Abstract
HIV-1 group M subtype B was the first HIV discovered and is the predominant variant of AIDS virus in most countries outside of sub-Saharan Africa. However, the circumstances of its origin and emergence remain unresolved. Here we propose a geographic sequence and time line for the origin of subtype B and the emergence of pandemic HIV/AIDS out of Africa. Using HIV-1 gene sequences recovered from archival samples from some of the earliest known Haitian AIDS patients, we find that subtype B likely moved from Africa to Haiti in or around 1966 (1962–1970) and then spread there for some years before successfully dispersing elsewhere. A “pandemic” clade, encompassing the vast majority of non-Haitian subtype B infections in the United States and elsewhere around the world, subsequently emerged after a single migration of the virus out of Haiti in or around 1969 (1966–1972). Haiti appears to have the oldest HIV/AIDS epidemic outside sub-Saharan Africa and the most genetically diverse subtype B epidemic, which might present challenges for HIV-1 vaccine design and testing. The emergence of the pandemic variant of subtype B was an important turning point in the history of AIDS, but its spread was likely driven by ecological rather than evolutionary factors. Our results suggest that HIV-1 circulated cryptically in the United States for ≈12 years before the recognition of AIDS in 1981.
Keywords: evolution, pandemic, phylogeny, archival, Haiti
Viral gene trees can deliver powerful insights into ecological and evolutionary processes (1). Population-level phylogenetic patterns reflect both transmission dynamics and genetic change, which in turn can accumulate because of selection (driven, for example, by host immunity) or drift. In this study, we use a phylogenetic approach and HIV-1 gene sequences recovered from early victims of AIDS to investigate when, where, and how HIV-1 emerged from Africa and spread worldwide. Although it accounts for fewer infections than subtype C, which dominates the HIV-1 epidemics in southern Africa and India and is spreading elsewhere (2), HIV-1 group M subtype B is arguably the most widespread HIV variant. No other subtype or circulating recombinant form predominates in as many countries around the world (3).
Our aim here is to combine phylogenetic, molecular evolutionary, historical, and epidemiological perspectives in an attempt to reconstruct the history of the subtype B pandemic. Such retrospective knowledge can clarify the past but also potentially can be of value for rational vaccine design that takes into account the genetic diversity of the virus (4) and for predicting the future complexity of regional and global HIV-1 genetic diversity. This is a function of how frequently HIV-1 strains disperse to, then successfully colonize, new geographic ranges and host populations, a question we address here.
The idea that Haiti might have played a special role in the unfolding of the AIDS pandemic predates the discovery of HIV. Soon after the initial recognition of AIDS (5), evidence of a high prevalence of the syndrome among Haitian immigrants in the United States (6) helped fuel speculation that Haiti may have been the source of the mysterious newly identified syndrome (7). It has since become clear that the causative agent, HIV-1 group M, actually originated not in Haiti but in central Africa, apparently sometime around 1930 (8, 9).
Nevertheless, the possibility remains that Haiti was the stepping-stone for the emergence of the exceptionally widespread subtype B lineage, and this idea has implications that extend beyond historical interest. Some researchers have noted that Haitian HIV-1 sequences tend to occupy basal positions on the subtype B phylogeny, suggestive of the epidemic originating there (9–11). Others argue vigorously that the Haitian HIV/AIDS epidemic was seeded from the United States, perhaps after Haiti became a popular sex tourism destination in the mid-1970s (12–14). However, these competing hypotheses have never been rigorously tested, despite their importance for understanding the global spread and vaccine-relevant genetic diversity of HIV-1.
To test these hypotheses, we recovered complete HIV-1 env and partial gag gene sequences from archival specimens collected in 1982–1983 from five Haitian AIDS patients, all of whom had recently immigrated to the United States and were among the first recognized AIDS victims (6). Being independent of and much older than the few previously published Haitian HIV-1 full-length env strains, these archival sequences offer a unique opportunity for resolving the origin and emergence of subtype B. They provide direct insight into Haitian HIV-1 genetic diversity at an exceptionally early time point and an unbiased sample for testing the a priori specified phylogenetic hypotheses addressed here.
Results and Discussion
The Geographical Origin of Subtype B.
Under the “Haiti-first” model, non-Haitian subtype B strains are expected to be phylogenetically nested within an older and hence more extensive range of Haitian genetic variation, with Haitian lineages branching off closest to the B subtype ancestor. To test whether it is this or an alternative pattern that characterizes this HIV-1 subtype [supporting information (SI) Fig. 3], we conducted a detailed Bayesian Markov chain Monte Carlo (MCMC) phylogenetic analysis using an alignment of the Haitian archival sequences plus 117 previously published subtype B env sequences from a total of 19 countries. Five African strains of subtype D (the closest relative to subtype B) served as the outgroup.
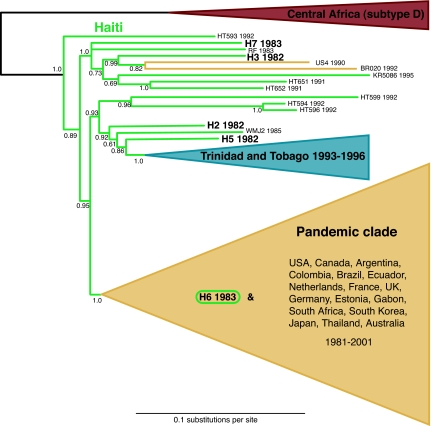
On the env gene phylogeny, the archival sequences occupy basal positions within subtype B (Fig. 1 and SI Fig. 4); there is extremely strong statistical support for a Haitian origin of subtype B, in that the probability of a U.S. or other non-Haitian origin [i.e., the posterior probability of any non-Haitian sequence(s) occupying the most basal position in the B clade] is <0.001. These results indicate, with high statistical significance, that subtype B arrived and began spreading in Haiti before it spread elsewhere and was not originally introduced to Haiti from the United States. In addition, the unequivocal support for the monophyly of subtype B as a whole (P = 1.0) supports the previous suggestion of a single (epidemically successful) introduction of this subtype from central Africa to Haiti (10).
Fig. 1.
The abridged majority-rule consensus tree summarizing the results from the MrBayes analysis of complete env genes. The branch lengths represent the mean value observed for that branch among the postburnin sampled trees. Posterior probabilities are indicated for each node. As expected under a “Haiti-first” model, the non-Haitian subtype B strains are phylogenetically nested within an older and more diverse range of Haitian viral variants. The 11 sequences of the Trinidad and Tobago clade and the 96 sequences of the pandemic clade are schematically represented by the blue and yellow triangles, respectively. Haitian or Haitian-linked sequences are shown in green, with the archival sequences labeled in larger bold text. The unabridged tree is available as SI Fig. 4.
Drummond et al. (15) found that relaxed-clock models are more accurate and precise at estimating phylogenetic relationships than unrooted methods. In other words, when data have evolved in a somewhat clock-like fashion, incorporating knowledge of the tempo of evolution can make topology estimation more reliable compared with methods that ignore this information. Therefore, we were also interested in what the relaxed-clock results (described below) revealed with respect to where subtype B originated (i.e., the topological relationships among subtype B sequences, regardless of timing/branch lengths). Under the relaxed-clock model, the posterior probability of a U.S. origin was 0.00003, and the probability of a Haitian origin was 0.9979.
The inference that subtype B reached Haiti before spreading to other countries does not depend on a dating analysis. One of the advantages of a Bayesian statistical framework is that it yields direct estimates of the probability of phylogenetic hypotheses, and in this case there is strong evidence to reject a U.S. or other non-Haitian origin of subtype B. This means that even if there is some uncertainty regarding precisely when HIV-1 entered Haiti or the United States (see below), there is little doubt about the sequence of events; the clear-cut topological information implies that the entry to Haiti occurred first. Moreover, our sampling bias in favor of non-Haitian subtype B makes the “Haiti-first” inference conservative; the Haitian strains occupy the basal positions within subtype B even though there were many more opportunities for recovering non-Haitian basal strains, if they existed.
The Dispersal of HIV-1 Out of Haiti.
We next investigated how many times the older Haitian HIV-1 epidemic has seeded detectable secondary outbreaks elsewhere. We found evidence for only three such events, despite the fact that our data set comprised 109 non-Haitian subtype B strains from the Caribbean, North and South America, Europe, Africa, Asia, and Australia. One instance is confined to Trinidad and Tobago (Fig. 1). All of the HIV-1 sequences from this country form a distinct monophyletic grouping with the strongest possible support (posterior probability of 1.0). Previous studies have asserted homosexual/bisexual contact with North American foreigners in the late 1970s or early 1980s as the alleged route of introduction (16, 17). Our results indicate this was not the case; the Trinidad and Tobago clade is unequivocally nested among Haitian strains, not North American ones, clear evidence that the predominantly heterosexual epidemic in this country can be explained by a single introduction linked to Haiti.
The next, and most important, secondary epidemic accounts for all but three of the remaining non-Haitian strains, encompassing 96 sequences representing every other country in our data set (Fig. 1). This “pandemic” clade forms another unequivocally monophyletic cluster (P = 1.0) nested within the basal Haitian strains. As for the Trinidad and Tobago clade, the most parsimonious explanation for this pattern is that all these subtype B infections from across the world emanated from a single founder event linked to Haiti. This most likely occurred when the ancestral pandemic clade virus crossed from the Haitian community in the United States to the non-Haitian population there.
The only other Haiti-linked outbreak we detected is comparatively insignificant, here comprising a single Brazilian sequence (BR020) and one American one (US4). Additional rare chains of transmission may have emerged from Haiti but remained undetectable in this study. Likewise, pandemic clade viruses may have reentered Haiti but were undetectable. Regardless, it is evident from this large international sample that the subtype B epidemics in most afflicted countries and the bulk of subtype B infections worldwide are caused by viruses belonging only to the pandemic clade.
Three additional nominally non-Haitian strains are noteworthy for falling among the basal Haitian lineages rather than within the three non-Haitian clades (Fig. 1). Each one provides further support for the notion that strains linked to Haiti occupy the deepest branches within subtype B. RF was sampled in 1983 from a Haitian immigrant to the United States. Like our five newly sequenced strains, it represents a Haitian strain that entered the United States via an immigrant host (10). WMJ2 was sampled in 1985 from a perinatally infected infant born to an HIV-positive Haitian immigrant mother (18). KR5086 was recovered in 1995 from a South Korean sailor infected in the Dominican Republic (19), a country directly linked to Haiti in terms of both geography and HIV-1 epidemiology (20). These cases, plus the additional ones considered in our study, show that the virus moved out of Haiti on many separate occasions but did so mostly as dead-end infections that evidently failed to ignite successful epidemics. A key question for future research will be to determine why Haiti-to-U.S. epidemic outbreaks have apparently become established so rarely since the initial introduction in or around 1969, despite presumably frequent movement of the virus because of rising HIV-1 prevalence, continued migration, and the once-thriving sex tourism industry linking Haiti and the United States.
Several additional analyses corroborated these findings. First, we reanalyzed our env gene data by replacing the African subtype D outgroup sequences with a variety of alternatives from different subtypes and geographical regions. This had no impact on the basal position of the Haitian strains within subtype B (SI Fig. 5a); their ancestral position cannot be dismissed as an artifact of convergent evolution with African subtype D sequences. Second, the gag sequences also unambiguously place the Haitian sequences in the most ancestral positions within subtype B (P = 0.9930) (SI Fig. 5b). We also inspected the nucleotide substitutions that mapped specifically onto the branch leading to the pandemic clade. For env, all eight such changes were silent at the amino acid level. For gag, only one of the six changes on the relevant branch indicated a change in amino acid between the Haitian strains and the pandemic clade strains. The paucity of amino acid substitutions along this clade-defining branch suggests that the ancestor of the pandemic clade probably possessed no selective advantage over other Haitian strains; its remarkable epidemic success may simply reflect ecological factors rather than evolutionary ones (chance colonization of a new population, as opposed to competitively superior transmission fitness). However, analysis of complete genomes would be necessary to definitively rule out selection.
The Timing of the Emergence of Subtype B.
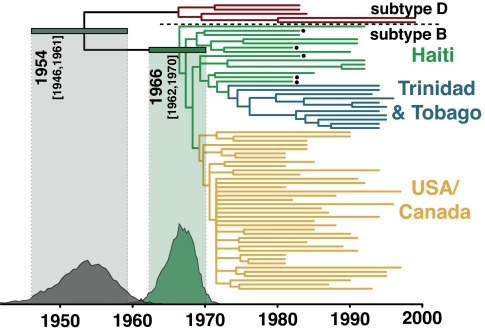
We used a second Bayesian MCMC method that simultaneously estimates phylogenetic relationships and times of most recent common ancestors (15) to perform a supplementary phylogenetic analysis on a reduced data set. This method uses a “relaxed molecular clock” model, so-called because it relaxes the need to assume a constant rate of molecular evolution across the tree to obtain date estimates from gene sequences. We estimated the time of the most recent common ancestor (TMRCA) of subtype B at 1966 (1962–1970) (Fig. 2), a date that suggests its arrival in Haiti may have occurred with the return of one of the many Haitian professionals who worked in the newly independent Congo in the 1960s (21).
Fig. 2.
The consensus tree of the relaxed molecular clock analysis, with the Haitian archival sequences bulleted. The tips of the tree correspond to year of sampling, and the branch lengths reflect the mean of the posterior probability density. The posterior probability density for the TMRCA for subtype B in Haiti is depicted in dark green, and the 95% highest probability density (HPD) is shown by the horizontal bar and light-green shading. The TMRCA means and 95% HPDs for the other key nodes were as follows: subtype B/D ancestor = 1954 (1946–1961); subtype D ancestor = 1966 (1961–1971); Trinidad and Tobago subtype B ancestor = 1973 (1970–1976); and U.S./Canada subtype B ancestor = 1969 (1966–1972). This analysis resolves the position of the archival sequence H6 as basal to the pandemic clade. Under the relaxed molecular clock, Pnon-Haitian-origin = 0.00003, Psimultaneous-origin = 0.0021, and PHaitian-origin = 0.9979.
The TMRCA of the U.S. epidemic is estimated to be 1969 (1966–1972) (Fig. 2), suggesting that HIV-1 was circulating cryptically in the United States for ≈12 years before the initial recognition of AIDS in 1981. The evidence of a single origin of the pandemic variant of subtype B allowed us to date the beginning of the actual U.S. HIV-1 epidemic, rather than the ancestor of multiple viruses introduced from Haiti; if multiple introductions from Haiti had occurred, the “U.S. MRCA” would actually correspond to a Haitian virus that predated the initial entry of HIV-1 to the United States (11). Serological evidence of an ≈5% prevalence of HIV-1 by 1978 in men who have sex with men (MSM) populations in both San Francisco (22) and New York City (23) suggests that several thousand individuals in the United States would already have been infected by then. Even assuming the fastest-documented growth rates for HIV-1 (24), this implies that the virus had been spreading in the MSM population for several years before this point, consistent with our findings here. The virus may well have been spreading slowly for an extensive period, perhaps in the heterosexual population, before entering the highest-risk MSM subpopulation, where it spread explosively enough to finally be noticed. We contend that the phylogenetic estimate, with appropriate confidence intervals, provides more reliable information on the date of the origin of the U.S. epidemic than the available epidemiological data, which cannot resolve this question.
Nevertheless, although our relaxed-clock methods should be reasonably robust to rate variation among lineages and uncertainty in phylogenetic inference, some caution is always warranted when such inferences are made. For example, the relatively sparse sampling of both Haitian and Trinidadian sequences means it is conceivable that more intensive future sampling could recover deeper-branching lineages and push back these TMRCA estimates slightly. This is unlikely to apply to the U.S. TMRCA, on the other hand, because of the already dense sampling of this HIV-1 population. If obtainable, additional archival sequences should help clarify the early spread of subtype B with greater precision.
The three-decade gap between the estimated timing of the HIV-1 M group ancestor (9) and the earliest evidence of HIV/AIDS in Africa (25, 26) seems unexceptional in comparison to the U.S. cryptic period, especially because a good deal of tuberculosis-caused mortality in Africa must have gone unrecognized as AIDS-related, then as now. Taken together with our dating analysis, including the subtype B/D ancestor dated to 1954 (1946–1961) (Fig. 2), the extensive cryptic period in the United States, therefore, provides compelling corroboration of an early 20th century M group ancestor (9).
Conclusion
Our findings imply that Haiti has the oldest-known HIV/AIDS epidemic outside of sub-Saharan Africa, which helps explain the high prevalence of AIDS and HIV-1 among Haitians in the early 1980s. Because of its 40-year history, the HIV-1 epidemic in Haiti exhibits a greater range of viral genetic diversity than the rest of the world's subtype B strains combined, much as the HIV-1 epidemic in the Democratic Republic of the Congo does for group M as a whole (27). This raises the possibility that subtype B strains in Haiti or elsewhere might exhibit distinct or more diverse antigenic properties compared with pandemic clade viruses. Vaccines derived from consensus or other central sequences should perhaps be based on extensive sampling of Haitian HIV-1 if they are intended to cover both Haitian subtype B strains as well as the pandemic clade.
Although it has long been clear that population bottlenecks and founder effects are a feature of the unfolding HIV/AIDS pandemic, the series of bottlenecks that punctuated the global emergence of subtype B is remarkable. The lack of evidence for selection associated with the spread of the pandemic clade of subtype B, moreover, points to the importance of chance events and ecological interactions in driving what was perhaps the most explosive worldwide dispersal of HIV-1.
Our phylogenetic estimates of timing anchor previous epidemiological observations that, on their own, cannot reliably date the origin of regional epidemics. Taken together, these sources of information suggest that HIV-1 was circulating in one of the most medically sophisticated settings in the world for more than a decade before AIDS was recognized.
Methods
The Archival Samples.
Peripheral blood mononuclear cell (PBMC) samples were collected in 1982 and 1983 at Jackson Memorial Hospital in Miami, FL, during one of the first investigations establishing that Haitians in Haiti and elsewhere were at risk for AIDS (6). One of the six PBMC samples obtained for this study failed to yield any amplifiable HIV-1 PCR products. As described in Pitchenik et al. (6), all of the patients were Haitian immigrants who had entered the United States after 1975 and progressed to AIDS by 1981 and hence were presumably infected with HIV-1 before entering the United States. The position of these sequences on the subtype B phylogeny (distinct from and basal to the dominant U.S. variant of subtype B) is consistent with this sequence of events.
Amplification and Sequencing of Archival HIV-1 DNA.
DNA was extracted from 10 μl of peripheral blood mononuclear cells by using QIAamp DNA micro kits (Qiagen, Valencia, CA), following the manufacturer's instructions for extractions from blood. After extraction, DNA was eluted into 100 μl of elution buffer AE and stored frozen at −20°C until required for DNA analyses. DNA was PCR-amplified from the extracts by using a nested PCR approach (28). First-round PCRs were undertaken in 25 μl of final volume reactions, using 0.1 μl of Platinum Taq HiFi enzyme (Invitrogen, Carlsbad, CA)/0.1 μl of 25 mM dNTP mix/2.5 mM (final concentration) MgSO4/10× PCR buffer/1–5 μl of DNA extract. Second-round amplifications were performed on 1 μl of the first-round PCR product by using the same reagent concentrations. Enzyme activation, dissociation, and extension temperatures followed the manufacturer's guidelines, with an extension time of 3 min. Annealing temperatures varied by extract in response to initial amplification success rates.
After amplification, the PCR products were visualized on 0.8% agarose gels stained with ethidium bromide and then purified by using QIAquick spin columns (Qiagen). Purified products were sequenced by using several overlapping primer pairs (28) by the University of Arizona Genomic Analysis and Technology Core Facility with ABI Big Dye 3.1 chemistry (Applied Biosystems, Foster City, CA) on Applied Biosystem 3730xl DNA Analyzers. Each sample was extracted, PCR-amplified, and sequenced twice to ensure that the sequences generated were not modified through low template copy number. We recovered five full-length env sequences and five partial (0.7- to 1.2-kb) gag sequences.
Sequence Alignments.
We used the Los Alamos National Laboratory HIV sequence database (29) to download all full-length published env and gag gene sequences of subtypes B and D. We then subjected the resulting sequence set to strict quality control measures to remove (i) incomplete sequences; (ii) sequences not published in a peer-reviewed journal; (iii) multiple sequences from the same patient; (iv) sequences suspected a priori of possibly anomalous evolutionary patterns (from long-term nonprogressors with nef deletions, laboratory workers infected accidentally, sequences exhibiting evidence of hypermutation, etc.); (v) sequences with midpeptide stop codons, frame-shift mutations, or nonnucleotide characters; or (vi) sequences for which there was any uncertainty regarding which subtype they belonged to. To search for such sequences, which might include unidentified intersubtype recombinants, we screened all sequences using the REGA HIV-1 Subtyping Tool (30) and then removed any sequence with bootstrapping support <100% or bootscanning support <1.0 for clustering with subtype B or D.
To ensure that no very-early-diverging subtype B strains were removed by this procedure, we inferred additional phylogenies that included the “cleaned” sequences (data not shown). For env, the only such strains that were positioned basal to the pandemic clade were from Trinidad and Tobago, and these clustered with the other sequences from the Trinidad and Tobago clade. Similarly, for gag, the only nonpandemic clade sequences that were removed (US4 and RF) were ones that had already been identified as basal in the env analysis; all other sequences with bootstrap or bootscan support <100% or 1.0 were from the pandemic clade. Unlike intersubtype recombinants, the possible nonexclusion of intrasubtype recombinants is not expected to affect inferences regarding the geographical origin and emergence of subtype B because intrasubtype recombination cannot plausibly lead to strains from one locality systematically falling basal to all of the others. Moreover, although unidentified intrasubtype recombination might increase the variance of dating estimates, it is unlikely to systematically bias these dates in one direction or the other in an exponentially growing population (31).
The resulting data sets were codon-aligned and then adjusted by eye in Squint Ver. 1.0 (M. Goode, University of Auckland, Auckland, New Zealand), and regions of ambiguous alignment were removed. We constructed three additional env alignments replacing the D subtype outgroup with a subtype C sequence from India, a subtype A sequence from Kenya, or a CRF01 (A/E) sequence from Thailand. All previously published sequences (see the taxon labels in SI Figs. 4 and 5b) are available from the Los Alamos National Laboratory HIV sequence database. All alignments are available from the authors upon request.
The Bayesian MCMC Phylogenetic Analysis and Estimation of the Probability of a Haitian or non-Haitian Origin of Subtype B.
We used MrBayes, Ver. 3.1 (32), to perform two independent runs of 20 million steps (for env). Examination of the MCMC samples with Tracer, Ver. 1.3 [A. Rambaut (University of Edinburgh) and A. J. Drummond (University of Auckland, Auckland, New Zealand); http://beast.bio.ed.ac.uk], indicated adequate mixing of the Markov chain. We discarded the first 2 million steps from each run as burn-in and combined the resulting MCMC samples (n = 36,002) for subsequent estimation of posteriors. Fewer steps (5 million) were required for convergence and adequate mixing in the gag analyses.
We used tree filtering in PAUP, Ver. 4.0b10 (33), to calculate the posterior probabilities of a Haitian, non-Haitian, or effectively simultaneous Haitian/non-Haitian origin of subtype B. Briefly, we removed from the posterior sample any tree with a Haitian sequence(s) in the most basal position. Only 4 of 36,002 env trees had a non-Haitian basal sequence (Pnon-Haitian-origin = 0.00011), and 24 others placed the Haitian and non-Haitian sequences in reciprocally monophyletic clades (Psimultaneous-origin = 0.00066), which left 35,974 with a Haitian sequence or group of sequences, in the most-ancestral position(s) within subtype B (PHaitian-origin = 0.9992). A similar approach was followed for the estimation of the probability of a Haitian or non-Haitian origin of subtype B for the gag and the relaxed-clock analyses.
For both the env and gag data sets, we used a parsimony approach as implemented in MacClade, Ver. 4.08 (34), to identify the nucleotide substitutions that mapped onto the branch leading to the pandemic clade of subtype B. We then determined whether these changes were synonymous or nonsynonymous.
The Relaxed Molecular Clock Analysis.
To infer the timescale of HIV-1 group M subtype B evolution, we used a Bayesian molecular clock method (15), as implemented in BEAST (http://beast.bio.ed.ac.uk), under an uncorrelated log-normal relaxed molecular clock model with a Bayesian Skyline coalescent tree prior. For this analysis to run in a reasonable time, we considered only subtype B sequences from Haiti, Trinidad and Tobago, and the United States (plus one from Canada), and we removed some pandemic clade sequences from overrepresented years and localities. The maximum a posteriori tree from the MrBayes analysis (available upon request) revealed that the sequences were scattered across the entire pandemic clade, consistent with a U.S. entry of the founding virus. We ran 10 independent MCMC analyses, with each run consisting of 100 million steps, and then discarded the first 5 million steps from each run as burn-in and combined the resulting postburn-in MCMC samples for subsequent estimation of posteriors.
Supplementary Material
Acknowledgments
We thank David Robertson and Adam Bjork for comments. This work was supported by grants from the National Institutes of Health and the Packard Foundation (to M.W.). A.R. is supported by a Royal Society University Research Fellowship.
Abbreviations
- TMRCA
time of the most recent common ancestor
- MCMC
Markov chain Monte Carlo.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF159970–EF159974 and EF362773–EF362777).
See Commentary on page 18351.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705329104/DC1.
References
- 1.Grenfell BT, Pybus OG, Gog JR, Wood JLN, Daly JM, Mumford JA, Holmes EC. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 2.Walker PR, Pybus OG, Rambaut A, Holmes EC. Infect Genet Evol. 2005;5:199–208. doi: 10.1016/j.meegid.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Ariën KK, Vanham G, Arts EJ. Nat Rev Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb MS, Schanker HM, Fan PT, Saxon A, Weisman JD. Morbid Mortal Wkly Rep. 1981;30:250–252. [Google Scholar]
- 6.Pitchenik AE, Fischl MA, Dickinson GM, Becker DM, Fournier AM, O'Conell MT, Colton RM, Spira TJ. Ann Intern Med. 1983;98:277–284. doi: 10.7326/0003-4819-98-3-277. [DOI] [PubMed] [Google Scholar]
- 7.Moses P, Moses J. Ann Intern Med. 1983;99:565. [Google Scholar]
- 8.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 10.Li W-H, Tanimura M, Sharp PM. Mol Biol Evol. 1988;5:313–330. doi: 10.1093/oxfordjournals.molbev.a040503. [DOI] [PubMed] [Google Scholar]
- 11.Robbins KE, Lemey P, Pybus OG, Jaffe HW, Youngpairoj AS, Brown TM, Salemi M, Vandamme A-M, Kalish ML. J Virol. 2003;77:6359–6366. doi: 10.1128/JVI.77.11.6359-6366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson WD, Pape JW. In: AIDS: Pathogenesis and Treatment. Levy JA, editor. New York: Dekker; 1989. pp. 65–78. [Google Scholar]
- 13.Farmer P. AIDS and Accusation. Berkeley: Univ of California Press; 2006. [Google Scholar]
- 14.Cohen J. Science. 2006;313:470–473. doi: 10.1126/science.313.5786.470b. [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Ho SYW, Phillips MJ, Rambaut PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartholomew C, Saxinger WC, Clark JW, Gail M, Dudgeon A, Mahabir B, Hull-Drysdale B, Cleghorn F, Gallo RC, Blattner WA. J Am Med Assoc. 1987;257:2604–2608. [PubMed] [Google Scholar]
- 17.Cleghorn FR, Jack N, Carr JK, Edwards J, Mahabir B, Sill A, McDanal CB, Connolly SM, Goodman D, Bennetts RQ, et al. Proc Natl Acad Sci USA. 2000;97:10532–10537. doi: 10.1073/pnas.97.19.10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn BH, Shaw GM, Taylor ME, Redfield RR, Markham PD, Salahuddin SZ, Wong-Staal F, Gallo RC, Parks ES, Parks WP. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 19.Daniels RS, Kang C, Patel D, Xiang Z, Douglas NW, Zheng NN, Cho HW, Lee JS. AIDS Res Hum Retrov. 2003;19:631–641. doi: 10.1089/088922203322280847. [DOI] [PubMed] [Google Scholar]
- 20.Cohen J. Science. 2006;313:473–475. doi: 10.1126/science.313.5786.473. [DOI] [PubMed] [Google Scholar]
- 21.Piot P, Quinn TC, Taelman H, Feinsod FM, Minlangu KB, Wobin O, Mbendi N, Mazebo P, Ndangi K, Stevens W, et al. Lancet. 1984;2:65–69. doi: 10.1016/s0140-6736(84)90241-1. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe HW, Darrow WW, Echenberg DF, O'Malley PM, Getchell JP, Kalyanaraman VS, Byers RH, Drennan DP, Braff EH, Curran JW, et al. Ann Intern Med. 1985;103:210–214. doi: 10.7326/0003-4819-103-2-210. [DOI] [PubMed] [Google Scholar]
- 23.Stevens CE, Taylor PE, Zang EA, Morrison JM, Harley EJ, Rodriguez de Cordoba S, Bacino C, Ting RC, Bodner AJ, Sarngadharanet MG, et al. J Am Med Assoc. 1986;255:2167–2172. [PubMed] [Google Scholar]
- 24.May RM, Anderson RM. Nature. 1987;326:137–142. doi: 10.1038/326137a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhu TF, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 26.Sonnet J, Michaux JL, Zech F, Brucher JM, de Bruyere M, Burtonboy G. Scand J Infect Dis. 1987;19:511–517. doi: 10.3109/00365548709032415. [DOI] [PubMed] [Google Scholar]
- 27.Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Nature. 2001;410:1035–1036. [Google Scholar]
- 28.Sanders-Buell E, Salminen MO, McCutchan F. In: The Human Retroviruses and AIDS 1995 Compendium. Myers G, Korber B, Hahn BH, Foley B, Mellors JW, et al., editors. Los Alamos, NM: Los Alamos National Laboratory; 1995. pp. III-15–III-21. [Google Scholar]
- 29.Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors JW, Wolinsky S, Korber B, editors. HIV Sequence Compendium. Los Alamos, NM: Los Alamos National Laboratory; 2005. [Google Scholar]
- 30.de Oliveira T, Deforche K, Cassol S, Salminem, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AM, van de Vijver DA, et al. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 31.Lemey P, Pybus OG, Rambaut A, Drummond AJ, Robertson DL, Roques P, Worobey M, Vandamme AM. Genetics. 2004;167:1059–1068. doi: 10.1534/genetics.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 33.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2003. Ver. 4.0b10. [Google Scholar]
- 34.Maddison DR, Maddison WP. MacClade 4: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2001. Ver. 4.08. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.