Abstract
Macrophages use phagocytosis to control the spread of pathogens in the body, to clear apoptotic cells, and to aid in tissue remodeling. The phagosomal membrane is traditionally thought to originate from the plasmalemma and then go through a series of maturation steps involving sequential fusion with endosomal compartments, leading to the formation of a phagolysosome. A recent model suggests that the endoplasmic reticulum (ER) is involved in the maturation as well. Here we use stable isotope labeling and multiple quantitative proteomic approaches to follow the dynamic composition of the maturing phagosome in RAW 264.7 macrophage cells to a greater depth and higher temporal resolution than was previously possible. Analysis of the results suggests that the traditional model of a linear sequence of fusion events with different compartments is more complex or variable than previously thought. By concomitantly measuring the degree to which each component is enriched on phagosomes, our data argue that the amount of ER involved in phagocytosis is much less than predicted by the model of ER-mediated phagocytosis.
Keywords: innate immunity, organelle, phagocytosis, stable isotope labeling, latex bead vacuoles
Phagocytosis is the process by which cells engulf particles. Primitive eukaryotes use phagocytosis primarily to obtain nutrients (1), whereas in more complex organisms it serves additional functions such as the clearance of apoptotic cells and various pathogens (2). Internalized objects are contained in a membrane-bound vacuole called the phagosome, and current models have the phagosome traversing a complex maturation process involving sequential fusion with early endosomes (EE), late endosomes (LE), and ultimately with lysosomes (LS) (3, 4). In this model, full maturation is characterized by luminal acidification and acquisition of hydrolytic enzymes, which serve to degrade or kill the cargo, typically microbes, within 2–4 h after internalization (5). Previous studies have aimed to characterize the phagosome proteomes in cell lines from mouse and fruit fly, as well as from Dictyostelium discoideum, Entamoeba histolytica, and Tetrahymena thermophila (6–10). Of particular note, the studies of Desjardins and colleagues (6) in mouse macrophages led them and others to propose a role for the endoplasmic reticulum (ER) in phagocytosis (11, 12), although this model has been challenged recently (13). Here we apply stable isotope labeling by amino acids in cell culture (SILAC) to develop a comprehensive, quantitative model of phagosome maturation that allows us to test several hypotheses suggested by current models of the process.
Results and Discussion
Maturation into a phagolysosome is absolutely crucial for eventual destruction of phagocytosed objects, whereas the success of pathogens such as Salmonella enterica and Mycobacterium tuberculosis depends largely on their ability to avoid phagolysosomal killing. To gain a better understanding into the process of phagosome maturation we used latex beads to model phagocytosis in RAW 264.7 mouse macrophage cells and SILAC with an LTQ-Orbitrap to measure the dynamics of the maturing phagosomal proteome with unparalleled accuracy.
IgG was chosen as an opsonin rather than whole serum (6) to reduce potentially confounding effects of phagocytosis through different receptor systems. IgG-opsonized latex beads were allowed to internalize for 10 min, and then latex bead-containing vacuoles (LBVs) were harvested at seven different time points (Fig. 1). We focused on events within 2 h of phagocytosis because most internalized objects are dead or destroyed by this time and there is no apparent physiological relevance to allowing vacuoles containing inert latex to mature for longer periods. From at least three biological replicates of each time point we identified 505 proteins associated with LBVs, 382 of which could be reliably quantified across various time points [supporting information (SI) Table 1]. Based on current models of phagosome maturation (4), we expected to find a large influx of EE markers, followed by LE markers and ending with a bolus of LS proteins. Certain markers of these compartments did peak at the expected time points, but endosomal proteins and the vesicle trafficking machinery as a whole did not arrive in three discrete packages, as predicted.
Fig. 1.
Application of SILAC to phagosome maturation. (a) IgG-opsonized, 0.8-μm latex beads were added to RAW 264.7 cells (T = 0) and allowed to phagocytose for 10 min, at which point external beads were washed away and LBVs were allowed to mature for the times indicated by dashed gray lines. Because SILAC is limited to a maximum of three conditions per experiment, one time point in each experiment was used to scale the other time points across experiments as described in Materials and Methods. For instance, the 90-min time point was used to scale between experiments 2 and 3 in this example. (b) Measured spectra for the [M + 2H]2+ ion of ATIGADFLTK from Rab7 covering all seven time points in three experiments are shown. The abscissa scale for each peak cluster is m/z, but note that the three clusters in each row are not ordered by m/z but rather by the time point that they represent. The ordinate axes on each have been corrected by the isotope enrichment factor measured for each experiment. (c) The overall profile that would be calculated for ATIGADFLTK based on only the three experiments shown. In reality the profile for a protein was the averaged, normalized intensity at each time point for each peptide identified from that protein.
Biochemical enrichment of an organelle is never perfect, because complete separation from all other organelles is essentially never achieved. Given what is known about phagosome development, one can easily imagine a situation where the actin mesh surrounding LBVs at certain stages could entrap pieces of other organelles, leading to their apparent time-dependent copurification with LBVs. In our experimental approach, one would expect an unchanging SILAC profile over time if proteins were enriched at the same time as LBVs but independent of the LBV maturation itself. However, such is not the case for most markers of other organelles that we have measured here (SI Table 2). One possible explanation for this could be that some organelles get trapped to varying degrees with the LBVs (e.g., caught in the surrounding actin mesh) at certain time points. An alternative explanation could be that the cell lysis procedure disrupts LBVs themselves, leading to some nonspecific association of cytoplasmic proteins with the beads. Proteins from other organelles should generally not affect profiles of LBV proteins, however, except where a protein is shared between LBVs and the copurifying organelle.
Multiple Visits to the LBV.
One by one the movement of most known endosomal markers has been followed on maturing phagosomes using Western blotting (14). Our data set contains the majority of proteins these early studies focused on, and the profiles measured by SILAC largely agree with those reported previously. For instance, EE antigen 1 is most abundant at 30 min, Niemann–Pick C1 and charged multivesicular body protein 6 (CMVB6) peak around 60 min, and the LS-associated membrane proteins (LAMPs) and cathepsins peak at 90 or 120 min (Fig. 2). However, we also observed that many proteins, including LAMP-1, heterotrimeric G proteins from the plasma membrane, and some cathepsins, display a biphasic profile with a smaller peak earlier in the time course. Although not explicitly defining a biphasic profile, different studies have found LAMP-1 on phagosomes very early after phagocytosis (15) or at later times (4). Many of the other proteins we measured also display a similar pattern (Fig. 2 and SI Table 2), suggesting that the EE, LE, and LS designations are too simplistic and that the conceptualized EE, LE, and LS compartments contain significant heterogeneity. Although there is undoubtedly some “spillage” or leakiness of even classical marker proteins such as LAMP-1 into different compartments, this alone is insufficient to explain the patterns we observe. The biphasic profiles observed are unlikely to be artifacts of nonsynchronous internalization, the LBV isolation procedure, or the quantitative MS approach for several reasons: (i) timing of washes and harvesting varied by no more than 3%, (ii) some of the profiles are confirmed by Western blots (Figs. 2 and 3), (iii) the peaks and troughs of all of the profiles do not align, and (iv) not all proteins show the biphasic profile.
Fig. 2.
Endosomal proteins repeatedly move on and off LBVs. LBV profiles of several endosomal and plasma membrane proteins identified in this study are shown. Cath, cathepsins; NPC1, Niemann–Pick C1 protein; CMVB6, charged multivesicular body protein 6. Shown are average profiles from at least three replicates of each time point. (Inset) Western blot of CathA across the time points indicated.
Fig. 3.
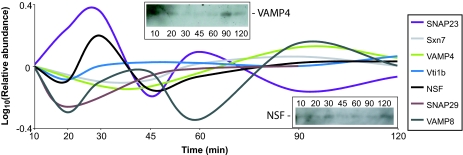
Dynamic profiles of SNAREs. Shown are LBV profiles of several SNARE and SNARE-associated proteins identified in this study. Sxn7, syntaxin 7; SNAP23 and SNAP29, synaptosome-associated protein of 23 or 29 kDa. Shown are average profiles from at least three replicates of each time point. (Insets) Western blots of VAMP4 and NSF across the time points indicated.
One of the hallmarks of phagosome maturation is the steady decline in the luminal pH, brought about by the acquisition of the vacuolar ATPase (vATPase). Many subunits of the vATPase were measured in this study, and as a group they peak at 90 min (SI Fig. 6), as predicted. However, the vATPase subunits all show a biphasic profile as well, with an earlier peak or plateau at 30 or 45 min. This suggests that the maturing phagosome initially fuses with an LE- or LS-like compartment, which is then followed by a larger influx of LE/LS proteins.
These conclusions are also supported by the LBV profiles of the proteins with a mechanistic role in vesicle traffic, the SNARE and Rab families of proteins. Participation of SNAREs such as syntaxin 7 and vesicle-associated membrane protein 8 (VAMP8)/endobrevin in LE/LS fusion is well known (16), and their LBV profiles reflect this action. Intriguingly, though, several SNAREs (Fig. 3 and SI Table 2), including VAMP8, also show a biphasic profile. There is still much debate as to whether SNAREs alone are sufficient to dictate fusion of two specific membranes (17); regardless, the biphasic profile of several SNARE proteins suggests that certain classes of compartments are fusing with the maturing LBV more than once.
Rabs.
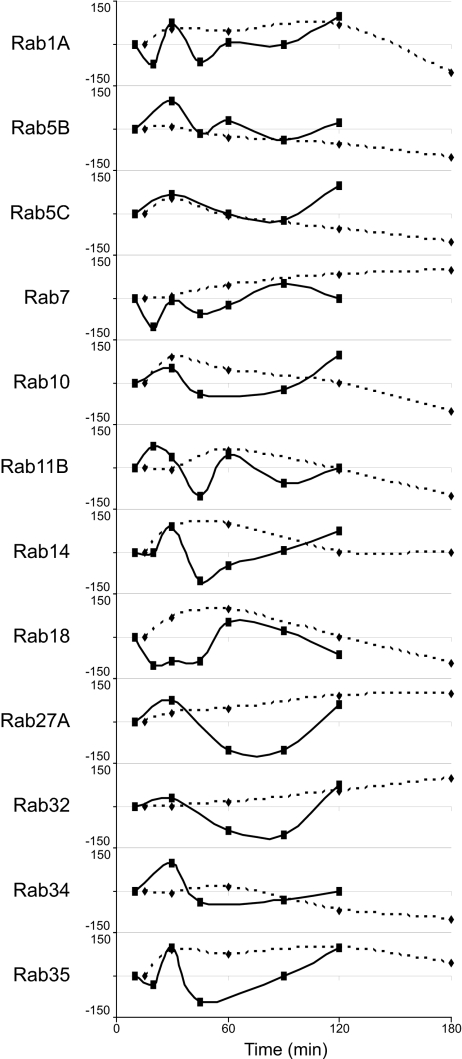
A number of Rab guanine triphosphatases are known to localize to specific compartments in the endocytic pathway and regulate membrane traffic (18). Our proteomic approach identified 20 Rabs on LBVs, 16 of which could be quantified sufficiently to construct a time course. Of these 16, 12 were also among the 48 Rabs examined for their phagosomal association by Smith et al. (19). As shown in Fig. 4, the mass spectrometry and microscopy profiles were largely congruous, at least with respect to whether a protein was increasing or decreasing in abundance at a certain time, if not in the actual magnitude. There were a couple of notable exceptions, particularly Rabs 14, 27, and 35, where our approach detected a decreased level of association in the middle time points relative to that measured by Smith et al. (19), possibly because of differences in the Salmonella-containing vacuole versus IgG-opsonized LBVs or epithelial cells versus macrophages. Nonetheless, the LBV profiles of these and the other Rabs measured here support the general conclusion that maturation involves more than just three discrete fusion events. At least one Rab protein peaked in abundance at each time measured, except for 45 min, suggesting that there are at least five discrete events or, more likely, a continuum of fusion.
Fig. 4.
Dynamic profiles of Rabs. Shown are profiles of the 12 Rab proteins common to the LBVs in our study and the Salmonella-containing vacuoles of Smith et al. (19). The data for our data (solid lines) and Smith et al.'s data (dashed lines) plots are normalized to the first time point (10 min for our data and 15 min for the data of Smith et al.), log10-transformed, and then expressed as a percentage of the highest absolute measurement. Our data profiles are from at least three replicates of each time point.
Unknown Players in Phagosome Maturation.
Although we undertook this study to try to address some hypotheses about phagosome maturation, the depth and temporal dimension of our data set also generate testable hypotheses by describing the phagosomal dynamics of several proteins not previously predicted to play a role in this process. To pick two of many examples, annexins A2 and A7 were enriched on LBVs at 60 min (SI Table 2), suggesting that they arrive with LE or play a role in their fusion with LBVs. Annexin A2 was previously shown to facilitate endosome fusion (20) and biogenesis (21), whereas A7 may be regulated by Mycobacterium avium (22). Because Mycobacterium is known to prevent phagolysosomal fusion, our data suggest that annexin A7 could be tested as a potential host target of Mycobacterium effectors. Likewise, annexin A2 could be tested as a more general regulator of LE traffic.
The Contribution of the ER.
ER proteins in biochemically enriched phagosomes were typically written off as contaminants until Desjardins and colleagues (11) used electron microscopy to argue that the ER itself could phagocytose particles and that successive waves of ER interact with the maturing phagosome (11). The role of the ER in phagocytosis is far from resolved, however, because more recently Grinstein and colleagues (23) proposed a diametrically opposed model where the ER plays at most a very small role in phagosome maturation, leaving no apparent middle ground to incorporate both theories. Several testable hypotheses come out of this controversy; here we use proteomic analysis of LBV preparations to address some of them.
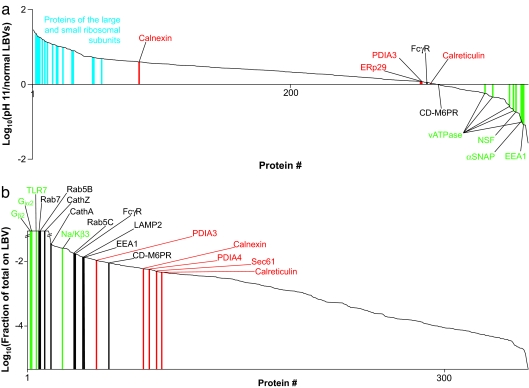
The ER is typically quite dense (24), so it should not migrate at all close to latex beads in a density gradient. However, the ER forms a reticular network throughout the cytoplasm, so if ER membrane is not contiguous with the phagosomal membrane then it is plausible that some of the ER gets trapped in the actin mesh surrounding the LBV or is otherwise physically but not functionally associated with vacuoles. To address the possibility that ER proteins are not integral to the phagosomal membrane, we quantified the effect of a pH 11.5 buffer on components of the LBV purification. High pH is commonly used to strip away nonintegral membrane proteins (25), so if pieces of the ER were somehow contaminating the LBV preparation and not part of the LBVs themselves then ER proteins would be expected to be depleted in LBVs isolated in high pH. However, most common ER marker proteins, with the exception of calnexin, were present at equal levels in LBVs isolated at physiological pH and at pH 11.5, as were integral LBV membrane proteins such as the Fcγ receptor and the cation-dependent mannose 6-phosphate receptor (Fig. 5a). This observation supports the conclusions of Garin et al. (6) that ER proteins are integral to the vacuolar membrane. That calnexin is increased with high pH suggests that a fraction of it may reside on some ER membrane not integral to the phagosomal membrane.
Fig. 5.
ER proteins on LBVs are integral to the membrane but constitute only a small fraction of the total ER. (a) Ranked relative abundances of proteins on LBVs isolated from RAW cells at pH 11.5 versus at pH 7.2. Specific ribosomal proteins (blue), ER markers (red), integral LBV membrane proteins (red), and membrane-associated proteins (green) are indicated. PDIA3, protein disulfide isomerase 3; αSNAP, soluble NSF attachment protein; FcγR, Fcγ receptor; CD-M6PR, cation-dependent mannose 6-phosphate receptor. (b) The amount of each protein found on LBVs at 10 min, expressed as a fraction of the total amount of that protein in the cell normalized for the fraction of cells taking up beads. Specific ER markers (red), endosomal/phagosomal markers (black), and plasma membrane markers (green) are indicated. TLR7, Toll-like receptor 7; Cath, cathepsin; Na/Kβ3, Na+/K+-ATPase β3 subunit. The ratios are cut off at 100-fold up or down because the linearity of the signal in the Orbitrap starts to drop off beyond this.
The surface area of an actively phagocytosing macrophage stays approximately constant, or, if anything, it increases, despite needing to use an area of membrane equal to or greater than its surface to engulf the particles (26, 27). This suggests that an intracellular source of membrane is used in replenishing the plasma membrane (PM) or in actually forming the phagosome. Endosomal and post-Golgi membranes are known to be such sources (14, 28), and Desjardins and colleagues (11) have proposed that the ER may be directly involved in phagocytosis. The area of the ER membrane is approximately twice that of the PM in hepatic phagocytes (Kupfer cells) (29), so it is reasonable to assume that the ratio would be similar in RAW cells. For each of >300 proteins found on LBVs, we measured the portion of the total complement of that protein actually found associated with the vacuole. Approximately 10% of the PM was found on LBVs, based on the enrichment of several PM markers (Fig. 5b), and, given the assumed 2:1 ratio of ER:PM membrane, the ER-mediated phagocytosis model would predict at least 20% of the ER membrane to be present on LBVs. However, LBV preparations contained only ≈0.3% of the cellular total of five ER marker proteins used by others (11, 23): protein disulfide isomerases, calnexin, calreticulin, and Sec61 (Fig. 5b). These data argue against the ER being a major source of membrane for newly forming phagosomes, whether it is through direct internalization into the ER (11) or as a source for replenishing lost PM. Our data do not, however, address the observation that the ER may be involved in engulfing only large particles (30) or the possibility that replenishment from the ER is required only after long, sustained periods of phagocytosis.
Conclusions
The dynamic profiles of 382 proteins associated with biochemically enriched phagosomes from RAW cells described here provide a comprehensive view of the process of phagosome maturation at a temporal resolution exceeding that of most previous studies focusing on even single phagosomal proteins. Our data are largely congruous with the few known markers of the process, but they also suggest that maturation does not just proceed as three discrete fusion events with EE, LE, and then LS. By directly measuring enrichment of proteins on LBVs, our data shed light on the current controversy surrounding the role of the ER in phagocytosis (11, 13) by arguing strongly against the ER-mediated phagocytosis model. Our data enhance the knowledge of the latex bead model of phagocytosis and will hopefully open new avenues for understanding pathogen-containing vacuoles.
Materials and Methods
Cell Culture and SILAC.
RAW 264.7 mouse macrophage-like cells were maintained as described (23) and split at a 1:4 dilution into one of three SILAC media formulations: (i) normal isotopic abundance arginine (42 mg/liter) and lysine (73 mg/liter), (ii) [13C6]arginine (43.5 mg/liter; Cambridge Isotope Laboratories, Andover, MA) and [2H4]lysine (75 mg/liter), or (iii) [13C6,15N4]arginine (44.5 mg/liter) and [13C6,15N2]lysine (77 mg/liter). All SILAC media were based on arginine- and lysine-free Dulbecco's modified Eagle's medium (Caisson Labs, North Logan, UT) supplemented with 10% heat-inactivated, dialyzed FBS (Invitrogen), 1% l-glutamine, and 1% penicillin/streptomycin (ThermoFisher Scientific, Bremen, Germany). Cells were passaged three more times in the above media at a 1:4 dilution each time before use. In our experience these labeling conditions led to 100% incorporation in most cell types, but in RAW cells we found inconsistent levels of heavy isotope incorporation from experiment to experiment. We attribute this phenomenon to the phagocytic capacity of these cells, which likely allows them to scavenge substantial levels of amino acids from the unlabeled proteins in serum. To compensate for incomplete labeling we analyzed 2 μg of the combined lysates from all three label sets and used the incorporation levels measured in the lysate to correct the ratios measured in each experiment individually. This incorporation pattern meant that the ion intensity for the light form of each SILAC triplet was typically higher than for the two heavier forms. Accordingly, to avoid a bias in the data-dependent acquisition the time point used for the light form was rotated from experiment to experiment.
LBV Isolation.
Six 14-cm plates of RAW cells were used per time point in each experiment. To initiate internalization the growth media were removed, the cells were washed once in PBS, and then 10 ml of serum-free and arginine/lysine-free DMEM was added to each plate. Mouse Ig-opsonized 0.8-μm latex beads (Sigma-Aldrich) were added to the cells at a 1:400 dilution and allowed to incubate for 10 min, at which time the bead solution was aspirated, the cells were washed once with PBS, and then 12 ml of the appropriate SILAC media was added back for various lengths of time. Each time point was measured between three and six times. For normal time courses LBVs were isolated essentially as described, using only the 10%, 25%, and 35% layers on the discontinuous sucrose density gradient (31). The same protocol was used to test the effects of high pH on LBVs except that 100 mM Na2CO3 (pH 11.5) was used to lyse the cells instead.
LC/MS.
Protein samples were solubilized in 1% sodium deoxycholate and 50 mM NH4Cl, heated to 99°C for 5 min, and then reduced, alkylated, digested, and analyzed on a linear trapping quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific) as described (32).
Formaldehyde Labeling.
Formaldehyde isotopologues (33) were used in place of SILAC to quantify the effects of high pH on LBVs and the degree of enrichment of LBV components. Briefly, peptides purified with C18 Stage Tips (34) were resuspended in 5 μl of 200 mM formaldehyde or deuterated formaldehyde (Cambridge Isotope Laboratories) and 0.5 μl of 1 M sodium cyanoborohydride and incubated for 30 min at room temperature away from light. The reaction mixture was then adjusted to pH 7.5, and a further 5 μl of the respective formaldehyde isotopologue plus 1 μl of 1 M cyanoborohydride was added. The reaction was allowed to continue for a further 30 min before being quenched by addition of 6 μl of 2.5 M NH4Cl for 10 min at room temperature.
Data Analysis.
Fragment spectra were extracted as described (32) and searched against the mouse International Protein Index database supplemented with the sequences of all human keratins and abundant bovine serum proteins (v3.25; 52,434 sequences) using Mascot (v2.1; Matrix Science) and allowing only tryptic peptides with up to one missed cleavage. MSQuant (http://msquant.sourceforge.net) was used to parse Mascot result files, to recalibrate mass measurements, and to extract quantitative ratios (SI Table 2). The final list of proteins was generated by using finaList.pl, an in-house script available upon request. A false discovery rate for protein identifications based on two or more peptides (SI Table 1) with a measured mass accuracy <5 ppm (the overall average was 0.56 ppm), a Mascot score ≥26, and length of at least eight residues was estimated to be <0.5% using reversed database searching. For Fig. 4, previously reported phagosomal association curves for Rab proteins with ΔinvA/Inv-containing vacuoles were extracted from the original publication (19) using Photoshop (Adobe Systems) to measure the density of the gray boxes for each time point. Rabbit antibodies against CathA and VAMP4 and mouse antibodies against N-ethylmaleimide-sensitive factor (NSF) were purchased from Abcam.
Supplementary Material
Acknowledgments
We thank Brett Finlay, Nat Brown, Erin Boyle, Carmen de Hoog, and Brian Coombes for helpful discussions and advice. This work was supported by Canadian Institutes of Health Research Operating Grant MOP-77688 (to L.J.F.). L.J.F. is a Michael Smith Foundation Scholar, a Peter Wall Institute for Advanced Studies Early Career Scholar, and the Canada Research Chair in Organelle Proteomics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705801104/DC1.
References
- 1.Cardelli J. Traffic. 2001;2:311–320. doi: 10.1034/j.1600-0854.2001.002005311.x. [DOI] [PubMed] [Google Scholar]
- 2.Underhill DM, Ozinsky A. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 3.Jutras I, Desjardins M. Annu Rev Cell Dev Biol. 2005;21:511–527. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- 4.Vieira OV, Botelho RJ, Grinstein S. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumell JH, Grinstein S. Curr Opin Microbiol. 2004;7:78–84. doi: 10.1016/j.mib.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. Mol Cell Proteomics. 2006;5:2228–2243. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Marion S, Laurent C, Guillen N. Cell Microbiol. 2005;7:1504–1518. doi: 10.1111/j.1462-5822.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- 9.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, et al. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs ME, DeSouza LV, Samaranayake H, Pearlman RE, Siu KW, Klobutcher LA. Eukaryot Cell. 2006;5:1990–2000. doi: 10.1128/EC.00195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnon E, Duclos S, Rondeau C, Chevet E, Cameron PH, Steele-Mortimer O, Paiement J, Bergeron JJ, Desjardins M. Cell. 2002;110:119–131. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- 12.Guermonprez P, Saveanu L, Kleijmeer M, Davoust J, Van Endert P, Amigorena S. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 13.Touret N, Paroutis P, Grinstein S. J Leukocyte Biol. 2005;77:878–885. doi: 10.1189/jlb.1104630. [DOI] [PubMed] [Google Scholar]
- 14.Desjardins M, Huber LA, Parton RG, Griffiths G. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitt A, Mayorga LS, Stahl PD, Schwartz AL. J Clin Invest. 1992;90:1978–1983. doi: 10.1172/JCI116077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullock BM, Smith CW, Ihrke G, Bright NA, Lindsay M, Parkinson EJ, Brooks DA, Parton RG, James DE, Luzio JP, Piper RC. Mol Biol Cell. 2000;11:3137–3153. doi: 10.1091/mbc.11.9.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paumet F, Rahimian V, Rothman JE. Proc Natl Acad Sci USA. 2004;101:3376–3380. doi: 10.1073/pnas.0400271101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerial M, McBride H. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 19.Smith AC, Heo WD, Braun V, Jiang X, Macrae C, Casanova JE, Scidmore MA, Grinstein S, Meyer T, Brumell JH. J Cell Biol. 2007;176:263–268. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayorga LS, Beron W, Sarrouf MN, Colombo MI, Creutz C, Stahl PD. J Biol Chem. 1994;269:30927–30934. [PubMed] [Google Scholar]
- 21.Mayran N, Parton RG, Gruenberg J. EMBO J. 2003;22:3242–3253. doi: 10.1093/emboj/cdg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pittis MG, Muzzolin L, Giulianini PG, Garcia RC. Eur J Cell Biol. 2003;82:9–17. doi: 10.1078/0171-9335-00293. [DOI] [PubMed] [Google Scholar]
- 23.Touret N, Paroutis P, Terebiznik M, Harrison RE, Trombetta S, Pypaert M, Chow A, Jiang A, Shaw J, Yip C, et al. Cell. 2005;123:157–170. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Howell KE, Palade GE. J Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holevinsky KO, Nelson DJ. Biophys J. 1998;75:2577–2586. doi: 10.1016/S0006-3495(98)77703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackam DJ, Rotstein OD, Sjolin C, Schreiber AD, Trimble WS, Grinstein S. Proc Natl Acad Sci USA. 1998;95:11691–11696. doi: 10.1073/pnas.95.20.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahraus A, Tjelle TE, Berg T, Habermann A, Storrie B, Ullrich O, Griffiths G. J Biol Chem. 1998;273:30379–30390. doi: 10.1074/jbc.273.46.30379. [DOI] [PubMed] [Google Scholar]
- 29.Blouin A, Bolender RP, Weibel ER. J Cell Biol. 1977;72:441–455. doi: 10.1083/jcb.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker T, Volchuk A, Rothman JE. Proc Natl Acad Sci USA. 2005;102:4022–4026. doi: 10.1073/pnas.0409219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills SD, Finlay BB. Eur J Cell Biol. 1998;77:35–47. doi: 10.1016/S0171-9335(98)80100-3. [DOI] [PubMed] [Google Scholar]
- 32.Chan QW, Howes CG, Foster LJ. Mol Cell Proteomics. 2006;5:2252–2262. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Hsu JL, Huang SY, Chow NH, Chen SH. Anal Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 34.Rappsilber J, Ishihama Y, Mann M. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.