), 0.5 μM (lane 3) or 2 μM (lane 4) JAK inhibitor I. Control 32D cells starved of serum/IL-3 in the presence of 0.1% DMSO for 3 h is shown in lane 1. Cell lysates were immunoblotted with antibodies that recognize the indicated proteins. Similar results were obtained with independently derived cell lines.
Expression of IL-27R on the Cell Surface of AML Cells.
Because we identified IL-27R as a transforming gene from the leukemic cells of an AML patient, we wanted to determine whether this gene was commonly expressed in AML patients. Using flow cytometry, we detected IL-27R on the cell surface of AML cells. Eight of 13 AML bone marrow samples tested had 2.5- to 10-fold more cells expressing IL-27R (ranging from 0.33% to 84.6%) than the average observed in normal bone marrow cells (ranging from 6.3% to 12.8%) (Fig. 1D and SI Fig. 7). This finding suggests IL-27R expression is retained in many AML patients. In all normal and AML samples tested, essentially all IL-27R-positive cells were CD33-positive, suggesting IL-27R is expressed in cells of the myeloid lineage in the bone marrow (SI Fig. 7 and data not shown).
IL-27R Requires JAK Activity to Transform Myeloid Cells.
We addressed the role of JAK family kinases in transformation by IL-27R by culturing 32D/IL-27R cells with the pan-JAK inhibitor, JAK inhibitor I (32). In the presence of this inhibitor the growth of these cells ceases (Fig. 2A). JAK inhibitor I induces a G1 and a G2 cell cycle arrest and a dramatic decrease in the number of cells in S-phase after 24 h of treatment (Fig. 2B). Cell viability also decreases in the presence of the inhibitor (Fig. 2C), because of the induction of apoptosis as determined by annexin V binding to the cells after 24 h of treatment with inhibitor (Fig. 2D). JAK inhibitor I not only blocks STAT1, STAT3, and STAT5 phosphorylation induced by IL-27R, but also prevents ERK1/2 activation (Fig. 2E). These experiments suggest the kinase activity of JAK family members is required for IL-27R-mediated cell growth and inhibition of apoptosis and that JAK activation is required for downstream activation of STATs and ERK1/2.
IL-27R Requires Its Box 1 Motif to Transform 32D Cells.
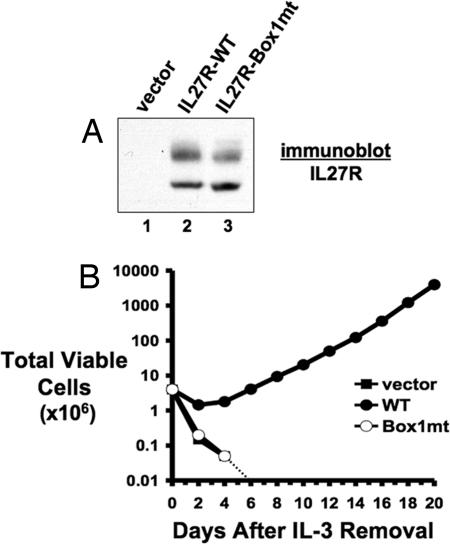
Within the cytoplasmic region of IL-27R is a Box 1 motif (17). This motif is often found in cytokine receptors and functions as an interaction motif with JAK proteins (33, 34). IL-27R has been shown to interact with JAK1 (35), although JAK2 is also activated after IL-27 stimulation of cells (20). Mutation of conserved prolines in Box 1 motifs generates a motif that is severely impaired in its ability to bind to JAK proteins (34). Therefore, we mutated these residues within the IL-27R Box 1 motif to alanine and expressed this mutant IL-27R in 32D cells (Fig. 3A). Removal of IL-3 from the growth medium of these cells results in complete cell death of the culture in a time course identical to cells expressing a control vector (Fig. 3B). Thus, IL-27R requires its Box 1 motif to transform myeloid cells. Collectively, our data suggest IL-27R-mediated transformation requires receptor-mediated activation of JAK family kinases.
Fig. 3.
IL-27R requires a functional Box 1 motif to transform 32D cells to cytokine independence. (A) 32D cells were infected with retrovirus encoding an empty vector (lane 1), WT IL-27R (lane 2), or IL-27R containing a mutant Box 1 motif (lane 3). After drug selection, cell lysates were immunoblotted for IL-27R expression. (B) 32D cells stably expressing control vector (■), WT IL-27R (●), or IL-27R-Box1mt (Box1mt) (○) were washed of IL-3 and cultured in RPMI medium 1640/10% FBS in the absence of IL-3 starting on day 0. The total number of viable cells was determined at each time point by trypan blue exclusion. The dotted line represents the total number of viable cells going below the limit of detection of a hemacytometer and to zero.
IL-27R Transforms BaF3 Cells to Cytokine-Independent Growth.
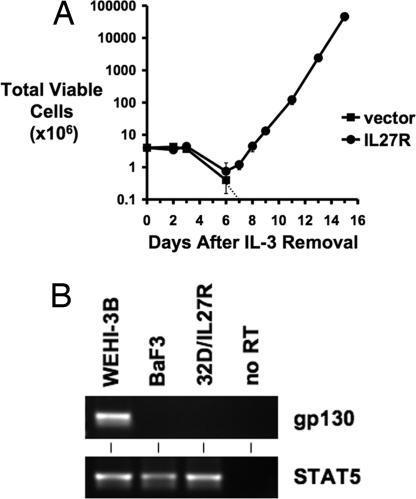
IL-27R requires the glycoprotein 130 (gp130) coreceptor to signal in response to IL-27 stimulation (18). BaF3 cells are IL-3-dependent pro-B cells that lack gp130 expression (18, 36). Through gp130 reconstitution studies, these cells have been used to show the requirement for gp130 to induce cell signaling in response to IL-27 (18). To test whether gp130 is required for IL-27R-mediated transformation of hematopoietic cells, we expressed IL-27R in BaF3 cells and cultured these cells in the absence of IL-3. Like 32D cells, BaF3 cells are transformed to cytokine independence by IL-27R (Fig. 4A), which suggests that IL-27R does not require gp130 expression to elicit a transforming signal in cells and also indicates that the transforming activity of IL-27R is not limited to 32D myeloid cells. RT-PCR analyses confirmed the lack of gp130 expression in BaF3 cells (Fig. 4B). Also, we could not detect gp130 expression in 32D cells transformed by IL-27R, suggesting that gp130 is not required for transformation of these cells and is not up-regulated to facilitate IL-27R-mediated signaling during transformation (Fig. 4B). Analyses of signaling pathways in BaF3 cells transformed by IL-27R indicate similar JAK/STAT and ERK pathways are activated as in 32D cells transformed by IL-27R (SI Fig. 8). Finally, whereas IL-27R-mediated transformation is gp130-independent, our observations still support the paradigm that IL-27-mediated signaling requires gp130 (18). IL-27-induced signaling (as measured by STAT1 phosphorylation) is not observed in IL-27R-expressing 32D or BaF3 cells, which lack gp130, but is seen in IL-27R-expressing 293T cells, which express gp130. siRNA depletion of gp130 in 293T cells inhibits IL-27-induced phosphorylation of STAT1 (SI Fig. 9).
Fig. 4.
IL-27R transforms IL-3-dependent BaF3 cells to cytokine independence. (A) BaF3 cells infected with control vector retrovirus or retrovirus designed to express IL-27R were washed of IL-3 and plated in RPMI medium 1640/10% FBS on day 0. The total number of viable cells expressing control vector (■) or IL-27R (●) was determined at each time point by trypan blue exclusion. The dashed line represents the total viable number of cells going below the limit of detection of a hemacytometer and to zero. (B) RT-PCR analysis for gp130 expression in WEHI-3B cells (as a positive control), BaF3 cells, and 32D cells transformed by IL-27R. RT-PCR for STAT5 served as a control for cDNA synthesis.
IL-27R Activates JAK2-V617F.
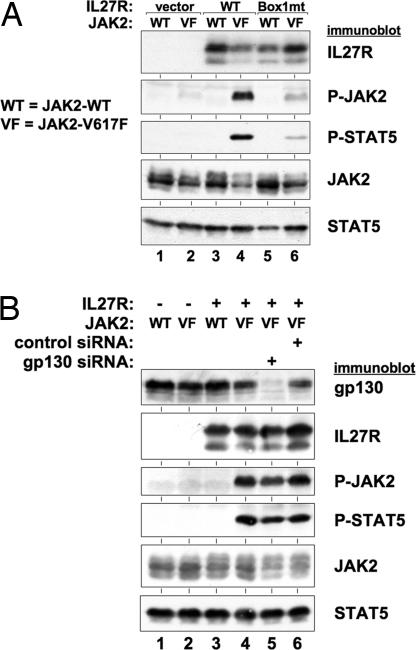
Homodimeric type I cytokine receptors have been shown to be required for the full activation of JAK2-V617F (37), a JAK2 mutant found in a variety of MPDs and AML (24–30, 38). It is believed homodimeric cytokine receptors (e.g., EpoR) provide a scaffold upon which JAK2-V617F proteins can bind and become activated by transphosphorylation (37). Such activation of mutant JAK2 is independent of ligand for the receptor. IL-27R is a type I cytokine receptor that normally functions as a heterodimeric partner with gp130 (18, 39). However, the ability of IL-27R to transform BaF3 and 32D cells, which lack gp130 expression, suggests IL-27R does not require gp130 to transform cells. Because the transforming properties of IL-27R do not depend on its heterodimeric partner gp130, we thought IL-27Rs could function as homodimeric receptors and therefore be able to activate JAK2-V617F (37). We found IL-27R activates JAK2-V617F, and its downstream target STAT5, in 293T cells (Fig. 5A). Expression of IL-27R along with WT JAK2 did not lead to activation of JAK2 or STAT5. IL-27R containing a mutated JAK-binding Box 1 motif (as in Fig. 3) was impaired in activation of JAK2-V617F and STAT5 compared with WT IL-27R (Fig. 5A). Because 293T cells express gp130, it is possible activation of JAK2-V617F by IL-27R requires gp130. However, depletion of gp130 by siRNA had no effect on IL-27R-mediated activation of JAK2-V617F and STAT5, demonstrating this effect is gp130-independent (Fig. 5B). Although we have not been able to coimmunoprecipitate IL-27R and JAK2 to demonstrate a presumed interaction, we are able to detect a complex formation between IL-27R and JAK2, as well as JAK1, in vitro (SI Fig. 10), suggesting IL-27R may complex with JAK2 in cells. Together, our data suggest that IL-27R can function in an analogous manner as homodimeric type I receptors to activate JAK2-V617F.
Fig. 5.
Activation of JAK2-V617F by IL-27R. (A) 293T cells were transfected with control vector (lanes 1 and 2), WT IL-27R (lanes 3 and 4), and IL-27R-Box1mt (lanes 5 and 6) along with WT JAK2 (lanes 1, 3, and 5) or JAK2-V617F (lanes 2, 4, and 6). Forty-eight hours after transfection, cells were lysed and analyzed by immunoblotting with antibodies against the indicated proteins. The combination of IL-27R and JAK2-V617F led to high levels of activated JAK2 and STAT5 (lane 4). IL-27R-Box1mt was impaired in its ability to activate JAK2-V617F and STAT5 (lane 6). (B) 293T cells were transfected with control vector (lanes 1 and 2) and IL-27R (lanes 3–6) along with JAK2 (WT, lanes 1 and 3), JAK2-V617F (VF, lanes 2 and 4–6), gp130 siRNA (lane 5), and control nonsilencing siRNA (lane 6). Forty-eight hours after transfection, cell lysates were analyzed by immunoblotting with antibodies that recognize the proteins indicated. Depletion of gp130 did not affect the ability of IL-27R to activate JAK2-V617F and STAT5 (lane 5).
Discussion
In an effort to identify genes that contribute to myeloid cell transformation, we used functional genetic screens of genes expressed in the leukemic cells of AML patients. Through our approach, we uncovered unique cell-transforming properties of IL-27R, the ligand-binding component of the receptor for IL-27 (18). Importantly, we have determined IL-27R is frequently expressed on the cell surface of a greater number of bone marrow cells of AML patients than of cells of normal bone marrow, suggesting it may play a role in leukemogenesis (Fig. 1D).
IL-27R is a member of the IL-6/IL-12 receptor family (40). A heterodimeric receptor complex of IL-27R/gp130 is required to activate signaling pathways in response to IL-27 stimulation of cells (18). This includes activation of JAK1, JAK2, Tyk2, STAT1, STAT2, STAT3, STAT4, and STAT5 (20, 21). IL-27 regulates various aspects of immune responses, including T cell-mediated immunity (21, 35, 39–42), and can also regulate the activity of B cells, mast cells, and monocytes (18, 39, 40, 43). Interestingly, IL-6 can function as a growth factor for various cancer cells (44, 45), including AML blast cells (46), suggesting signaling by members of this cytokine receptor family can promote oncogenic cell growth. In support of this notion, Takeda et al. (47) have observed hyperproliferation of T cells designed to overexpress IL-27R.
In this article, we show that expression of IL-27R induces IL-3-independent growth of 32D myeloid and BaF3 pro-B cells (Figs. 1A and 4A). Because BaF3 cells lack gp130 expression (18, 36), IL-27R-mediated transformation does not depend on gp130. In addition, we were unable to detect expression of gp130 in 32D cells (Fig. 4B), demonstrating IL-27R does not require gp130 to elicit a transforming signal in myeloid cells. 32D cells transformed by IL-27R have constitutively activated JAK/STAT family members (Fig. 1C), the JAK-binding Box 1 motif of IL-27R is required for its transforming activity (Fig. 3B), and JAK inhibition blocks cell transformation by IL-27R (Fig. 2). These data suggest IL-27R-mediated activation of JAK family members is critical for its transforming capacity.
JAK family proteins play a major role in myeloid disorders as highlighted by the discovery of the JAK2-V617F point mutation. JAK2-V617F likely contributes to the pathogenesis of various MPDs, including polycythemia vera, essential thrombocytosis, and myelofibrosis (24–28). Homodimeric type I cytokine receptors have been shown to be required for JAK2-V617F-mediated activation and transformation. Homodimeric receptors, including EpoR, TpoR, and GCSFR, support the activation of JAK2-V617F in a ligand-independent manner (37). These receptors likely provide a scaffold upon which mutant JAK2 proteins can interact, which is believed to be necessary for the activation of the mutant kinase. Although IL-27R is a component of a heterodimeric cytokine receptor with gp130, we show that it is capable of activating JAK2-V617F in cells in a gp130-independent manner (Fig. 5). This finding suggests IL-27R can functionally replace homodimeric cytokine receptors to support the activation of JAK2-V617F. It is still to be determined whether or not IL-27R functions as a homodimer in the context of its ligand-independent transforming properties.
Recently, point mutations in Mpl, the gene for the type I cytokine receptor TpoR, have been found in JAK2-V617F-negative myelofibrosis and essential thrombocythemia patients (48, 49). These mutations were identified under the hypothesis that patients who lack a JAK2 mutation may have other mutations that lead to JAK2 activation, such as mutations in upstream activators of JAK2, including cytokine receptors. The identification of point mutations in Mpl in myeloid disorders suggests mutations in other type I cytokine receptors may also contribute to diseases of the myeloid system. Our data suggest that contribution of heterodimeric cytokine receptors to JAK2-V617F pathogenesis, as well as JAK2-V617F-negative myeloid disorders, should be considered.
We suggest that a nonmutated single chain of a heterodimeric type I cytokine receptor has the ability to transform hematopoietic cells and that a single component of a heterodimeric type I cytokine receptor can functionally replace a homodimeric type I receptor as an activator of JAK2-V617F. In light of these findings, our data suggest that heterodimeric type I cytokine receptors may play unappreciated roles in mediating activation of signaling pathways in myeloid disorders and, like TpoR, such receptors may contribute to JAK2-V617F-negative MPDs. This contribution may be through altered expression or mutation of the receptor and can be investigated further by performing sequencing and expression studies in patients with MPDs as well as AML.
Materials and Methods
Cell Culture and Retrovirus Production.
293T cells were maintained in DMEM supplemented with 10% FBS. 32D cells and BaF3 cells were grown in RPMI medium 1640 supplemented with 10% FBS and 5% WEHI-3B conditioned medium as a source of IL-3. Ecotropic retrovirus was made in 293T cells by using the pVPack system (Stratagene). Stable cell lines were generated by retroviral infection as described in SI Text.
cDNA Library Construction.
Under Institutional Review Board approval, AML samples were obtained from the Moffitt Cancer Center Tissue Core Facility as viably frozen mononuclear cells from the bone marrow of untreated patients. mRNA was isolated by using a FastTrack 2.0 mRNA Isolation Kit (Invitrogen). Double-stranded cDNA was prepared with a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and purified in two fractions of two sizes by using the Geneclean III kit (Q-Biogene). cDNA fractions were ligated into the pEYK3.1 retroviral vector (14), and ligations were transformed into E. cloni electrocompetent cells (Lucigen). The library contained ≈3.3 million bacterial colonies with a cloning efficiency of ≈90%.
Screening of cDNA Library and Isolation of cDNA from Cells.
32D cells expressing exogenous Bcl2 were infected with retrovirus made from the AML cDNA library. Four independent infections were done for each cDNA fraction. Two days after infection, cells were plated in the absence of IL-3 to select for IL-3-independent transformants. Genomic DNA was isolated from IL-3-independent cells and treated with Cre recombinase (NEB) to excise pEYK3.1 plasmids, containing putative transforming cDNAs, which were then isolated by bacterial transformation.
Cell Growth Analysis.
To assay 32D and BaF3 cell response to IL-3 deprivation, cells were washed twice with RPMI medium 1640/10% FBS. Cells were plated at a concentration of 4 × 105/ml in RPMI medium 1640/10% FBS, and cell growth and viability were monitored by trypan blue exclusion.
Immunoblot Analyses.
Cells were washed in PBS and lysed in lysis buffer [25 mM Tris (pH 7.4), 150 mM NaCl, 25 mM NaF, 1% Triton X-100, 1 mM sodium vanadate, 2 mM sodium pyrophosphate, 10 μg/ml leupeptin, 2 μg/ml aprotinin, and 1 mM PMSF]. Protein concentrations were determined with a BCA protein assay kit (Pierce Biotechnology), and equal amounts of protein were analyzed by SDS/PAGE. Primary antibodies used in this study include: IL-27R (TCCR) (C-term) (Sigma), phospho-(P-) STAT1(Y701), P-STAT3(Y705), P-JAK1(Y1022/1023), P-JAK2(Y1007/1008), P-ERK(T202/Y204), JAK1, JAK2 (Cell Signaling Technology), P-STAT5(Y694) (BD Transduction Laboratories), and STAT1, STAT3, STAT5, and ERK1 (Santa Cruz Biotechnology). Immunoblots were developed by using ECL Western Blotting Substrate (Pierce Biotechnology).
Flow Cytometry for IL-27R Expression.
Expression of IL-27R was determined by flow cytometry using an anti-human TCCR/WSX-1 antibody (R&D Systems) labeled with Alexa Fluor-647 (Invitrogen/Molecular Probes). Details are provided in SI Text.
JAK Inhibitor I Studies.
32D cells were washed twice with RPMI medium 1640 containing 0.1% BSA and incubated in the same medium with either 0.1% DMSO or 0.5 μM or 2 μM JAK inhibitor I (EMD Biosciences/Calbiochem) for 3 h before lysis. For cell growth and viability, cells were plated at a concentration of 2 × 105/ml in RPMI medium 1640/10% FBS and either 0.1% DMSO or 0.5 μM JAK inhibitor I. Cell growth and viability were determined by trypan blue exclusion. After 24 h of JAK inhibitor I treatment, the percentage of apoptotic cells and cell cycle profiles were determined by flow cytometry. See SI Text.
Construction of IL-27R Box 1 Mutant.
IL-27R Box1 mutant was generated by site-directed mutagenesis (Stratagene). See SI Text.
JAK2-V617F Activation Studies.
293T cells were transfected with combinations of pBabe-puro, pBabe-puro-IL-27RWT, or Box1mt, and MSCVneo-JAK2-WT or V617F (a gift of Ross Levine, Memorial Sloan-Kettering Cancer Center, New York), and SMARTPool siRNA against gp130 (Dharmacon) or a nonsilencing control siRNA (Qiagen) by using calcium phosphate precipitation. Cells were lysed and analyzed by immunoblotting 2 days after transfection.
Supplementary Material
Acknowledgments
We thank George Daley for the pEYK3.1 plasmid, Ross Levine for the JAK2-WT and V617F expression plasmids, Amy Hazen for technical advice, and William Kerr for helpful discussions. This work was supported in part by the Flow Cytometry and Molecular Biology Core Facilities at the H. Lee Moffitt Cancer Center and Research Institute, National Institutes of Health/National Cancer Institute Grant K01CA098330, the American Cancer Society, and The V Foundation for Cancer Research (G.W.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702388104/DC1.
References
- 1.Frohling S, Scholl C, Gilliland DG, Levine RL. J Clin Oncol. 2005;23:6285–6295. doi: 10.1200/JCO.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Gilliland DG. Curr Opin Hematol. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Gilliland DG. Semin Hematol. 2002;39:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- 4.Deguchi K, Gilliland DG. Leukemia. 2002;16:740–744. doi: 10.1038/sj.leu.2402500. [DOI] [PubMed] [Google Scholar]
- 5.Gilliland DG, Tallman MS. Cancer Cell. 2002;1:417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 6.Carnicer MJ, Nomdedeu JF, Lasa A, Estivill C, Brunet S, Aventin A, Sierra J. Leuk Res. 2004;28:19–23. doi: 10.1016/s0145-2126(03)00125-5. [DOI] [PubMed] [Google Scholar]
- 7.Reuther GW, Lambert QT, Caligiuri MA, Der CJ. Mol Cell Biol. 2000;20:8655–8666. doi: 10.1128/mcb.20.23.8655-8666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reuther GW, Lambert QT, Rebhun JF, Caligiuri MA, Quilliam LA, Der CJ. J Biol Chem. 2002;277:30508–30514. doi: 10.1074/jbc.M111330200. [DOI] [PubMed] [Google Scholar]
- 9.Meyer J, Rhein M, Schiedlmeier B, Kustikova O, Rudolph C, Kamino K, Neumann T, Yang M, Wahlers A, Fehse B, et al. Leukemia. 2007;21:2171–2180. doi: 10.1038/sj.leu.2404882. [DOI] [PubMed] [Google Scholar]
- 10.Greenberger JS, Sakakeeny MA, Humphries RK, Eaves CJ, Eckner RJ. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 12.Laneuville P, Heisterkamp N, Groffen J. Oncogene. 1991;6:275–282. [PubMed] [Google Scholar]
- 13.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, et al. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 14.Koh EY, Chen T, Daley GQ. Nucleic Acids Res. 2002;30:e142. doi: 10.1093/nar/gnf142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baffy G, Miyashita T, Williamson JR, Reed JC. J Biol Chem. 1993;268:6511–6519. [PubMed] [Google Scholar]
- 16.Nunez G, London L, Hockenbery D, Alexander M, McKearn JP, Korsmeyer SJ. J Immunol. 1990;144:3602–3610. [PubMed] [Google Scholar]
- 17.Sprecher CA, Grant FJ, Baumgartner JW, Presnell SR, Schrader SK, Yamagiwa T, Whitmore TE, O'Hara PJ, Foster DF. Biochem Biophys Res Commun. 1998;246:82–90. doi: 10.1006/bbrc.1998.8576. [DOI] [PubMed] [Google Scholar]
- 18.Pflanz S, Hibbert L, Mattson J, Rosales R, Vaisberg E, Bazan JF, Phillips JH, McClanahan TK, de Waal Malefyt R, Kastelein RA. J Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- 19.Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, Hibbert L, Churakova T, Travis M, Vaisberg E, et al. Immunity. 2002;16:779–790. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 20.Kamiya S, Owaki T, Morishima N, Fukai F, Mizuguchi J, Yoshimoto T. J Immunol. 2004;173:3871–3877. doi: 10.4049/jimmunol.173.6.3871. [DOI] [PubMed] [Google Scholar]
- 21.Lucas S, Ghilardi N, Li J, de Sauvage FJ. Proc Natl Acad Sci USA. 2003;100:15047–15052. doi: 10.1073/pnas.2536517100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin TS, Mahajan S, Frank DA. Oncogene. 2000;19:2496–2504. doi: 10.1038/sj.onc.1203486. [DOI] [PubMed] [Google Scholar]
- 23.Sternberg DW, Gilliland DG. J Clin Oncol. 2004;22:361–371. doi: 10.1200/JCO.2004.10.124. [DOI] [PubMed] [Google Scholar]
- 24.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 25.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 26.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 27.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 28.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R, Chase AJ, Grand FH, et al. Blood. 2005;106:2162–2168. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 29.Lee JW, Kim YG, Soung YH, Han KJ, Kim SY, Rhim HS, Min WS, Nam SW, Park WS, Lee JY, et al. Oncogene. 2006;25:1434–1436. doi: 10.1038/sj.onc.1209163. [DOI] [PubMed] [Google Scholar]
- 30.Levine RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E, Berger R, Clark JJ, Willis SG, Nguyen KT, et al. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towatari M, Iida H, Tanimoto M, Iwata H, Hamaguchi M, Saito H. Leukemia. 1997;11:479–484. doi: 10.1038/sj.leu.2400617. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JE, Cubbon RM, Cummings RT, Wicker LS, Frankshun R, Cunningham BR, Cameron PM, Meinke PT, Liverton N, Weng Y, et al. Bioorg Med Chem Lett. 2002;12:1219–1223. doi: 10.1016/s0960-894x(02)00106-3. [DOI] [PubMed] [Google Scholar]
- 33.Ihle JN. Nature. 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 34.Tanner JW, Chen W, Young RL, Longmore GD, Shaw AS. J Biol Chem. 1995;270:6523–6530. doi: 10.1074/jbc.270.12.6523. [DOI] [PubMed] [Google Scholar]
- 35.Takeda A, Hamano S, Yamanaka A, Hanada T, Ishibashi T, Mak TW, Yoshimura A, Yoshida H. J Immunol. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- 36.Nandurkar HH, Hilton DJ, Nathan P, Willson T, Nicola N, Begley CG. Oncogene. 1996;12:585–593. [PubMed] [Google Scholar]
- 37.Lu X, Levine R, Tong W, Wernig G, Pikman Y, Zarnegar S, Gilliland DG, Lodish H. Proc Natl Acad Sci USA. 2005;102:18962–18967. doi: 10.1073/pnas.0509714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter CA. Nat Rev Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- 40.Villarino AV, Huang E, Hunter CA. J Immunol. 2004;173:715–720. doi: 10.4049/jimmunol.173.2.715. [DOI] [PubMed] [Google Scholar]
- 41.Artis D, Villarino A, Silverman M, He W, Thornton EM, Mu S, Summer S, Covey TM, Huang E, Yoshida H, et al. J Immunol. 2004;173:5626–5634. doi: 10.4049/jimmunol.173.9.5626. [DOI] [PubMed] [Google Scholar]
- 42.Chen Q, Ghilardi N, Wang H, Baker T, Xie MH, Gurney A, Grewal IS, de Sauvage FJ. Nature. 2000;407:916–920. doi: 10.1038/35038103. [DOI] [PubMed] [Google Scholar]
- 43.Larousserie F, Charlot P, Bardel E, Froger J, Kastelein RA, Devergne O. J Immunol. 2006;176:5890–5897. doi: 10.4049/jimmunol.176.10.5890. [DOI] [PubMed] [Google Scholar]
- 44.Frassanito MA, Cusmai A, Iodice G, Dammacco F. Blood. 2001;97:483–489. doi: 10.1182/blood.v97.2.483. [DOI] [PubMed] [Google Scholar]
- 45.Molnar EL, Hegyesi H, Toth S, Darvas Z, Laszlo V, Szalai C, Falus A. Semin Cancer Biol. 2000;10:25–28. doi: 10.1006/scbi.2000.0304. [DOI] [PubMed] [Google Scholar]
- 46.Saily M, Koistinen P, Zheng A, Savolainen ER. Eur J Haematol. 1998;61:190–196. doi: 10.1111/j.1600-0609.1998.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 47.Takeda A, Hamano S, Shiraishi H, Yoshimura T, Ogata H, Ishii K, Ishibashi T, Yoshimura A, Yoshida H. Int Immunol. 2005;17:889–897. doi: 10.1093/intimm/dxh268. [DOI] [PubMed] [Google Scholar]
- 48.Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, Steensma DP, Elliott MA, Wolanskyj AP, Hogan WJ, et al. Blood. 2006;108:3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- 49.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G, Moore S, Galinsky I, et al. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.