Abstract
The importance of sympatric speciation (the evolution of reproductive isolation between codistributed populations) in generating biodiversity is highly controversial. Whereas potential examples of sympatric speciation exist for plants, insects, and fishes, most theoretical models suggest that it requires conditions that are probably not common in nature, and only two possible cases have been described for tetrapods. One mechanism by which it could occur is through allochronic isolation—separation of populations by breeding time. Oceanodroma castro (the Madeiran or band-rumped storm-petrel) is a small seabird that nests on tropical and subtropical islands throughout the Atlantic and Pacific Oceans. In at least five archipelagos, different individuals breed on the same islands in different seasons. We compared variation in five microsatellite loci and the mitochondrial control region among 562 O. castro from throughout the species' range. We found that sympatric seasonal populations differ genetically within all five archipelagos and have ceased to exchange genes in two. Population and gene trees all indicate that seasonal populations within four of the archipelagos are more closely related to each other than to populations from the same season from other archipelagos; divergence of the fifth sympatric pair is too ancient for reliable inference. Thus, seasonal populations appear to have arisen sympatrically at least four times. This is the first evidence for sympatric speciation by allochrony in a tetrapod, and adds to growing indications that population differentiation and speciation can occur without geographic barriers to gene flow.
Keywords: Oceanodroma castro, phylogeography, genetic isolation, seasonal populations, storm-petrel
Speciation—the evolution of reproductive isolation between populations—is thought generally to involve the gradual accumulation of genetic differences between geographically isolated (allopatric) populations through selection or genetic drift (reviewed in ref. 1). Although this “allopatric” model of speciation is widely accepted, it does not provide a satisfactory mechanism for the origin of many species, such as sympatric sister species, and several alternatives have been proposed (reviewed in ref. 1). According to various “sympatric” models, reproductive isolation could evolve in the absence of geographic isolation through polyploidization (e.g., in flowering plants) or nonrandom mating [e.g., according to host in phytophagous insects or brood parasitic birds, migratory route in birds, or breeding time (allochrony) in salmon and periodical cicadas] (1–4). Sympatric speciation was first proposed by Darwin but was refuted by Mayr and has been controversial ever since (reviewed in refs. 1 and 3). Although sympatric speciation is possible in theory, most models require conditions that are probably uncommon in nature, such as an appropriate balance between selection and recombination, linkage between genes involved in ecological specialization and reproductive isolation, and/or a small number of loci controlling local adaptation, and habitat and mate preference. It also requires populations to be able to coexist ecologically during and after the evolution of reproductive isolation (1, 3, 5, 6).
Sympatric speciation is difficult to demonstrate in nature. Potential examples exist for plants, insects, and fishes (1–7), but only two possible cases have been described for tetrapods: host-specific races of brood parasitic indigobirds (Vidua spp.) (8, 9) and ecologically segregated races of Nesospiza buntings on Tristan da Cunha (10). Coyne and Orr (ref. 1, p. 142) delineate four criteria that must be met to provide a convincing case: “1. The species must be largely or completely sympatric. 2. The species must have substantial reproductive isolation, preferably based on genetic differences. 3. The sympatric taxa must be sister groups. 4. The biogeographic and evolutionary history of the groups must make the existence of an allopatric phase very unlikely.” Unfortunately, the geographic and phylogenetic signatures of sympatric speciation are easily obscured by range expansion, extinction, and lineage sorting (1, 11). However, modern methods of molecular analysis are enabling increasingly rigorous tests of speciation models.
Oceanodroma castro (the Madeiran or band-rumped storm-petrel; Procellariiformes: Hydrobatidae) provides a useful test model for sympatric speciation. This small pelagic seabird forages throughout tropical and subtropical regions of the Atlantic and Pacific Oceans and breeds on several islets in Japan, Hawaii, the Galapagos, and the northeast and central Atlantic (12) (Fig. 1). The timing of breeding varies considerably among colonies [supporting information (SI) Table 2]: at one extreme, some populations have a single, compact breeding season (e.g., Japan); at other islands, the nesting season is much more protracted (e.g., Cape Verde); at the other extreme, several colonies have two distinct laying periods separated by an interval with no breeding (specifically, the Azores, Desertas, and the Galapagos). Evidence from breeding and molting phenology, feather mercury concentrations, band (ring) returns, morphology, vocalizations, and mtDNA sequences suggests that different populations nest in the same areas—even in the same burrows—at some of these colonies (12–20). Differences in all these characters between seasonal populations within the Azores indicate that they represent reproductively isolated species (16, 18, 20) and thus meet the first of Coyne and Orr's criteria for sympatric speciation (1). Seasonal populations of O. castro may therefore represent cases of sympatric divergence and speciation through habitat preference, specifically, breeding season (allochrony). Here, we compare variation in five microsatellite loci and a portion of the mitochondrial control region among 17 populations of O. castro, including four known pairs of sympatric seasonal populations and samples from two seasons from Cape Verde (which has protracted breeding) to test whether seasonal populations are genetically isolated and originated sympatrically.
Fig. 1.
Breeding locations of O. castro (circles). Black symbols represent locations with breeding during the Northern Hemisphere hot season; white symbols represent locations with breeding during the Northern Hemisphere cool season; black and white symbols represent locations with sympatric seasonal populations; gray symbol represents protracted (aseasonal) breeding with two seasonal peaks. Archipelago names are in standard type; islands that are mentioned in the text are in parentheses. Baixo and Praia are 5 km apart; Branco and Raso are 7 km apart.
Results and Discussion
Global Population Genetic Structure.
Comparison of variation in microsatellites and mtDNA among O. castro sampled throughout the breeding range support previous observations (19, 20) that gene flow is generally restricted in O. castro and that population genetic structure is strong (Fig. 2 and SI Tables 3–5): Only 6 of 184 estimates of migration rates (m, as a proportion of the population) between population pairs generated from microsatellite variation by using the program BayesAss were significantly >0 (SI Table 3). Estimates of population divergence time (T, in Nf generations, where Nf is female effective population size) derived from mitochondrial control region sequences by using the program MDIV were significantly >0 for most (43 of 49) comparisons (SI Table 4). Few estimates of gene flow (M, in females per generation) from MDIV were significantly >1 (SI Table 4). Finally, most indices of population differentiation (FST for microsatellites and ΦST and δ for control region sequences) were significantly >0 (Fig. 2 and SI Table 5). Most exceptions to these patterns involved comparisons among cool-season populations within the northeast Atlantic (mainland Portugal, the Azores, Desertas, Selvagem, and the Canaries). Notably, both types of genetic markers indicate that O. castro from different geographic regions (Japan, the Galapagos, and the Atlantic) are genetically isolated and probably diverged >250,000 years ago (Fig. 3). Although few birds were sampled from Hawaii and Ascension, these individuals also differ from those elsewhere and are most closely related to Japanese and northeast Atlantic cool populations, respectively (20).
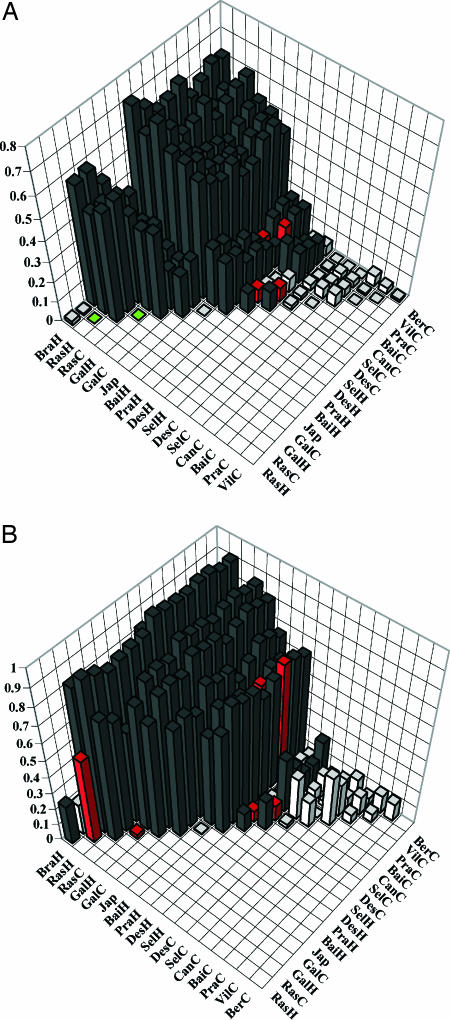
Fig. 2.
Pairwise estimates of FST (based on microsatellite variation; A) and ΦST (based on control region sequences; B) for pair-wise comparisons of O. castro colonies. Red bars represent estimates that are significantly >0 at α = 0.05 after sequential Bonferroni corrections for sympatric seasonal populations; dark gray bars represent estimates that are significantly >0 for allopatric populations; green bars represent estimates that are not significantly different from 0 for sympatric seasonal populations; white bars represent estimates that are not significantly different from 0 for allopatric populations. Data from SI Table 5. Populations are arranged in approximate order of increasing divergence for clarity of presentation. Colony codes are the first three letters from Fig. 1; H, hot-season population; C, cool-season population.
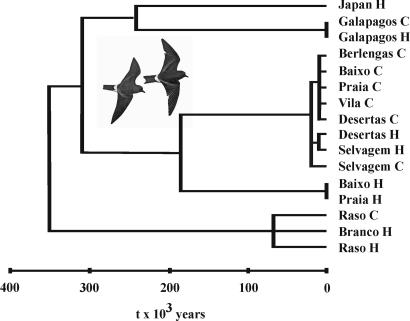
Fig. 3.
Approximate timing of divergence of O. castro populations, inferred from the minimum evolution tree for estimates of t (divergence time in years) derived either from T [time in Nf generations, from MDIV (SI Table 4); comparisons within the Galapagos, northeast Atlantic, and Cape Verde] or from δ [Kimura-two-parameter corrected net sequence divergence (SI Table 5); comparisons between archipelagos] from mtDNA sequence variation. C, populations breeding in the Northern Hemisphere cool season; H, populations breeding in the Northern Hemisphere hot season. The Canaries population was excluded because the small sample size made estimates of T unreliable.
In agreement with results of previous studies (20), significant genetic structure also exists within the Atlantic, and gene flow appears to be restricted (SI Tables 3–5). Samples from Cape Verde are especially divergent from those elsewhere (Fig. 3). Additionally, most hot-season populations from the Azores, Desertas, Selvagem, and Cape Verde differ from each other and do not appear to exchange genes, despite the geographic proximity of some colonies (SI Tables 3–5).
Relationships Among Sympatric Seasonal Populations.
Results of the present study indicate that seasonal populations of O. castro meet most or all of Coyne and Orr's (1) criteria for sympatric speciation.
Reproductive isolation.
Variation in microsatellites and mtDNA indicates that sympatric seasonal populations within two archipelagos—the Azores and Cape Verde—are genetically isolated: estimates of migration rate (m) based on microsatellite variation do not differ from 0; estimates of gene flow (M) based on mtDNA variation are <1 (although not always significantly so); and estimates of FST based on microsatellites and ΦST and δ based on mtDNA are significantly >0 for most comparisons (up to 0.16%, 0.69%, and 2.9%, respectively; Table 1 and Fig. 2). In addition, seasonal samples within the Azores are virtually reciprocally monophyletic on the mtDNA gene tree, i.e., most haplotypes from the two populations occur on separate branches, indicating prolonged genetic isolation (Fig. 4). Estimates of divergence time (T) from MDIV also differ from 0 and suggest that sympatric seasonal populations within these archipelagos have been genetically isolated for 73,000 to180,000 years (Fig. 2 and Table 1). Although these molecular results do not provide definitive evidence for reproductive isolation, they suggest that the populations have not been exchanging genes for thousands of generations.
Table 1.
Estimates of migration rate (m as a proportion of the population, from BayesAss), female-mediated gene flow (M in females per generation, from MDIV), population differentiation (FST, ΦST, and δ, from Arlequin), and time since divergence (T in 2Nf generations and t in years, from MDIV) for sympatric seasonal populations of O. castro in five archipelagos (summarized from SI Tables 3–5)
| Parameter | Azores |
Desertas | Selvagem | Cape Verde |
Galapagos |
|
|---|---|---|---|---|---|---|
| Baixo | Praia | Raso | Plaza Norte | |||
| Microsatellites | ||||||
| m (from cool to hot population) | 0.003 | 0.001 | 0.22* | 0.16 | 0.003 | 0.15* |
| m (from hot to cool population) | 0.005 | 0.003 | 0.001 | 0.001 | 0.030 | 0.26* |
| FST | 0.11** | 0.16** | 0.07* | 0.08** | 0.00 | 0.00 |
| mtDNA | ||||||
| M | 0.44 | 0.23† | 0.82 | 5.0‡ | 0.31 | na |
| T | 1.6* | 1.6* | 0.27* | 0.04* | 0.56* | 0.44 |
| t | 110 | 180 | 27 | 27 | 73 | na |
| ΦST | 0.58** | 0.69** | 0.07** | 0.09** | 0.43** | 0.02* |
| δ | 2.5* | 2.9* | 0.11** | 0.08** | 2.4** | 0.04* |
*, Significantly >0 before sequential Bonferroni correction;
**, significantly >0 after sequential Bonferroni correction;
†, significantly <1 at α = 0.05;
‡, significantly >1 at α = 0.05. na, could not be calculated (T did not differ from 0).
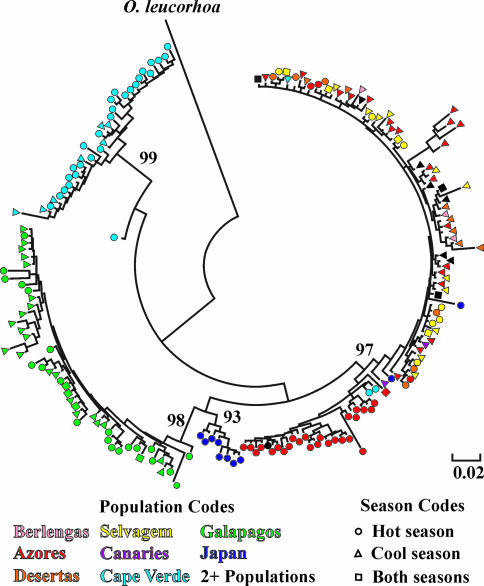
Fig. 4.
Neighbor-joining tree for control region haplotypes for O. castro. Symbols represent populations where haplotypes were found. Numbers on branches are support from interior branch tests for key groups. The scale represents Kimura two-parameter-corrected sequence divergence among haplotypes. O. leucorhoa, Leach's storm-petrel.
Sympatric seasonal populations in the Galapagos, Desertas, and Selvagem appear to represent earlier stages of genetic isolation: Most indices of population differentiation are significantly greater than 0, but gene flow (as indicated by m or M) appears to be ongoing (Table 1).
Thus, the five pairs of seasonal populations appear to represent three stages of divergence [sensu Avise (11), Tautz (21)]. Those from the Azores are genetically differentiated, presumably reproductively isolated, and virtually completely segregated on the mtDNA gene tree (Fig. 4); given that they are diagnosable by season as well as by morphology and vocalizations (16, 18), they constitute species by most definitions (1). Sympatric seasonal populations within Cape Verde appear to be genetically isolated; however, they are not yet reciprocally monophyletic on the gene tree or diagnosable by any character other than breeding season, so represent an earlier stage of speciation than the Azores populations. Sympatric seasonal populations in Desertas, Selvagem, and the Galapagos exhibit some genetic differentiation as well as slight morphometric differences (15, 17, 19) but are not genetically isolated.
In contrast, little differentiation exists either among cool season populations from different colonies within the northeast Atlantic, or between the two Azores hot-season colonies: Few estimates of population differentiation differ from zero (SI Table 5), and several estimates of migration and gene flow are significant (SI Tables 3 and 4).
Sympatric taxa are sister groups.
Two lines of evidence indicate that sympatric seasonal populations of O. castro are sister taxa. First, within the mtDNA gene tree, haplotypes from Galapagos seasonal populations occur together within a strongly supported monophyletic clade (Fig. 4). Most haplotypes from Cape Verde seasonal samples also group together within a strongly supported monophyletic lineage. Haplotypes from Azores seasonal populations occur together within a strongly supported but poorly resolved Atlantic clade, although this clade also includes haplotypes from other northeast Atlantic colonies; the Azores hot-season population is approaching monophyly within this lineage. Haplotypes from the Desertas and Selvagem seasonal populations also occur together within the northeast Atlantic clade.
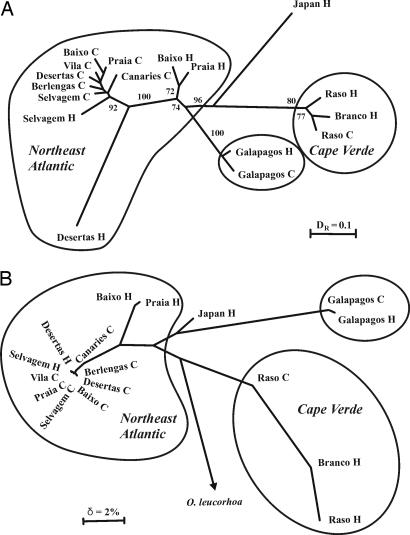
Second, on both the nuclear and mtDNA population trees, Galapagos seasonal populations form a monophyletic group, as do Cape Verde seasonal samples (Fig. 5); each of these lineages has strong support on the nuclear tree (Fig. 5A). Similarly, Azores seasonal populations occur within an unresolved but monophyletic clade that includes other northeast Atlantic cool-season populations; although they do not group together, note that a sister-taxon relationship would be quickly lost from the population tree if gene flow is ongoing among northeast Atlantic cool-season populations.
Fig. 5.
Unrooted neighbor-joining trees for O. castro populations based on microsatellite variation (A), and mitochondrial control region sequences (B). Numbers on branches for A are support from bootstrapping across individuals (values <70% not shown). O. leucorhoa, Leach's storm-petrel. C, populations breeding in the Northern Hemisphere cool season; H, populations breeding in the Northern Hemisphere hot season.
An historical allopatric phase is very unlikely.
If seasonal populations arose allopatrically, or arose only once sympatrically, evidence of historical fragmentation and secondary contact should be present in the form of deep branches in the gene tree (11, 22). This historical fragmentation should be apparent whether or not the original colonies still exist. No evidence was found for historical fragmentation or secondary contact between any sympatric seasonal populations either from the general shape of the gene tree (Fig. 4) or from nested clade analysis of the northeast Atlantic populations (20) or nested contingency analysis of the Galapagos clade (19).
Thus, seasonal populations of O. castro in the Azores and Cape Verde meet all of Coyne and Orr's criteria for sympatric speciation (1). Seasonal populations in Desertas, Selvagem and the Galapagos also fulfill three of the critera, but are probably not yet reproductively isolated.
At least two alternatives to sympatric origins are possible for seasonal populations of O. castro. One is that seasonal populations arose allopatrically, followed by sympatry and hybridization. This could result in the observed clustering of sympatric seasonal populations on the population trees. The possibility of hybridization is supported by 10 control-region haplotypes that are shared between seasonal populations (SI Table 6) and by polyphyly within the mtDNA gene tree (Fig. 4). However, three observations argue against this scenario. First, hybridization would have to be extensive to result in the observed population and gene trees, but results from BayesAss and MDIV indicate that there is no exchange of either nuclear or mitochondrial genes between seasonal populations within the Azores or Cape Verde (SI Tables 3 and 4). Second, as noted above, no evidence of historical fragmentation and/or secondary contact (hybridization) was found within the gene tree (Fig. 4). Finally, of the 10 haplotypes that are shared between sympatric seasonal populations (SI Table 6), most (eight) are located at or near the root of the gene tree (19, 20), indicating that they represent retained ancestral variation; only one haplotype each in the Galapagos and northeast Atlantic occurred on a branch tip, suggestive of possible contemporary gene flow or hybridization.
Another possible explanation is that seasonal populations originated on different islands within an archipelago (i.e., through microallopatry), followed by sympatry through range expansion (11). Although possible, this explanation is less parsimonious than sympatric origins, i.e., it requires more events: speciation plus range expansion. Furthermore, it must have happened multiple times to generate the population trees seen in Fig. 5. However, the possibility of microallopatric origins is difficult to preclude for the Azores seasonal populations: given that the Azores cool-season breeders appear to be part of a large, interbreeding metapopulation that includes colonies from the coast of mainland Portugal, Desertas, Selvagem, and the Canaries, cool-season breeders could have originated anywhere within the northeast Atlantic and expanded into the Azores after the origin of the Azores hot-season population. As noted previously, the phylogenetic and phylogeographic signals of sympatric speciation are quickly obscured (1, 11), and information regarding the precise site of origin may have been lost for the Azores hot-season population. Nonetheless, given the large foraging and nonbreeding distributions of this species (23), the origin of seasonal populations even on neighboring islands would still constitute sympatric speciation under most definitions (see ref. 6 for a summary of definitions; e.g., origin of reproductive isolation within the dispersal range of the species; speciation in the absence of geographic isolation; speciation under random mating with respect to geography).
Results from MDIV suggest that seasonal populations originated recently: 110,000–180,000 years ago in the Azores, ≈73,000 years ago in Cape Verde, ≈27,000 years ago in Desertas and Selvagem, and within the last few thousand years in the Galapagos (Fig. 3 and SI Table 4). All these dates correspond to interglacial or postglacial periods. Cold-water upwellings, oceanic mixing, and marine productivity at low latitudes are thought to have increased during glaciations (reviewed in ref. 20). This would have increased foraging opportunities for O. castro, enabling population growth. Correspondingly, Smith et al. (20) found evidence from mtDNA sequences of demographic expansion during the last glaciation (≈60,000–25,000 years ago). Presumably, decreased marine productivity and upwellings during interglacials would increase competition for food in O. castro, potentially favoring divergence in breeding seasons.
Results of the present study also suggest that the cool-season populations were derived from the hot-season populations in the Galapagos, Cape Verde, and the Azores [contra Monteiro and Furness (16)]. Specifically, if breeding season is mapped onto any of the population or gene trees (Figs. 4 and 5), hot-season breeding is indicated to be the ancestral state. However within Desertas, migration from the cool- into the hot-season population is significantly higher than from the hot into the cool population (Table 1), suggesting that the Desertas hot-season population may have been derived secondarily from the cool-season population. Alternatively, seasonal populations may have originated through disruptive selection on an ancestral population that had a protracted breeding season (2), such as occurs at some colonies of O. castro [e.g., Cape Verde and possibly Selvagem (SI Table 2)] and closely related species [e.g., ashy storm-petrel O. homochroa; Polynesian storm-petrel Nesofregetta fuliginosa (12)]. Both theoretical and empirical evidence indicates that, if the timing of reproduction is variable and heritable, populations may diverge and adapt to different breeding seasons because of isolation by time (2). Results for Cape Verde are especially interesting in this respect: Although breeding occurs year round, O. castro sampled during different seasons differ in their mtDNA and appear to be genetically isolated (SI Tables 3–5). The heritability of breeding time has not been studied in this species, but breeding time is known to be heritable in snow geese (Chen caerulescens) and a variety of passerine species (reviewed in ref. 2).
High levels of genetic diversity in all O. castro populations (SI Tables 6 and 7) suggest that the origins of seasonal populations were not associated with severe genetic bottlenecks [thus, not founder-induced divergence (reviewed in ref. 1)]. Given the tight annual scheduling of breeding and molting, a sudden shift of adults from one season to the other also seems unlikely. More probably, the change involved juveniles or failed breeders or gradual divergence of populations over many generations [e.g., because of isolation by time (above)]. Competition for nest sites may have caused displacement of birds from one season to another (16, 24). Alternatively or additionally, a change in breeding season may have enabled individuals to exploit new food resources and thus to reduce intraspecific competition for food. Note, however, that the origin of seasonal populations from an aseasonal ancestor does not necessarily require selection for different seasons but, theoretically, can occur solely through restricted gene flow if the timing of breeding is heritable (2).
Conclusions
The present results strongly suggest that allochronic populations of O. castro are genetically isolated within at least two archipelagos (the Azores and Cape Verde) and have arisen sympatrically in at least four locations (within Desertas, Cape Verde, Selvagem, and the Galapagos) and probably also within the Azores. The existence of breeding asynchrony within many species of plants and animals [(2, 6) e.g., ≈10% of seabird species have >1-month asynchronies in breeding seasons among colonies, and 20% have asynchronous breeding within colonies (25, 26)], evidence for heritability of breeding time (reviewed in ref. 2) and the existence of genetic divergence between temporally segregated populations in a diversity of species (1, 27, 28) suggest that, although probably not general, sympatric speciation by allochrony may not be unusual. These results add to growing evidence for population differentiation and speciation in sympatric and parapatric populations of birds (29). For example, morphologically and genetically differentiated sympatric seasonal populations have been reported in Leach's storm-petrels [O. leucorhoa (ref. 30 and V.L.F., P. Gulavita, A. Bailie, and T. Birt, unpublished data)], and significant population genetic structure has been found in the absence of geographic isolation in seabirds [e.g., Galapagos petrels Pterodroma phaeopgyia (31)], raptors [e.g., Eurasian kestrels Falco tinnunculus (27)], and passerines [e.g., indigobirds and buntings (8–10)]. Thus, speciation in the absence of geographic isolation may be more common than is currently recognized (4). It is also possible that some currently parapatric or allopatric species arose sympatrically, with speciation being followed by a range shift.
Materials and Methods
DNA samples were obtained from 562 O. castro from 17 populations, including 386 samples previously described by Smith et al. (20) and 176 additional samples from the coast of mainland Portugal, the Azores, Desertas, Selvagem, the Canaries, and Cape Verde (SI Tables 6 and 7). Samples comprise most colonies where O. castro breeds, and both hot- and cool-season populations from the Azores (two islets 5 km apart: Baixo and Praia), Desertas (Furna dos Rosques), Selvagem (Selvagem Grande, 265 km from Desertas), and the Galapagos (Plaza Norte), as well as two seasons from Cape Verde (where breeding is protracted; two islets 7 km apart: Raso and Branco; Fig. 1 and SI Table 2). Breeding season affiliation was confirmed by examination of the brood patch, morphometrics, and/or molt (16). All samples were collected under the appropriate permits.
PCR primers for nine microsatellite loci were developed by using standard protocols (V.L.F., and Z. Sun, unpublished data). Loci were amplified in three sets of multiplexed reactions (Mix 1: Oc49, Oc79, Oc84; Mix 6: Oc51, Oc63, Oc79–2; Mix 7: Oc28B, Oc64B, Oc87) using an optimized primer concentration (0.1 μM for all primers except 0.3 μM for Oc84F and Oc84R), 1× Multiplex MixR (Qiagen), and 0.45 μM Dye 4-labeled M13 forward primer. PCRs involved an initial denaturation at 94°C for 15 s, followed by 35 cycles of 94°C for 45 s, 56°C (Mixes 1 and 6) or 58°C (Mix 7) for 1 min, and 72°C for 1 min 30 s, and a final extension at 60°C for 30 s. Samples were tested for amplification in 1.3% agarose gels and then subjected to electrophoresis through a Beckman CEQ 8000R automated sequencer. Genotypes were confirmed and scored by eye. Populations were tested for deviations from both Hardy–Weinberg and linkage equilibrium by using Arlequin (version 3.1) (32).
Four loci (Oc63, Oc64B, Oc79, and Oc84) did not amplify reliably. Locus Oc79 also had significant heterozygote deficiencies within several populations, suggesting the existence of a null (nonamplifying allele).These loci were therefore excluded from subsequent analyses.
Mitochondrial control region sequences for 386 samples have been published previously (20). Sequences for an additional 122 samples were obtained by using primers OcL61 (20) and H530 (V.L.F. and P. Gulavita, unpublished data) by using the protocols of Smith et al. (20). Sequences were also obtained for two Leach's storm-petrels (Oceanodroma leucorhoa) to root the mtDNA trees. Nonoverlapping sequences were trimmed from the combined data set, generating 205 haplotypes (SI Table 6).
Genetic differentiation among populations was indexed by using pair-wise estimates of FST (microsatellites) and ΦST and δ (control region sequences using Kimura's two-parameter correction for multiple substitutions with γ = 0.45), with significance determined by 10,000 bootstrap replications using the program Arlequin. Estimation of contemporary gene flow from FST or its analogues involves several dubious assumptions, most notably, that populations are in equilibrium between migration and genetic drift (33). We therefore used molecular assignments to estimate gene flow from microsatellite variation by using the program BayesAss (34). Three million MCMC iterations were run, with a burn-in of 1,000,000 iterations and a sampling frequency of 2,000. Δ (the maximum amount a parameter can change each iteration) was set to 0.15 (the default value). Samples from the Galapagos and the Atlantic were analyzed separately because these populations form highly distinct lineages on the population trees; Japan was not included because it is highly divergent from the other populations and lacks seasonal populations. Female-mediated gene flow between population pairs (M in females per generation) was estimated from control region sequences by using a maximum-likelihood approach based on coalescent theory, by using the program MDIV (35); TMAX and MMAX were adjusted for each comparison to ensure that likelihood estimates equilibrated. Samples from the Galapagos, Cape Verde, and the northeast Atlantic were analyzed separately, and Japan was not included, as above. Canaries samples also were excluded because of small sample size. Estimates of M were tested for difference from 1 by using likelihood ratio tests.
MDIV also was used to test whether populations are genetically isolated. Specifically, likelihood ratio tests were used to determine whether divergence time (T, in Nf generations, where Nf is female effective population size) was >0. Divergence time in years (t) was estimated as Tθ/2μ, where θ = 2Nfμ, and μ is the per-fragment mutation rate [estimated at 3.3−5, assuming a divergence rate of 21% per million years for Domain I of the mitochondrial control region of birds (36)].
To visualize the genetic relationships among populations, a phenetic population tree was generated from microsatellite data by neighbor-joining on Roger's genetic distances by using the program Populations (version 1.2.3beta, www.bioinformatics.org/project/?group_id = 84); confidence for branches was determined by bootstrapping across individuals. Other distance measures [Nei's minimum genetic distance (which increases linearly with time), Wright's linearized FST, and Reynold's unweighted distance] gave similar results. A phenetic population tree was generated from control-region sequences by neighbor-joining on Kimura-two-parameter corrected distances (δ) between population samples by using Mega (version 3.1) (37). A gene tree was generated for the control region haplotypes by using statistical parsimony, but several branch lengths exceeded the connection limit so that relationships among key populations could not be determined (20). Therefore, a neighbor-joining tree was generated for Kimura-two-parameter corrected distances between haplotypes by using Mega; use of other tree-construction algorithms or distance measures made only minor changes to branch tips. Support for branches was determined by interior branch tests (38).
A rejection level (α) of 0.05 was used throughout, with sequential Bonferroni corrections applied to minimize Type I errors.
Supplementary Material
Acknowledgments
We thank E. Astudillo Sánchez, J. Bried, P. Calabuig, M. Carvalho, O. Hasegawa, V. Neves, A. Paterson, T. Steeves, T. Telfer, S. Torres, Y. Watanuki, R. White, and F. Zino for help with sample collections. Laboratory support was provided by T. Birt, M. Angel Peinado, S. (Scott) Taylor, and Z. Sun. P. Beerli, L. Bernatchez, T. Birt, L. Ratcliffe, P. Palsboll, S. (Sherry) Taylor, and three anonymous referees provided helpful discussions and/or comments on drafts. MDIV analyses were performed by using the Computational Biology Service Unit from Cornell University (Ithaca, NY), which is partially funded by Microsoft Corporation. Funding was provided by the American Museum of Natural History, the American Ornithologists' Union, the Cooper Ornithological Society, the LIFE program of the European Union, the Portuguese Fundação para a Ciência e a Tecnólogia, Fondo FEDER, Fundación Banco Bilbao Vizcaya Argentaria, Generalitat de Catalunya, Ministerio de Educacion y Ciencia of Spain, Natural Sciences and Engineering Research Council of Canada (Postgraduate Scholarship and Discovery Grant Programs), and Queen's University School of Graduate Studies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mitochondrial sequences reported in this paper have been deposited in the GenBank database (accession nos. AY600297, AY771004, AY771005, DQ178703–DQ178869, and EU252035–EU252071).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700446104/DC1.
References
- 1.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 2.Hendry AP, Day T. Mol Ecol. 2005;14:901–916. doi: 10.1111/j.1365-294X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 3.Gavrilet S. Fitness Landscapes and the Origin of Species. Princeton: Princeton Univ Press; 2004. [Google Scholar]
- 4.Edwards SV, Kingan SB, Calkins JD, Balakrishnan CN. Proc Natl Acad Sci USA. 2005;102:6550–6557. doi: 10.1073/pnas.0501846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gavrilets S, Vose A, Marluenga M, Salzburger A. Mol Ecol. 2007;16:2893–2909. doi: 10.1111/j.1365-294X.2007.03305.x. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilets S, Vose A. Mol Ecol. 2007;16:2910–2921. doi: 10.1111/j.1365-294X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- 7.Barluenga A, Stolting KN, Salzburger W, Muschick M. Nature. 2004;439:719–723. doi: 10.1038/nature04325. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen MD, Sefc KM, Payne RB. Nature. 2003;424:928–931. doi: 10.1038/nature01863. [DOI] [PubMed] [Google Scholar]
- 9.Sefc KM, Payne RB, Sorenson MD. Mol Ecol. 2005;14:1407–1419. doi: 10.1111/j.1365-294X.2005.02492.x. [DOI] [PubMed] [Google Scholar]
- 10.Ryan PG, Bloomer P, Moloney CL, Grant TJ, Delport W. Science. 2007;315:1420–1423. doi: 10.1126/science.1138829. [DOI] [PubMed] [Google Scholar]
- 11.Avise JC. Phylogeography. Cambridge, MA: Harvard Univ Press; 2000. [Google Scholar]
- 12.Brooke M. Albatrosses and Petrels Across the World. New York: Oxford; 2004. [Google Scholar]
- 13.Snow DW, Snow BK. Ibis. 1966;108:283–284. [Google Scholar]
- 14.Harris MP. Proc Calif Acad Sci. 1969;37:95–165. [Google Scholar]
- 15.Faria BF. Boletim do Museu Municipal do Funchal. 1998;(Suppl 5):167–176. [Google Scholar]
- 16.Monteiro LR, Furness RW. Philos Trans R Soc London Ser B. 1998;353:945–953. [Google Scholar]
- 17.Nunes M. Arquipélago Life Mar Sci. 2000;(Suppl 2 Part A):175–179. [Google Scholar]
- 18.Bolton M. Ibis. 2007;149:255–263. [Google Scholar]
- 19.Smith AL, Friesen VL. Mol Ecol. 2007;16:1593–1603. doi: 10.1111/j.1365-294X.2006.03154.x. [DOI] [PubMed] [Google Scholar]
- 20.Smith AL, Monteiro L, Hasegawa O, Friesen VL. Mol Phylogen Evol. 2007;43:755–773. doi: 10.1016/j.ympev.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Tautz D. In: Adaptive Speciation. Dieckmann U, Doebeli M, Metz JAJ, Tautz D, editors. Cambridge, UK: Cambridge Univ Press; 2004. pp. 305–321. [Google Scholar]
- 22.Templeton A. Mol Ecol. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith JL, Hyrenbach KD. Mar Ornithol. 2003;31:155–166. [Google Scholar]
- 24.Bolton M, Medeiros R, Hothersall B, Campos A. Biol Conserv. 2004;116:73–80. [Google Scholar]
- 25.del Hoyo J, Elliott A, Sargatal J. Handbook of the Birds of the World. Vol 1. Barcelona: Lynx Edicions; 1992. [Google Scholar]
- 26.del Hoyo J, Elliott A, Sargatal J. Handbook of the Birds of the World. Vol 3. Barcelona: Lynx Edicions; 1992. [Google Scholar]
- 27.Casagrande S, Dell'Omo G, Costantini D, Tagliavini J. Evol Ecol Res. 2006;8:1029–1038. [Google Scholar]
- 28.Maes GE, Pujolar JM, Hellemans B, Volckaert FAM. Mol Ecol. 2006;15:2095–2107. doi: 10.1111/j.1365-294X.2006.02925.x. [DOI] [PubMed] [Google Scholar]
- 29.Friesen VL, Burg T, McCoy K. Mol Ecol. 2007;16:1765–1785. doi: 10.1111/j.1365-294X.2006.03197.x. [DOI] [PubMed] [Google Scholar]
- 30.Ainley D. Auk. 1980;97:937–953. [Google Scholar]
- 31.Friesen VL, González JA, Cruz-Delgado F. Conserv Genet. 2006;7:105–115. [Google Scholar]
- 32.Excoffier L, Laval G, Schneider S. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 33.Hedrick PW. Evolution (Lawrence, Kans) 1999;53:313–318. [Google Scholar]
- 34.Wilson GA, Rannala B. Genetics. 2005;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen R, Wakeley J. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenink PW, Baker AJ, Tilanus MGJ. Proc Natl Acad Sci USA. 1993;90:94–98. doi: 10.1073/pnas.90.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular Evolutionary Genetics Analysis Software, Version 2.1. Tempe, AZ: Arizona State Univ; 2001. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.