Abstract
Heart formation requires the coordinated recruitment of multiple cardiac progenitor cell populations derived from both the first and second heart fields. In this study, we have ablated the Bmp receptor 1a and the Wnt effector β-catenin in the developing heart of mice by using MesP1-cre, which acts in early mesoderm progenitors that contribute to both first and second heart fields. Remarkably, the entire cardiac crescent and later the primitive ventricle were absent in MesP1-cre; BmpR1alox/lox mutants. Although myocardial progenitor markers such as Nkx2-5 and Isl1 and the differentiation marker MLC2a were detected in the small, remaining cardiac field in these mutants, the first heart field markers, eHand and Tbx-5, were not expressed. We conclude from these results that Bmp receptor signaling is crucial for the specification of the first heart field. In MesP1-cre; β-cateninlox/lox mutants, cardiac crescent formation, as well as first heart field markers, were not affected, although cardiac looping and right ventricle formation were blocked. Expression of Isl1 and Bmp4 in second heart field progenitors was strongly reduced. In contrast, in a gain-of-function mutation of β-catenin using MesP1-cre, we revealed an expansion of Isl1 and Bmp4 expressing cells, although the heart tube was not formed. We conclude from these results that Wnt/β-catenin signaling regulates second heart-field development, and that a precise amount and/or timing of Wnt/β-catenin signaling is required for proper heart tube formation and cardiac looping.
Keywords: BmpR1a, first heart field, heart looping, mesoderm patterning, second heart field
The vertebrate multichambered heart is the first organ that forms during development (1). The heart forms from two separate progenitor cell lineages, the first heart field (FHF) and the second heart field (SHF), which arise from a common progenitor at gastrulation (2). At embryonic day (E) 6.5, these progenitors express the essential transcription factor MesP1 (mesoderm posterior1; ref. 3), which is the earliest transcription factor required for the movement of mesoderm-derived cardiac precursors to the future heart region. The FHF progenitors form the cardiac crescent, whereas SHF progenitors are found medially to the crescent. The cells of both heart fields then move to the midline, where the FHF progenitors form a linear heart tube that later contributes to the left ventricle. Cells of the SHF proliferate, migrate, and join with cardiomyocytes of the FHF, resulting in the rightward looping of the cardiac tube. Finally, cells of the SHF contribute to the future right ventricle and the inflow and outflow tracts (4–7).
From E7.5 onward, cells of both progenitor heart fields can be distinguished by the expression of specific transcription factors and signaling molecules: FHF progenitors express genes such as Tbx-5, eHand, and Bmp10, whereas SHF progenitors express Isl1, dHand, and Fgfs (4, 5, 8–10). Wnt ligands also act as regulators of early cardiogenesis and are involved in the conversion of the linear heart tube to the multichambered heart (11). Canonical Wnt ligands activate the expression of target genes through a pathway that is controlled by β-catenin (12). Explant cultures in frog and chicken revealed that a zone of low Wnt activity is necessary to initiate cardiogenesis; for this, Wnt ligands secreted from the dorsal neural tube are inhibited by antagonists, Dkk-1, Crescent, and FrzB, which are expressed by the anterior endoderm, the neural tube, and the neural crest (12–15). In contrast, activation of canonical Wnt signaling in pluripotent mouse cells was shown to induce cardiogenic differentiation, whereas Wnt inhibitors blocked cardiogenesis (16). Conventional deletion of β-catenin in mice prevented both anterior-posterior axis formation and subsequent mesoderm induction, resulting in the absence of heart structures (17). Conditional ablation of β-catenin in neural crest cells by using Wnt1-cre affected outflow tract remodelling, whereas ablation of β-catenin in mouse embryonic endoderm by using Ker19-cre caused cell-fate changes from endoderm to precardiac mesoderm, which resulted in ectopic formation of multiple hearts (18, 19). Until now, the controversy of whether activation or repression of canonical Wnt signaling is required during early cardiogenesis has not been resolved. It is also not known how canonical Wnt signals control the heart-specific transcriptional machinery (1, 2).
Bmp signals also participate in the formation of the heart. Bmp ligands activate the expression of target genes through Smad1/5/8 (20). In chicken embryos, Bmp signaling acts early in cardiogenesis and promotes differentiation of FHF and SHF progenitors (6, 21, 22). In the mouse, conventional ablation of the Bmp receptor 1a abolished mesoderm formation at the onset of gastrulation, and thus no heart was formed (23). Conditional deletions of Bmp2, Bmp4, and the Bmp receptor 1a in the heart and the neural crest by using Nkx2-5-cre, MHC-cre, and Wnt1-cre, respectively, caused late effects on cardiogenesis, i.e., defects in trabeculation, septation, and cushion formation (20, 24–26). However, the question of whether Bmp signaling is also required for early cardiogenesis in mice remained open. It is also not known how Bmp signals control the heart-specific transcriptional machinery (1, 20).
Results
To define cells that receive canonical Wnt signals in the developing heart, we used our Axin2lacZ allele in mice (27). Axin2 (Conductin) is a well characterized β-catenin/Tcf target gene. Enhanced Wnt/β-catenin signaling (β-gal expression) can be detected at E8.35 in the SHF progenitors, but not in the cardiomyocytes of the FHF [supporting information (SI) Fig. 6 A–D]. We also assessed the phosphorylation of Smad1/5/8 as readout for Bmp signaling: Phospho-Smad1/5/8 was detected in cells of both the FHF and the SHF (SI Fig. 6 E and F). These results indicate that Bmp signaling is activated in both heart fields, whereas canonical Wnt signaling is activated in the SHF.
Loss of Bmp Signaling Prevents the Formation of the FHF.
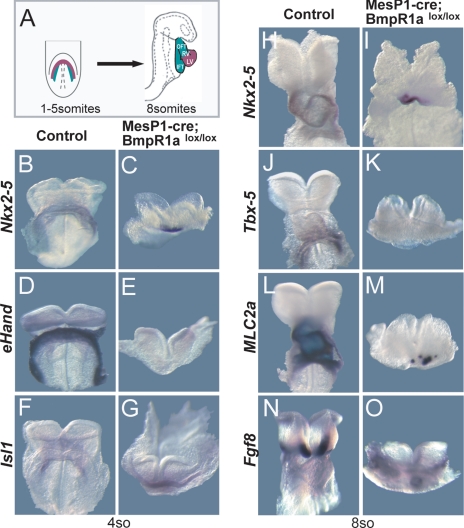
We performed a conditional knockout of the BmpR1a in heart progenitors by using MesP1-cre (28, 29). Strong expression of BmpR1a is observed in the forming cardiac crescent at the zero- to three-somite stages (see SI Fig. 7 A and B), whereas at the six- to eight-somite stage, the expression of the BmpR1a is restricted to the SHF (SI Fig. 7 C and D). MesP1 is the earliest known gene that is expressed in all mesodermally derived structures of the FHF and SHF from E6.5 onward (3, 30, 31). We have assessed cre activity by using the Z/AP indicator mice, and MesP1-cre-mediated recombination of the Z/AP reporter gene was indeed observed in the early developing heart (data not shown and ref. 32). Remarkably, in MesP1-cre; BmpR1alox/lox mutants, the entire cardiac crescent and the entire primitive ventricle were absent, and only a small cardiac field remained (see Fig. 1 A–C, H, and I for the four- and eight-somite stages; Nkx2–5 marks myocardial progenitors and differentiating cardiomyocytes of the FHF and SHF; ref. 33). Moreover, genes expressed in the FHF such as eHand, Gata6, and Tbx-5 (8, 9) were completely absent in MesP1-cre; BmpR1alox/lox mutants, whereas those of the SHF such as Isl1 and Fgf8 (4, 31) were still expressed in the small remaining cardiac field (Fig. 1 D–G, J, K, N, and O; see also SI Fig. 7 I–L and O–P1 and data not shown). We also examined Mef2c, which is expressed in the SHF driven by a specific enhancer (34). Mef2c expression is reduced but not lost in BmpR1a loss-of-function mutants (SI Fig. 7 M and N). At the two-somite stage, Nkx2–5 was initially broadly expressed in the lateral plate mesoderm of the BmpR1alox/lox mutants (SI Fig. 7 E–F1), but it is not properly maintained at later stages. These data indicate that, in the BmpR1a mutants, cardiac precursors are initially formed but that they are not maintained and can therefore not differentiate and form the cardiac crescent. We did not observe a general growth retardation effect in the MesP1-cre; BmpR1alox/lox embryos. For instance, the somite numbers increased as expected from E8.0 to E9.0 (2- to 20-somite stages), although the morphology of the mutant embryos was perturbed. However, we observed expression of the cardiomyocyte marker MLC2a in a small central region at the eight-somite stage (Fig. 1 L and M), but not at the three- to four-somite stage (SI Fig. 7 G and H) (35). This indicates that, whereas FHF derivatives such as the primitive ventricle are absent in the BmpR1alox/lox mutants, some cardiomyocytes of the SHF still formed. These results suggest that Bmp receptor 1a signaling controls the generation of cardiomyocyte progenitors in the FHF.
Fig. 1.
Conditional ablation of the Bmp receptor 1a prevents FHF formation. (A) Scheme of mouse embryos showing FHF (red) and SHF (green) in frontal and side views at crescent and looping stages. (B–O) Frontal views of whole-mount in situ hybridizations for Nkx2-5, eHand, Tbx-5, MLC2a, Fgf8, and Isl1 in controls (B, D, F, H, J, L, and N) and mutants (C, E, G, I, K, M, and O) at crescent (B–G) and looping (H–O) stages. Note that the crescent structure and the expression of FHF genes eHand and Tbx-5 are lost in MesP1-cre; BmpR1alox/lox mice.
Wnt/β-Catenin Signaling Is Required for the Generation of the SHF and for Looping Morphogenesis.
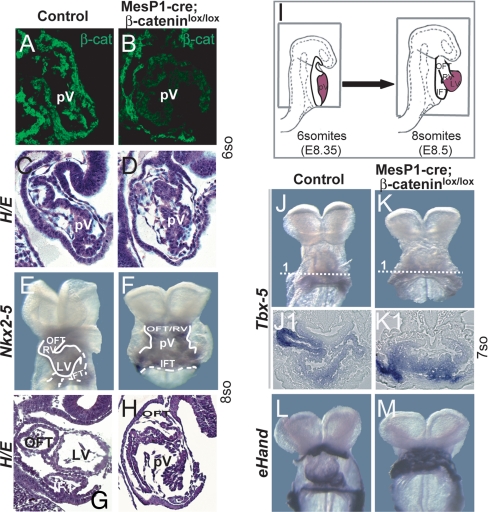
Because Wnt/β-catenin signaling is activated specifically in the SHF, we used our floxed β-catenin mice (36) to conditionally inactivate the β-catenin gene in this tissue, again by using MesP1-Cre mice. The mutant β-catenin allele contains two loxP sites that flank exons 3–6; upon recombination, essential parts of the β-catenin gene are deleted and a null mutation is generated (36). Immunofluorescence analysis of the conditional MesP1-cre; β-cateninlox/lox mutants demonstrated that at the six-somite stage, β-catenin was indeed absent in the primitive ventricle (pV) and SHF (Fig. 2 A and B and SI Fig. 8 A–B1), although heart morphology was not significantly affected at this stage (Fig. 2 C and D). In contrast, cardiac looping was completely disrupted and the outflow tract was shortened at the eight-somite as well as at later stages in the conditional loss-of-function mutations as assessed by staining for Nkx2–5 and MLC2a and by H&E staining (Fig. 2 E–H and SI Fig. 9 C–F1). The left ventricle as well as inflow and outflow tracts were readily seen in the control; only the pV and a shortened outflow tract could be seen in the mutant, as shown in sagital sections of the heart (Fig. 2 G and H; see also the scheme of the sections in Fig. 2I, squares). Thus, the cardiac structures at the eight-somite stage of the mutant grossly resembled those of the six-somite stage in the control (compare Fig. 2C with Fig. 2H). We did not observe MesP1-cre; β-cateninlox/lox mutants in which cardiogenesis proceeded beyond this stage. There was no general growth retardation effect in the MesP1-cre; β-cateninlox/lox embryos, because the somite numbers increased from E8.0 to E9.75. Whole-mount in situ hybridizations and transverse sections demonstrated that markers of the FHF such as Tbx-5 and Gata4 were normally expressed in MesP1-cre; β-cateninlox/lox mice, whereas eHand was expressed throughout the primitive ventricle (Fig. 2 J–M and SI Fig. 9 A and B), indicating the formation of atria and left ventricle but the reduction of right ventricle.
Fig. 2.
Conditional loss-of-function mutation of β-catenin disrupts cardiac looping. (A and B) Immunofluorescence for β-catenin shows loss of β-catenin in the myocard and unaltered expression in the body wall that overlies the pericardial cavity in MesP1-cre; β-cateninlox/lox mice (B, compare with controls in A). (C and D) H&E staining of sagittal sections of the heart tubes at the six-somite stage. (E and F) Whole-mount in situ hybridization for Nkx2-5 at the eight-somite stage. (G and H) H&E staining of sagittal sections of the heart at the eight-somite stage. Note the loss of cardiac looping in MesP1-cre; β-cateninlox/lox mice at the eight-somite stages, while the inflow tract (IFT) and a shortened outflow tract (OFT)/right ventricle (RV) are formed. (I) Scheme of mouse embryos highlight the FHF in red and the sagital section planes (squares) at the six- and eight-somite stages. (J–M) Whole-mount in situ hybridizations for FHF genes show normal expression of Tbx-5 in the atria and expanded expression of eHand in the primitive ventricle (pV) in MesP1-cre; β-cateninlox/lox mice. LV, left ventricle.
We next examined the expression of SHF markers in MesP1-cre; β-cateninlox/lox mutants (Fig. 3). Isl1 marks cardiac precursors of the SHF (4) and was indeed strongly reduced at both the anterior and posterior regions in MesP1-cre; β-cateninlox/lox mice, in particular in the outflow tract and the splanchnic mesoderm (Fig. 3 B and C; see also transverse sections in Fig. 3 B1–B4 and C1–C4; note arrowheads and arrows in B2 and C2). Transverse sections showed that Isl1 is also expressed in the foregut endoderm of control embryos and that it is not reduced in the β-catenin loss-of-function mutants (SI Fig. 8 G–I). Wnt11 has been shown to be expressed in the myocardium of the outflow tract (37), and Wnt11 was expressed in this region in both control and mutant embryos, although the outflow tract in the mutant was shortened (Fig. 3 D and E). The expression of dHand, Bmp7, Fgf8, and Fgf10 was not changed in the SHF of MesP1-cre; β-cateninlox/lox mice (Fig. 3 F–K; sections showing dHand expression are in Fig. 3 F1–G2; see also SI Fig. 9 G and H; refs. 5, 10, and 31). Moreover, Bmp2 expression appeared elevated in the remaining outflow tract myocardium in MesP1-cre; β-cateninlox/lox mice (SI Fig. 9 K–L1; ref. 21). Together, these data show that β-catenin signaling is required for the proper generation of Isl1+ progenitor cells in the SHF.
Fig. 3.
Conditional loss-of-function of β-catenin affects SHF formation. (A) Scheme of mouse embryo indicates SHF in green with a transverse section plane at E8.5. (B–K) Whole-mount in situ hybridization for Isl1, dHand, Fgf8, and Fgf10 in controls (B, F, J, and L) and MesP1-cre; β-cateninlox/lox embryos (C, G, I, and K) at the eight-somite stage and for Wnt11 (compare control in D with mutant in E) at the 10-somite stage. (B1–C4 and F1–G2) Transverse sections in four section planes (indicated with dotted lines in B and F). (B1 and C1) Pharyngeal arch. (B2, C2, F1, and G1) Outflow tract. (B3, C3, F2, and G2) Splanchnic mesoderm, right and left ventricle. (B4 and C4) Inflow tract. Arrows and arrowheads in whole mounts and sections of Isl1 in situ hybridizations indicate the reduced expression in splanchnic mesoderm and outflow tract.
MesP1-Induced Gain-of-Function Mutation of β-Catenin Prevents Heart Tube Formation.
We next characterized MesP1-cre; β-cateninloxEx3/+ mice that carry a conditional gain-of-function mutation of β-catenin (Fig. 4; refs. 29 and 38). Recombination of the β-cateninloxEx3 allele leads to the expression of a constitutively active β-catenin that lacks N-terminal phosphorylation sites essential for its degradation (38). Immunofluorescence analyses demonstrated that β-catenin was located nuclearly in cardiac mesoderm of FHF and SHF at the five-somite stage in MesP1-cre; β-cateninloxEx3/+ mutants (SI Fig. 8 A1 and C1). The cardiac crescent in MesP1-cre; β-cateninloxEx3/+ mutants was initially formed normally, as shown by in situ hybridization for Nkx2-5, Gata4, Tbx-5, and eHand (Fig. 4 A–D and SI Fig. 8 J–O). However, the expression of Isl1 appears enhanced and broadened at both crescent and looping stages (Fig. 4 E, F, M, and N and SI Fig. 8 G–I). There was no general growth retardation effect in the MesP1-cre; β-cateninloxEx3/+ embryos, because the somite numbers increased from E8.0 to E8.75. Remarkably, the formation of a linear heart tube was severely disrupted in the gain-of-function β-catenin mutants (Fig. 4 G–N). Instead, two separate clusters of cardiomyocytes were formed laterally, suggesting a defect in migration of FHF and SHF progenitors as indicated by in situ hybridization for MLC2a or Mef2c (Fig. 4 G, H, K, and L).
Fig. 4.
Sustained activation of β-catenin signaling inhibits heart tube formation. (A–F) Whole- mount in situ hybridizations for the indicated genes in controls (A, A1, C, E, G, I, K, and M) and in MesP1-cre; β-cateninloxEx3/+ embryos (B, B1, D, F, H, J, L, and N) at the four- to five-somite stage and the eight-somite stage. Note that Isl1 expression is increased in mutants, and heart tube formation is abolished in MesP1-cre; β-cateninloxEx3/+ mice at the eight-somite stage.
Bmp4 Is a Target of Wnt/β-Catenin Signaling in the SHF.
The expression of Bmp4 in the SHF has been previously reported in both mouse and chicken embryos (6, 25). In our study, we found a complete absence of Bmp4 expression in the heart region of conditional β-catenin loss-of-function mutant mice. In contrast, a strong up-regulation of Bmp4 could be observed in the gain-of-function mutation at the crescent and heart-tube stages (Fig. 5 A–C and SI Figs. 8 D–F and 9 I and J). In situ hybridization for Axin2, which indicates the strength of canonical Wnt signaling (27, 39), revealed a strong up-regulation in the cardiac crescent of the gain-of-function mutation, overlapping with the pattern of Bmp4 expression (compare Fig. 5 D–F with Fig. 5 A–C). Furthermore, we assessed whether strong Wnt/β-catenin signaling affects proliferation and apoptosis in FHF and SHF by scoring for phospho-histone H3 (pHH3) and TUNEL staining in Isl1+ and Isl1− heart cells at the four- to six- and eight-somite stages. In the loss-of-function mutants, a 41% reduction of SHF proliferation was observed at the four- to six-somite stage, but not at the eight-somite stage (P = 0.048; see Fig. 5G and SI Fig. 10). However, in the gain-of-function mutants, SHF proliferation was unaffected, whereas the proliferation rates of FHF were elevated by 2.1-fold (P = 0.001). Moreover, we were able to demonstrate that the increase in SHF proliferation correlated to an increase in the number of Isl1+ cells (33.5%, P = 0.01) from four- to six-somite stages to eight-somite stages in controls, whereas the MesP1-cre; β-cateninlox/lox mutants' Isl1+ cell count remained unaffected (Fig. 5I). Importantly, the difference in SHF proliferation did not correlate with apoptotic defects in loss-of-function embryos; apoptotic rates were unaffected, whereas apoptosis in the SHF of gain-of-function embryos was induced (2.7-fold; P = 0.007) as shown by TUNEL analysis (Fig. 5H). These findings suggest that Wnt/β-catenin signaling is required to promote proliferation of Isl1+ cells. However, direct transcription of Isl1 was not affected. The data therefore suggest that Bmp4 rather than Isl1 is a target of β-catenin signaling in the SHF.
Fig. 5.
Bmp and Wnt signaling regulate region-specific heart morphogenesis. (A–F) Whole-mount in situ hybridization for Bmp4 and Axin2 in controls (A and D) and MesP1-cre; β-cateninlox/lox (B and E) and MesP1-cre; β-cateninloxEx3/+ (C and F) mice at the five-somite stage. Note that the expression of Bmp4 and Axin2 is lost in the cardiac crescent of MesP1-cre; β-cateninlox/lox mice, whereas Bmp4 and Axin2 are overexpressed in MesP1-cre; β-cateninloxEx3/+ cardiac crescents. (G and H) Quantification of proliferation and apoptosis of controls (solid light-colored bars) and MesP1-cre; β-catenin loss-of-function (striped dark-colored bars) and gain-of-function (dotted medium-colored bars) mutants, as assessed by proportion of phospho-histone H3 (pHH3)- and TUNEL-positive nuclei in FHF (red) and SHF (green). Significant statistical differences between control and mutants are indicated as follows: *, P < 0.05; **, P ≤ 0.01. (I) Average over the total Isl1+ cell numbers per section at the four- to six-somite stage and the eight-somite stage. (J) (Left) schemes of mouse embryos show the location and contribution of FHF (red) and SHF (green) cells at crescent (frontal view), heart tube, and looping stages (lateral views). The time points are indicated at which Bmp receptor 1a and Wnt/β-catenin signaling function in heart development. Bmp receptor 1a signaling (red) affects the FHF (red) (a role in SHF development could not be examined), whereas β-catenin signaling (green) affects the SHF (green). (Right) the magnification of the looped heart shows how β-catenin or Bmp receptor 1a signaling might affect heart-specific gene expression. The scheme also indicates where these factors are expressed in the FHF (red) and SHF (green). [Scheme reproduced and modified with permission from ref. 1 (Copyright 2006, Cell) and ref. 2 (Copyright 2005, Nat Rev Genet).]
Discussion
We conditionally ablated the Bmp receptor 1a and the canonical Wnt effector β-catenin in the developing heart by using MesP1-cre, which is already functional in mesenchymal cardiac precursors that contribute to cells of the FHF and SHF. Ablation of the BmpR1a by MesP1-cre resulted in the complete absence of FHF specification. In contrast, loss-of-function mutations of β-catenin induced by MesP1-cre affected specifically the SHF, but not the FHF.
Conditional ablation of the Bmp receptor 1a in cardiac precursors resulted in the absence of the cardiac crescent and the loss of the expression of FHF specific genes such as Gata6, Tbx-5, and eHand (see scheme in Fig. 5J). Apparently, Bmp signaling is crucial in the mesodermal precursors and/or the early FHF cells, which is a previously undescribed finding in heart morphogenesis of mice. We also show that Nkx2-5, a gene essential for cardiac precursor formation and differentiation (33), is initially present in the forming crescent but is strongly reduced at later stages. This phenomenon results in a block of early cardiac differentiation in the BmpR1a mutant, as has been shown in Xenopus (40). Overexpression of Bmp2 and implantation of Noggin-expressing cells in chicken embryos had previously been shown to induce and prevent early heart generation, respectively, but FHF and SHF were not distinguished at that time (21). It is known that Nkx2-5 controls the expression of the essential transcription factor eHand in FHF cells (2). However, Nkx2-5 is also expressed in differentiating cardiomyocytes of the SHF (1, 2), and the minute amounts of residual Nkx2-5 in the BmpR1a mutants at later stages might indeed be expressed in this SHF. Together, our data suggest that, in the absence of BmpR1a signaling, heart progenitor cells fail to progress toward the heart-specific lineages, i.e., myocardial progenitor cell markers are expressed and myocardial differentiation is compromised, with only a few myocardial cells appearing later and thus no heart is formed. In a recent publication, Smad1 had been shown to be required at later stages of cardiogenesis in mice (41). We show that loss of BmpR1a results in an earlier and stronger phenotype. This discrepancy may be explained by compensation by Smad5 in the Smad1 loss-of-function mutants (41). Because defects in FHF specification were so dominant in BmpR1a mutants, a role of Bmp receptor signaling in SHF development could not be examined in this mutant.
In contrast, conditional ablation of β-catenin by MesP1-cre resulted in absence of cardiac looping, reduced right ventricle, and a lack of Isl1+ progenitor cells in the SHF (Fig. 5J). It had previously been shown that Isl1+ precursors of the heart are particularly important for outflow tract and right ventricle formation and that the atrial myocardium seemed to be reduced in the absence of Isl1 (4). Loss-of-function β-catenin prevented Bmp4 expression in SHF cells, which acts upstream of Isl1 in the chicken and controls outflow tract septation in mice (6, 20, 25). Along this same line, gain-of-function mutation of β-catenin resulted in an expansion of Isl1+ progenitors and in the strong promotion of Bmp4 expression. The expression of other essential genes for the generation of the SHF was not changed, indicating that their expression is independent of β-catenin function. It is known that the generation of SHF progenitors requires cell proliferation, cell migration, and differentiation, which are essential processes for cardiac looping and right ventricle formation (2, 4). Our cell proliferation analyses in control and mutant embryos suggest that β-catenin signaling is required for SHF proliferation at crescent stages, which mainly resulted in the reduction of Isl1-expressing cells at later stages. Additionally, lack of dispersion of the progenitors into the right ventricle and the formation of two separate fields of differentiated cardiomyocytes were observed, suggesting that cell migration and/or differentiation are defective in our β-catenin mutant hearts. The expression of Bmp2 in the outflow tract myocardium also appeared elevated in loss-of-function β-catenin mice, which is in agreement with recently published work, showing that Bmp2 via Smad1 signaling is required for suppression of proliferation (41).
Recent work has identified several steps in which Wnt activation and Wnt inhibition are required in early embryogenesis (12). Canonical and noncanonical Wnt signaling are essential for the formation of embryonic axes and mesoderm formation and patterning (17, 42, 43). In contrast, inhibition of canonical Wnt signaling is required for head formation (44, 45). Both, canonical and noncanonical Wnt signaling are also crucial for heart formation (11). Experiments with explant cultures from chicken and frog embryos indicated that repression of canonical Wnt signaling is required for early cardiogenesis (13–15, 46). Experiments in mice were not conclusive because (i) in pluripotent mouse cells, Wnt inhibitors blocked the differentiation of cardiomyocytes (16, 47); (ii) conditional ablation of β-catenin by using cre that acted later caused defects in outflow tract remodelling (18); and (iii) several canonical Wnts are expressed in the precardiac mesoderm, in the outflow and inflow tracts of the linear heart tube, or within the developing myocardium. These data indicated that some Wnt signaling may be required for proper heart formation (11). With the MesP1-cre, we removed competence to respond to Wnt signaling before specification of FHF and SHF progenitors. However, we did not observe defects in FHF development, indicating that Wnt signaling is not required at this step. In contrast, SHF specification was severely disturbed, resulting in a lack of cardiac looping and proper outflow tract and right ventricle formation. It was recently shown that defects in right ventricle and outflow tract formation are also observed when competence to Wnt signaling was removed during and after SHF specification (48, 49). Our results using both loss- and gain-of-function mutations of β-catenin suggest that it may be the amount of Wnt/β-catenin signaling that is important for early cardiogenesis. High levels of functional β-catenin in precardiac mesoderm prevented the fusion of cardiac crescent cells to the linear heart tube, whereas loss of β-catenin averted cardiac looping. However, as fusion of cardiac crescent cells and cardiac looping are temporally separate events, we might also consider that the timing of Wnt signaling is important.
The question arises of how the two developmental signaling pathways, Bmp and canonical Wnt signaling, crosstalk with the transcriptional networks that had been established to be essential for heart formation (Fig. 5J; ref. 2). Previous studies have shown that a positive role of Bmp signaling in the induction of cardiogenesis is conserved between species, mouse, chicken, frog, and fly, whereas the function of canonical Wnt signaling in heart induction has been discussed controversially (13–15, 18, 19, 50). Our results suggest that the Bmp receptor 1a signaling in the FHF directly or indirectly controls the expression of the transcription factors Gata6, Tbx-5, and Nkx2-5, which were previously shown to regulate the expression of eHand (Fig. 5J; ref. 2). Our observations agree with results on the early function of Bmp signaling in chicken cardiogenesis (6, 21, 22). In chicken embryos, Bmp2 that is located medially to the heart-forming region induced the expression of Nkx2-5 and Gata4/5/6, whereas repression of Bmp signaling by Noggin-expressing cells, which are implanted in precardiac mesoderm, abolished Nkx2-5 and eHand expression (21). The requirement of the Wnt/β-catenin pathway for Isl1+ SHF progenitors is consistent with its effect on Bmp4 expression; Bmp4 has been shown to play a major role during SHF specification upstream of Isl1 (6). In chicken embryos, Bmps were also shown to be required for the incorporation of cells into the outflow tract myocardium (7). Our data show that canonical Wnt signaling regulates specifically Bmp4, which is required for proliferation of Isl1+ cells of the SHF and for recruitment of these cells into the forming outflow tract. These findings could be the explanation for the observed heart defects in loss-of-function β-catenin mutants. Studies in the fly have shown that wingless, the Wnt1 ortholog, collaborates with the Bmp homologue, decapentaplegic (dpp), to specify the formation of the dorsal vessel, which is reminiscent of the linear heart tube in vertebrates (50, 51). Both, modulation of Wnt and Bmp signaling were shown to be required for cardiogenesis in chicken, but how and where these pathways can be linked remained unresolved (13). Thus, we have shown a previously undescribed connection of Bmp and canonical Wnt signaling in SHF specification in mice. The further unraveling of the intricate interplay of the developmental signaling pathways with the heart-specific transcription factor network requires the analysis of further mutant mice and, in particular, compound mutant mice.
Materials and Methods
Mouse Strains.
The different mouse strains, β-cateninlox, β-cateninloxEx3, BmpR1alox, Axin2LacZ, and MesP1-cre mice, the conditions for breading, and the genotyping primers were described in refs. 3, 27, 28, 36, and 38. Mutant embryos were identified by PCR using amnion or yolk sac.
In Situ Hybridization and Histological Techniques.
Embryos were prepared in cold PBS, age-matched by their somite numbers, and fixed in 4% formaldehyde. Immunofluorescence and H&E analyses were performed on 8 μm of paraffin, 14 μm of cryo, or 6 μm of plastic sections as described in ref. 17. The following primary antibodies were used: guinea pig anti-Isl1/2 (kindly provided by T. Jessell, Columbia University, New York, NY) and rabbit-anti-phospho-histone H3 (Upstate, Lake Placid, NY). Whole-mount in situ hybridization was performed using digoxygenin-labeled (DIG) RNA probes (Roche, Indianapolis, IN) (17), followed by plastic embedding and sectioning. The antisense transcripts of mouse cDNAs were as follows: Axin2 (BC057338); Bmp4 (X56848.1); Fgf8, dHand, eHand, and Tbx5 (kindly provided by M. Blum, University of Hohenheim, Stuttgart–Hohenheim, Germany); Fgf10 (NM_008002.3); Gata4 (NM_008092); Isl1 (NM_021459.2); Mef2c (BC026841.1); MLC2a (35); Nkx2–5 (AF091351.1) and Wnt11 (NM_009519.1).
For more information, also see SI Materials and Methods.
Proliferation, Apoptosis Assays, and Statistical Analysis.
Proliferation and apoptosis were assessed by immunostaining for phospho-histone H3(pHH3) and TUNEL (Chemicon, Temecula, CA). Three to six heart sections each of three embryos of each genotype and stage were used for quantification. Results are shown as means ± SD. Groups were compared by one-way ANOVA followed by a Bonferroni test, with P < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank R. R. Behringer (M. D. Anderson Cancer Center, Houston, TX) for kindly providing BmpR1alox/lox mice; M. Blum, (University of Hohenheim) for advice at an early stage of this work; C. Eichhorn, (Max Delbrueck Center) for statistical analysis; and T. Redmer and M. Rosario, (both at Max Delbrueck Center) for help in the course of the work. This work was supported in part by German Research Foundation Grant SFB366.
Abbreviations
- En
embryonic day n
- FHF
first heart field
- SHF
second heart field.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703113104/DC1.
References
- 1.Srivastava D. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M, Meilhac S, Zaffran S. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 3.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. Development (Cambridge, UK) 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 4.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly RG, Brown NA, Buckingham ME. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 6.Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Development (Cambridge, UK) 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- 7.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Development (Cambridge, UK) 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 8.Biben C, Harvey RP. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 9.Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Dev Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg LM, Eisenberg CA. Dev Biol. 2006;293:305–315. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Huelsken J, Birchmeier W. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 13.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider VA, Mercola M. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzahor E, Lassar AB. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura T, Sano M, Songyang Z, Schneider MD. Proc Natl Acad Sci USA. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. J Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 19.Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 20.van Wijk B, Moorman AF, van den Hoff MJ. Cardiovasc Res. 2006;72:244–255. doi: 10.1016/j.cardiores.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Schlange T, Andree B, Arnold HH, Brand T. Mech Dev. 2000;91:259–270. doi: 10.1016/s0925-4773(99)00311-1. [DOI] [PubMed] [Google Scholar]
- 22.Schultheiss TM, Burch JB, Lassar AB. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 23.Mishina Y, Suzuki A, Ueno N, Behringer RR. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 24.Gaussin V, Van de PT, Mishina Y, Hanks MC, Zwijsen A, Huylebroeck D, Behringer RR, Schneider MD. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Proc Natl Acad Sci USA. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. Development (Cambridge, UK) 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de WM, Clevers H, Schlag PM, Birchmeier W, et al. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Genesis. 2002;32:69–72. doi: 10.1002/gene.10038. [DOI] [PubMed] [Google Scholar]
- 29.Soshnikova N, Zechner D, Huelsken J, Mishina Y, Behringer RR, Taketo MM, Crenshaw EB, III, Birchmeier W. Genes Dev. 2003;17:1963–1968. doi: 10.1101/gad.263003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Development (Cambridge, UK) 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 31.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Development (Cambridge, UK) 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 33.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 34.Lin Q, Schwarz J, Bucana C, Olson EN. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubalak SW, Miller-Hance WC, O'Brien TX, Dyson E, Chien KR. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- 36.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 37.Pandur P, Lasche M, Eisenberg LM, Kuhl M. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 38.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Katsev S, Cai C, Evans S. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 41.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawakami Y, Capdevila J, Buscher D, Itoh T, Rodriguez EC, Izpisua Belmonte JC. Cell. 2001;104:891–900. doi: 10.1016/s0092-8674(01)00285-9. [DOI] [PubMed] [Google Scholar]
- 43.Moon RT, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 44.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 45.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 46.Foley AC, Mercola M. Genes Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF. Proc Natl Acad Sci USA. 2007;104:9319–9324. doi: 10.1073/pnas.0701212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, et al. Proc Natl Acad Sci USA. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park M, Wu X, Golden K, Axelrod JD, Bodmer R. Dev Biol. 1996;177:104–116. doi: 10.1006/dbio.1996.0149. [DOI] [PubMed] [Google Scholar]
- 51.Frasch M. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.