Abstract
Little is known regarding the functional effects of common autoimmune susceptibility variants on human immune cells. The SNP CT60 (rs3087243; A/G) located in the 3′ UTR of the CTLA4 gene has been associated with autoimmune diseases. We examined a cohort of healthy individuals stratified by genotypes at CTLA4 to gain insight into the functional effects of allelic variation on T cell signaling. Using phospho-site-specific mAbs, we tested the hypothesis that the CT60 genotype at CTLA4 is associated with altered T cell antigen receptor (TCR) signaling in naive and/or memory T cells. By normalizing for the extent of the initial TCR signaling event at CD3ζ, we observed that the relative responsiveness to TCR stimulation as assessed by phosphorylation levels of downstream signaling molecules was altered in naive (CD4+CD45RAhigh) and memory (CD4+CD45RAlow) T cells obtained from individuals with the disease-susceptibility allele at CTLA4. Thus, allelic variation associated with autoimmune disease can alter the signaling threshold of CD4+ T cells. These experiments provide a rational approach for the dissection of T cell-susceptibility genes in autoimmune diseases.
Keywords: genotype, human, T cell antigen receptor signaling, CTLA4, autoimmunity
The recent successful completion of several genome-wide association scans, together with the rapid advancement in high-throughput genotyping technologies, have resulted in the discovery of an ever-increasing number of genetic variants associated with susceptibility to human autoimmune diseases (1–5). However, because of the small genetic effects of these variants, compared with the strong genetic effects seen in Mendelian diseases, the study of genotypic variation in relation to phenotypes has been a substantial challenge. One well validated region associated with susceptibility to autoimmune disease harbors the CTLA4 gene on chromosome 2q33. An associated allele in this susceptibility region is the G allele of the SNP CT60 (rs3087243; A/G) in the CTLA4 gene region, which has been associated with risk to type 1 diabetes, Graves disease, autoimmune hypothyroidism, systemic lupus erythematosus, and Addison's disease (6–10).
The expression of mRNA isoforms of CTLA4 has been investigated in relation to CTLA4 genotype in healthy controls. Moreover, the genotype-dependent difference in the expression of soluble CTLA4 transcripts appears to be T cell-specific (6, 10). In the nonobese diabetic (NOD) mouse, the orthologous CTLA4 region also is associated with autoimmune diabetes, and, similar to humans, an allelic variant causes the increased expression of a major splice isoform. In the NOD model, the isoform is ligand-independent CTLA-4 (11) and results in strongly inhibited T cell responses, as well as increased susceptibility to disease (12). Together these investigations in both the experimental model and in humans suggest that genetic variation in the CTLA4 gene region may have an important effect on T cell function because of altered T cell signaling.
Although CTLA4 is important in autoimmune disease susceptibility, it is unknown how the autoimmune-associated allelic variant mechanistically influences the fundamental outcomes of T cell antigen receptor (TCR) engagement. A major difficulty of studying the potential relationship between a given genotype and T cell function has been the lack of high-throughput technologies in the study of signaling pathways. Multiparameter flow-cytometric analysis of phosphorylated proteins associated with TCR signaling allows both the processing of a large number of samples as well as more global analyses of signaling pathways, while requiring small numbers of cells (13). The ability to identify a small homogeneous cell population within the large heterogeneous mixture of cell populations found in human blood makes this technology particularly valuable in the field of immunogenetics.
To gain insight into the functional effects of CTLA4 allelic variants on T cell signaling, we examined a cohort of healthy individuals without self-reported symptoms of inflammatory disease stratified by their genetic variation in the CTLA4-susceptibility region. Specifically, by using phospho-site-specific mAbs, we tested the hypothesis that the autoimmune disease-associated allele at CTLA4 alters phosphorylation responses downstream of the TCR in naive and memory T cell subsets within human blood, which are reported to have different overall phosphorylation patterns upon TCR stimulation (14–18). Here we demonstrate that allelic variation found in autoimmune disease is associated with changes in the signaling threshold of CD4+ T cells. Although we cannot precisely identify how noncoding allelic variation at CTLA4 biochemically leads to altered immune function, these experiments provide a rational approach for the mechanistic examination of genotype/phenotype relationships in human inflammatory disease.
Results
Assay Development.
Characterization of CD4+CD45RAhigh and CD4+CD45RAlow T cells.
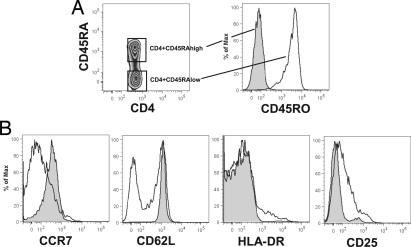
We distinguished naive and memory CD4+ T cells by the presence of high levels or lack of CD45RA expression, respectively (19, 20). We recognize that the absence or presence of CD45RA does not precisely divide T cells into memory and naive subsets (21, 22), and we also note that a small percentage of contamination of either subset is possible. Nevertheless, direct whole-blood staining demonstrated that the CD4+CD45RAhigh T cells are CD45RO−, CCR7high, CD62Lhigh, HLA-DR−, and CD25− (Fig. 1 A and B). Conversely, most CD4+CD45RAlow T cells were CD45ROhigh, CCR7low and were either low or high in CD62L expression. HLA-DR+ and CD25+ cells were only found within the CD4+CD45RAlow subset. Thus, our analyses were based on subsets of ex vivo T cells highly enriched for naive or memory populations of CD4+ T cells.
Fig. 1.
Phenotypic characteristics of naive CD4+CD45RAhigh and memory CD4+CD45RAlow T cells. (A) CD4+CD45RAhigh T cells are CD45RO−. (B) Expression difference of subset-identifying cell-surface markers between naive CD4+CD45RAhigh and memory CD4+CD45RAlow T cells. Extracellular staining was performed on whole blood within 30 min of blood donation. The open histogram represents data gated on CD4+CD45RAhigh T cells, and the shaded histogram shows data gated on CD4+CD45RAlow T cells. A representative example is shown.
Titration of anti-CD3 stimulation in whole-blood samples.
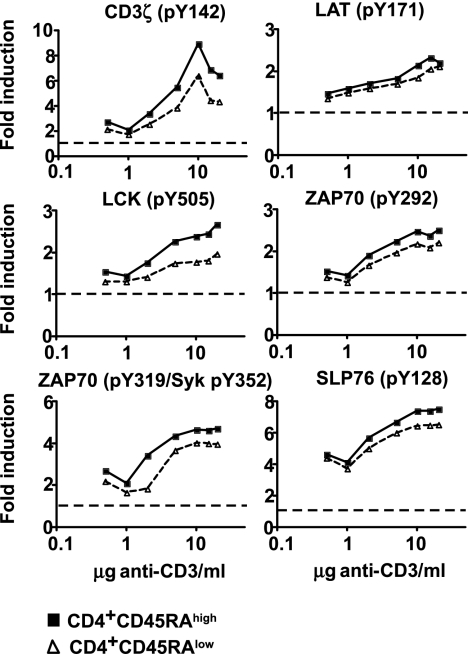
In an attempt to avoid experimental variation introduced by cell-purification procedures, which may induce phosphorylation events in cells, we opted to study TCR signaling in fresh whole blood. To determine the sensitivity of the flow cytometry-based technology used in whole blood, we performed titration experiments by stimulating whole blood with different concentrations of a soluble anti-CD3 mAb (UCHT1 clone). TCR stimulation by cross-linking CD3 over a range of anti-CD3 concentrations showed that 10 μg of anti-CD3/ml whole blood is a saturating dose for both CD4+CD45RAhigh and CD4+CD45RAlow T cells (Fig. 2). We also show that the system is highly sensitive because an anti-CD3 concentration of 1 μg/ml of whole blood already shows induction of phosphorylation above baseline at all markers studied. For all following experiments, signaling was induced with 10 μg of anti-CD3/ml of whole blood.
Fig. 2.
Induction of phosphorylation over a range of anti-CD3 concentrations. Phosphorylation was induced by incubation of whole blood at 37°C for 5 min. Titration results for both CD4+CD45RAhigh (filled squares) and CD4+CD45RAlow (open triangles) T cells are shown. Baseline levels are indicated by the dotted line. Donor-specific baseline levels are taken into account in the fold induction of phosphorylation.
Kinetics of phosphorylation events after anti-CD3 stimulation.
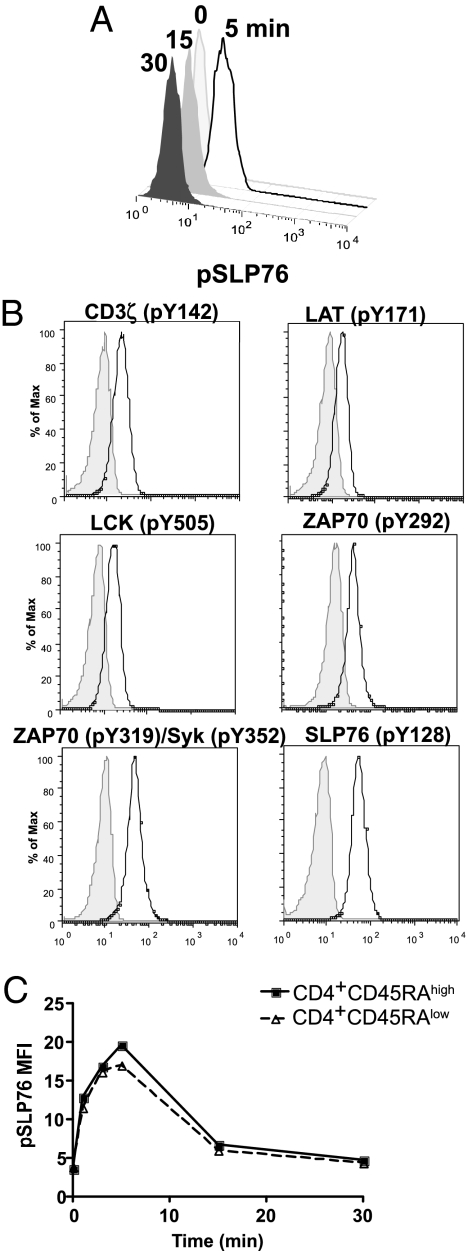
Having determined the saturating dose of anti-CD3 stimulation for whole blood, we then determined the kinetics of maximal phosphorylation after anti-CD3 stimulation for each site of phosphorylation that we targeted. To explore phosphorylation events distal to TCR activation, we selected six different tyrosine phosphorylation sites for study: CD3ζ (pY142), LAT (pY171), LCK (pY505), ZAP70 (pY292), ZAP70 (pY319/Syk Y352), and SLP76 (pY128). We measured each of these phosphorylation events at 0, 5, 15, and 30 min and followed changes in the level of phosphorylation over time, as demonstrated in Fig. 3A by a representative set of histograms showing raw cytometric data for SLP76 (pY128) over the 30-min time course in one blood sample. Representative histograms of the six phospho-site-specific Abs that we studied at the 5-min time point demonstrate that each of these six Abs clearly identifies a population of cells with phosphorylation events over the baseline levels (Fig. 3B). Both the CD4+CD45RAhigh and CD4+CD45RAlow T cell populations showed comparable kinetics (Fig. 3C). Thus, we were able to establish a common sampling time at which to evaluate all targeted tyrosine residues in both naive and memory T cells: A 5-min time point after whole-blood stimulation with 10 μg of soluble anti-CD3/ml of whole blood provided the optimal sampling point for the phosphorylation events studied here.
Fig. 3.
Time courses. (A) Histograms of raw flow-cytometric data gated on total CD4+ T cells. (B) Representative example of histograms showing the induction of phosphorylation with 10 μg/ml of soluble anti-CD3 at 5 min (shaded histogram, 0 min; open histogram, 5 min). (C) Representative example of a time course of pSLP76 phosphorylation. Mean values of three different healthy donors are shown.
Naive and memory CD4+ T cell populations differ in baseline phosphorylation states.
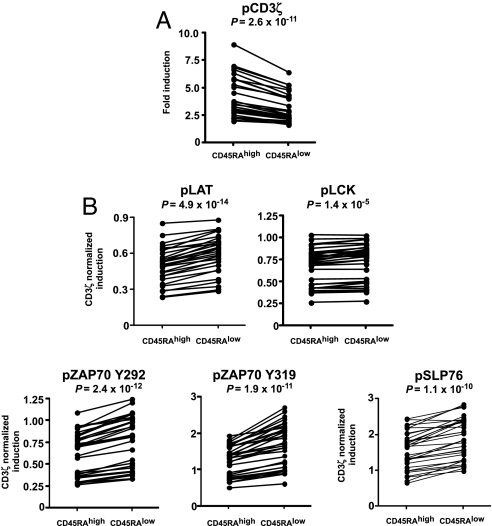
Given earlier observations of decreased tyrosine phosphorylation upon TCR stimulation in memory T cells (14–18), we first studied baseline phosphorylation levels of the six tyrosine residues in naive CD4+CD45RAhigh and memory CD4+CD45RAlow T cells. For all 32 subjects in our study, we collected the median fluorescence intensity for each of the six phospho-site-specific Abs in each of the two populations at time 0. We then compared the distribution of the median intensity values at each site of phosphorylation by using a paired t test. These analyses showed that, before stimulation, the extent of basal phosphorylation is significantly higher in ex vivo CD4+CD45RAlow memory T cells than in ex vivo CD4+CD45RAhigh naive T cells at all six sites of phosphorylation studied (P < 0.0005 for each of the six tyrosine residues) (Table 1).
Table 1.
Memory CD4+CD45RAlow T cells have a higher baseline phosphorylation level than CD4+CD45RAhigh T cells
| Variable | Mean difference between CD4+CD45RAlow and CD4+CD45RAhigh | 95% CI | P |
|---|---|---|---|
| pCD3ζ | 0.44 | 0.29–0.58 | 7.3 × 10−7 |
| pLAT | 0.65 | 0.49–0.82 | 3.7 × 10−9 |
| pLCK | 0.80 | 0.69–0.91 | 8.9 × 10−16 |
| pZAP70 (Y292) | 2.17 | 1.84–2.50 | 2.6 × 10−14 |
| pZAP (Y319) | 0.80 | 0.62–0.97 | 1.7 × 10−10 |
| pSLP76 | 0.57 | 0.28–0.86 | 3.8 × 10−4 |
Mean differences between naive CD45RAhigh and memory CD45RAlow subsets are shown for CD4+ T cells. Positive values indicate that the median fluorescence intensity was higher in the CD45RAlow subset. P values were obtained by paired t tests analyzing CD4+CD45RAhigh and CD4+CD45RAlow T cells from 32 healthy blood donors.
Increased overall tyrosine phosphorylation in memory CD4+CD45RAlow T cells upon pCD3ζ normalization.
Having shown that naive CD4+CD45RAhigh T cells display significantly lower baseline phosphorylation levels at all investigated tyrosine residues, we then examined the signaling events associated with TCR cross-linking with anti-CD3 mAb. Comparing the induction of phosphorylation in CD4+CD45RAhigh and CD4+CD45RAlow T cells 5 min after anti-CD3 stimulation (Fig. 4A), the CD4+CD45RAlow subset exhibited significantly less overall tyrosine phosphorylation, consistent with data obtained by others (14–18). This finding was apparent for all tyrosine residues examined, including the most proximal TCRζ residue (Fig. 4A).
Fig. 4.
Induction of phosphorylation in CD4+CD45RAhigh and CD4+CD45RAlow T cells. (A) CD4+CD45RAhigh T cells show greater induction of phosphorylation than CD4+CD45RAlow T cells, as shown by pCD3ζ phosphorylation. For all other studied proteins, this pattern was statistically significant too, except for ZAP70 (Y319/Syk Y352). (B) Normalization to pCD3ζ reveals increased phosphorylation in CD4+CD45RAlow T cells compared with CD4+CD45RAhigh T cells at all phospho-sites studied. Phosphorylation is shown at 5 min with 10 μg of anti-CD3/ml blood.
We then developed an analytical model to control for the strength of the initial stimulating signal through the TCR, which may differ among individuals/time points examined because of subtle differences in surface TCR expression. This transformation normalized each median intensity value for a particular phosphorylated protein by dividing it by the median intensity for CD3ζ (pY142) and allows us to compare the efficiency of the signaling cascade independent of alterations in the initial TCR stimulation event. Fig. 4B displays the results of these analyses, which demonstrate that the proportion of phosphorylated residues at the five signaling molecules distal to CD3ζ (pY142) in the signaling cascade is actually greater when normalized to initial CD3ζ (pY142) phosphorylation in memory CD4+CD45RAlow T cells, compared with naive CD4+CD45RAhigh T cells.
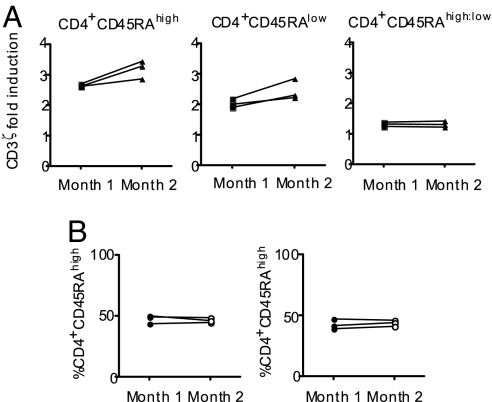
The ratio of CD4+ naive:memory TCR signaling is a stable phenotype.
To investigate the effects of CTLA4 genotypes on TCR signaling, it was important to select the most robust parameter for use in the comparison of a large number of subjects. First, we evaluated possible parameters on both the stability of the signaling phenotype over time (i.e., whether an individual's median intensity values for the phosphoAbs fluctuates longitudinally) and the sensitivity to variation in sample preparation and quantitation. Fixation, permeabilization, staining, and measurement on a flow cytometer may all affect the median fluorescence intensity of any given marker. We tested three donors in independent experiments performed 1 month apart. Although the raw values of TCR-induced protein phosphorylation for the CD4+ naive and memory subsets differed considerably when assessed 1 month apart, the ratio of the sensitivity to CD3-mediated signaling when comparing CD4+CD45RAhigh to CD4+CD45RAlow T cells remained constant over time (Fig. 5). These data indicate that, at any given time point, elevated or reduced T cell sensitivity to TCR signaling remains comparable among naive and memory populations of CD4+ T cells (i.e., coordinate regulation in the two subsets). In addition, this ratio is informative because it corrects for the fluctuation in median intensity values that were observed in the earlier longitudinally assessed samples, which are probably, to a large extent, because of technical variance in our measurements. These data, which are captured ex vivo from whole blood, (i) highlight the power of our approach to characterize subtle differences in phosphorylation states across a signaling pathway with minimal manipulation of cells, and (ii) provide a rationale to analyze specific T cell subpopulations in generating data for our analyses correlating immunophenotype with genotype.
Fig. 5.
The ratio of CD4+CD45RAhigh and CD4+CD45RAlow T cells is a stable phenotype over time. (A) Note that a CD4+CD45RAhigh:low ratio of >1 indicates that the induction of phosphorylation was greater in CD4+CD45RAhigh than in CD4+CD45RAlow T cells. Whole blood was stimulated with 10 μg of soluble anti-CD3 per milliliter of blood for 5 min at 37°C. These data are representative of all other phosphomarkers studied. Median fluorescence at 5 min is shown, with donor-specific background phosphorylation levels taken into account. (B) The percentages of CD4+CD45RAhigh and CD4+CD45RAlow T cells remained constant.
Genetic Analysis of TCR Signaling.
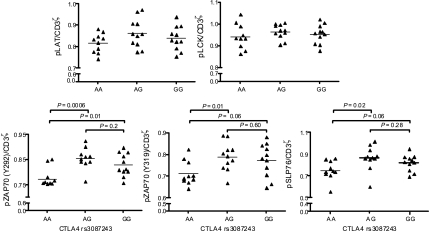
We investigated the phosphoprotein ratios of pLCK, pLAT, pZAP70, and pSLP76 in CD4+ naive:memory T cells after TCR signaling as the primary response variable for our analysis of the effect of the CTLA4 allelic variant on TCR signaling. To control for the initial TCR signal, we normalized this ratio to the pCD3ζ signal such that signal propagation distal to the TCR could be studied. Any effect that selectively influenced either the naive or memory subset would be seen as an altered ratio of CD4+ naive:memory TCR signaling. We analyzed T cell signaling data based on the categorization of the cohort into the three genotype classes: AA, AG, and GG. At ZAP70 (Y292), donors with the susceptible G allele showed a greater (pCD3ζ normalized) CD4+ naive:memory signaling ratio (Fig. 6). Quantitative trait analysis showed an association between the G allele and increased phosphorylation ratios (P = 0.01). In a pairwise comparison of genotype classes, donors with the AA genotype differed significantly from AG (P = 0.006) and GG (P = 0.01) donors (Fig. 6). The other ZAP70 (pY315/Syk pY352) and SLP76 (pY128) residues followed similar, statistically significant trends. We note that the association of the phosphorylation ratio was consistent between the markers because the extent of phosphorylation among these proteins is highly correlated [RZAP70(Y319) and ZAP70 (Y292)2 = 0.58, P < 0.0001; RSLP76 and ZAP70 (Y292)2 = 0.43, P < 0.0001; RZAP70(Y319) and SLP762 = 0.79, P < 0.0001].
Fig. 6.
CTLA4 genotype-dependent phosphorylation pattern. Subjects with the AG and GG CTLA4 genotype at rs3087243 show different signaling patterns in CD4+CD45RAhigh and CD4+CD45RAlow T cells, compared with the AA genotype. Data were normalized to the pCD3ζ signal. Whole blood was stimulated for 5 min with 10 μg/ml soluble anti-CD3. Donor-specific baseline phosphorylation was accounted for. n(AA), 10; n(AG), 11; n(GG), 11.
The observed association between CT60 genotype and the signaling phenotype could be due to an effect of the CT60 variant on the percentages of CD4+CD45RAhigh and CD4+CD45RAlow T cells. However, when the percentages of CD4+CD45RAhigh and CD4+CD45RAlow T cells were added as covariates in a regression analysis, no effect on the association (P > 0.05) was observed. Donor gender and age also did not affect this phenotype (P > 0.05).
To address the question of whether the amount of CTLA4 affects TCR signaling, total CTLA-4 levels in CD4+CD45RAhigh and CD4+CD45RAlow T cells ex vivo from 15 donors were measured (three AA, eight AG, and five GG), and no genotypic differences in expression were observed (data not shown). We also have examined whether blocking of extracellular CTLA-4 influences TCR signaling induced by CD3 cross-link. The addition of 10 μg of anti-CTLA-4 (clone 14D3) per ml of whole blood before CD3 cross-link did not result in altered phosphorylation of any of the tyrosine residues studied, compared with the addition of an irrelevant IgG or CD3 cross-link alone in three healthy subjects (data not shown).
Discussion
Allelic variation in the CTLA4 gene region has been clearly associated with risk for a number of autoimmune diseases, but the functional consequences of this variant are largely unknown. Here we present direct evidence for an association between this autoimmune disease variant and proximal TCR signaling events. Expression of this variant is phenotypically associated with phosphorylation changes for ZAP70 and SLP76 in CD4+ T cells. We hypothesize that this genotype is associated with a phenotype where autoreactive T cells are stochastically more likely to become activated as a risk of developing autoimmune disease.
T cell phenotypes in relationship to the CTLA4 genotype at the CT60 SNP have been previously examined. Expression of a soluble mRNA isoform of CTLA4 relative to a full-length form is correlated within healthy volunteers (10). Subjects with the susceptible genotype at CT60 have less sCTLA4 mRNA than subjects with the protective genotype. Atabani and colleagues (6) confirmed this observation in their study of CTLA4 isoforms in purified T cells and demonstrated an association of CT60 and sCTLA4 mRNA levels with the frequency of CD4+CD25+ regulatory T cells in healthy human volunteers. Importantly, the genotype-dependent difference in the expression of soluble CTLA4 transcripts may be T cell-specific because it is not observed when steady-state CTLA4 mRNA from peripheral blood mononuclear cells, rather than purified T cells, are studied (23). The orthologous CTLA4 region in the mouse also is associated with autoimmune diabetes in the NOD mouse. A SNP in exon 2 of the mouse gene causes the increased expression of a major splice isoform, called ligand-independent CTLA4 (11), and results in strongly inhibited T cell responses, as well as increased susceptibility to disease (12). Together these investigations in both experimental models and in humans suggest that genetic variation in the CTLA4 gene may have an important effect on T cell function because of altered T cell signaling.
CD45 has tyrosine phosphatase activity and may modulate early signaling in T cells by differential association with the TCR (24). It is paradoxical that memory T cells exhibit rapid and robust responses to antigen at low-activation thresholds in the context of decreased TCR-related tyrosine phosphorylation in both humans (16) and mice (17). It has been proposed that these observations may result from the ability of memory CD4+ T cells to more effectively process an activation signal downstream of the TCR (25). Our data are consistent with this hypothesis and suggest that CD4+ memory T cells more efficiently couple initial TCR signals to further downstream signals.
The relative responsiveness of the CD4+CD45RAhigh versus CD4+CD45RAlow subsets to TCR signaling was altered in donors with the susceptible G allele at CT60, whereas there was no statistical difference between the AG and GG genotype classes, indicating a dominant effect of the G allele. We note that only three of the five tyrosines show a genotype association. The observation that pLAT and pLCK did not show association with the G allele may be because the pLAT and pLCK signals only showed minor differences between CD4+CD45RAhigh and CD4+CD45RAlow T cells with CD3ζ normalization (Δ = 0.09 and 0.03, respectively), compared with those seen for pZAP70 (Y292), pZAP70 (Y319)/Syk (Y352), and pSLP76 (Y128) (Δ = 0.14, 0.42, and 0.35, respectively). However, an interesting alternative explanation may be that the CT60 genotype at CTLA4 may have a more profound effect at the less proximal TCR signaling events, including ZAP70 and SLP76.
Both ZAP70 Y292 and ZAP70 Y315/Syk Y352 phosphorylation patterns showed similar trends in the genotypic analysis, which was of interest because Y292 has been reported to play a role in attenuating the TCR signal and Y315 in the enhancement of ZAP70 function (26–28). The downstream signaling consequences of these two opposing regulatory effects of Y292 and Y315 is unclear, made more difficult because the phosphorylation signal obtained at Y315 also is capturing that of Syk Y352 because this anti-ZAP70 Ab cross-reacts with the Syk Y352 residue.
The mechanism by which the autoimmune susceptibility allele of CT60 at CTLA4 may cause an altered coordinate responsiveness of CD4+CD45RAhigh and CD4+CD45RAlow T cells will be difficult to dissect particularly because the mechanism for CTLA4 modulation of T cell inhibition is as yet unknown. Lee and colleagues (29) showed that CTLA4 is able to directly associate with the TCRζ chain and that this association resulted in ζ chain dephosphorylation. However, we do not propose that the level of CTLA4 expression, which may be indirectly related to the CT60 allele, is associated with the altered responsiveness to TCR stimulation of CD4+CD45RAhigh and CD4+CD45RAlow T cells because we have not observed any significant differences in total CTLA-4 expression in these two populations among a subset of subjects in the three genotype classes (data not shown). This observation may not be surprising because it is the mRNA species encoding a soluble CTLA-4 isoform that has been associated with the CT60 allele (6, 10). Its measurement at the protein level is, as yet, not possible because of the lack of suitable mAbs for this isoform.
It is difficult to conclude whether the major effect of the CT60 allele is to alter the threshold of activation of naive as compared with memory T cells. The CT60 allele may affect the memory T cell subset because these cells may be subject to greater regulation by CTLA-4 (30) and thus potentially their isoforms. Moreover, although autoreactive T cells are present in healthy subjects and those with autoimmune disease, those circulating in individuals with autoimmune disease are in an activated/memory state (31–33). However, it is tempting to speculate that the CT60 allelic variant in the CTLA4 region lowers the threshold of activation by naive, CD4+CD45RAhigh T cells, allowing self-antigen reactive T cells to enter into an activated state directly leading to autoimmune disease.
Although the CT60 variant may be used as a surrogate marker to capture autoimmune susceptibility in the CTLA4 region (10), we note that the true causal variant may be, as yet, unidentified. It is possible that the susceptibility allele may not only have an effect on CTLA4, but also on other genes in the region (such as an inducible costimulator), as has been shown in the orthologous region of the NOD mouse (11, 34).
In T cells obtained from peripheral blood of subjects with the autoimmune disease systemic lupus erythematosus, defective expression and tyrosine phosphorylation of the TCRζ chain has been reported (35–37). The molecular mechanisms of expression and phosphorylation defects in the TCRζ chain may involve distinct DNA sequences in the 3′ UTR of the CD247 gene encoding TCRζ. The 3′ UTR sequences have been proposed to lead to mRNA stability and translation of an alternatively spliced form of TCRζ mRNA that is correlated with the TCRζ defect (38, 39). We note that these signaling phenotypes, including the one we have studied, will be regulated by variation in not just a single gene, but will be orchestrated by many genetic variants present in an individual. The identification of these allelic variants will allow future studies to dissect the relative roles of one or a combination of alleles on TCR signaling.
Phosphorylation patterns have predominantly been analyzed by in vitro kinase assay and antityrosine immunoblotting that used cell lines or negative or positive selection procedures to purify specific cell types and study certain isolated populations. Here we used multicolor flow-cytometric technology to study phosphorylation events at a single cell level. Polychromatic flow cytometry has been successfully applied to complex cell populations in human peripheral blood (13, 40), murine immunological (41), and cancer cell signaling studies (42). We show that this highly sensitive technology allows the direct interrogation of fresh, unmanipulated ex vivo samples, which are more reflective of in vivo immune profiles. In addition, we believe that the study of the most proximal phenotype to the genotypic variant of interest will increase the likelihood to detect subtle differences dependent on the genotype. The ability to study phosphorylation events at a single-cell level also may be beneficial in the study of rare immune cell populations. Indeed, we have already successfully applied our multiparameter flow cytometry-based analysis to investigate signaling properties of certain regulatory T cell populations in whole blood (L.M.M., D.E.A., C. Baecher-Allan, and D.A.H., unpublished data).
In conclusion, we have used polychromatic flow cytometry to investigate the immune phenotype associated with allelic variation in the CTLA4 gene region. We find that the CT60 variant associated with autoimmune disease alters the signaling pattern of CD4+ T cells, demonstrating a direct mechanistic link between the allelic variation in the CTLA4-susceptibility region and T cell function. Finally, these studies describe a platform for the rapid, precise, and more comprehensive human genotype-to-phenotype investigations required for understanding how allelic variation mediates human inflammatory diseases.
Methods
Subjects.
All human blood samples were obtained with informed consent and according to the Institutional Ethics Review Board Protocols. The 32 healthy individuals of Caucasian origin used in this study were without self-reported symptoms of inflammatory disease and included 16 females and 16 males. The age range was 19–45 years, with a mean age of 29.7.
Blood Collection.
All blood samples were collected in sterile 10-ml lithium heparin Monoject tubes. Immediately upon blood collection, blood samples were put on ice and processed within 45 min of collection to prevent artifactual increases in baseline phosphorylation levels.
Assessment of T Cell Receptor Signaling.
Phosphorylation-state analysis was performed on human whole blood by using BD Phosflow technology according to the manufacturer's instructions (BD Biosciences, San Diego, CA). Two milliliters of whole blood were incubated with soluble anti-CD3 (UCHT1) mAb (BD Biosciences) in 50-ml polypropylene Falcon (Cowley, UK) conical tubes kept on ice for 15 min and washed with ice-cold PBS. After centrifugation, supernatants were removed, and goat anti-mouse Ig for CD3 cross-linking was added. After incubation on ice for 15 min, tubes were transferred to a 37°C water bath. After induction of phosphorylation at 37°C, cells were fixed using 10 ml of BD Phosflow Lyse/Fix Buffer and permeabilized with 1 ml of ice-cold BD Perm Buffer III (BD Biosciences) for 30 min on ice. Cells were washed twice with 1% FBS/PBS and incubated with 10% mouse serum for blocking for 20 min.
In the CTLA-4 blocking experiment, the previous protocol was used, with the exception that, during incubation with anti-CD3 mAb, 10 μg of anti-CTLA4 (clone 14D3) per ml of whole blood or an irrelevant IgG was added.
FACS Staining.
For four-color cell surface and intracellular staining, 250,000 cells were stained with CD4-PerCP, CD3-APC, and two phospho-site-specific mAbs conjugated with Alexa Fluor-488 or PE (BD Biosciences): PE mouse anti-CD3ζ (CD247) (pY142, clone K25–407.69), PE mouse anti-LAT (pY171, clone 158-1169), Alexa Fluor 488 Mouse anti-Lck (pY505, clone 4/LCK-Y505), PE mouse anti-ZAP70 (pY292), PE mouse anti-ZAP70 (pY319)/Syk (pY352, clone 17A/P-ZAP70), and Alexa-Fluor 488 mouse anti-SLP-76 (pY128, clone J141–668.36.58). We note that the anti-ZAP70 Y319 Ab cross-reacts with Syk Y352 because of sequence homology. Staining was performed at room temperature for 30 min, after which cells were fixed by using a 1% paraformaldehyde/PBS solution. All samples were acquired on a FACS Calibur (BD Biosciences) by using CellQuest software within 12 h after staining. The level of phosphorylation was measured by dividing the median fluorescence intensity given by cells at 5 min by the background median fluorescence intensity. Data were analyzed by using FlowJo version 6.4.7 (TreeStar, Ashland, OR).
For extracellular staining as shown in Fig. 1, the following Abs (all from BD Biosciences) were used: Alexa 700 anti-CD4 (clone RPA-T4), APC anti-CD62L (clone Dreg 56), PerCP anti-HLA-DR (clone L243), APC anti-CD25 (clone M-A241), FITC anti-CD45RA (clone HI100), APC anti-CD45RA (clone HI100), FITC anti-CD45RO (clone UCHL1), and PE anti-CCR7 (clone 3D12). Whole blood was stained with the recommended amount of mAbs, and RBC lysis was performed with BD FACS Lysing Solution (BD Biosciences).
Staining for total CTLA-4 was performed by using PE anti-CTLA-4 (clone BNI3) and the BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) according to the manufacturer's instructions.
Genotyping of the rs3087243 (CT60) Variant at CTLA4.
Genomic DNA was extracted from whole blood by using the Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN). All samples were genotyped in duplicate by using restriction fragment length polymorphism (10) and TaqMan (Applied Biosystems, Foster City, CA).
Statistical Analysis.
Pairwise comparison of genotype classes was performed by using Student's t tests. Quantitative trait analysis was performed by using the PLINK toolkit version 0.99r by S. Purcell (http://pngn.mgh.harvard.edu/purcell/plink/).
Acknowledgments
We thank Vissia Viglietta for performing phlebotomy from healthy controls, Khadir Raddassi for help with blood donor appointments, and all blood donors for their participation in this study. This work was supported by National Institutes of Health Grants R01 AI39229, R01 AI44447, and P01 AI39671 (to D.A.H.); the American Cancer Society (D.E.A.); the Wellcome Trust and Juvenile Diabetes Research Foundation (L.S.W.); a Juvenile Diabetes Research Foundation Postdoctoral Fellowship (to L.M.M.); and a Jacob Javits Merit Award (NS2427 to D.A.H.) from the National Institute of Neurological Disorders and Stroke.
Abbreviations
- NOD
nonobese diabetic
- TCR
T cell antigen receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.P.N. is a guest editor invited by the Editorial Board.
References
- 1.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, et al. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 3.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 5.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atabani SF, Thio CL, Divanovic S, Trompette A, Belkaid Y, Thomas DL, Karp CL. Eur J Immunol. 2005;35:2157–2162. doi: 10.1002/eji.200526168. [DOI] [PubMed] [Google Scholar]
- 7.Blomhoff A, Lie BA, Myhre AG, Kemp EH, Weetman AP, Akselsen HE, Huseby ES, Undlien DE. J Clin Endocrinol Metab. 2004;89:3474–3476. doi: 10.1210/jc.2003-031854. [DOI] [PubMed] [Google Scholar]
- 8.Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA. Diabetologia. 2007;50:741–746. doi: 10.1007/s00125-007-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres B, Aguilar F, Franco E, Sanchez E, Sanchez-Roman J, Jimenez Alonso J, Nunez-Roldan A, Martin J, Gonzalez-Escribano MF. Arthritis Rheum. 2004;50:2211–2215. doi: 10.1002/art.20347. [DOI] [PubMed] [Google Scholar]
- 10.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 11.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, et al. J Immunol. 2004;173:164–173. doi: 10.4049/jimmunol.173.1.164. [DOI] [PubMed] [Google Scholar]
- 12.Vijayakrishnan L, Slavik JM, Illes Z, Greenwald RJ, Rainbow D, Greve B, Peterson LB, Hafler DA, Freeman GJ, Sharpe AH, et al. Immunity. 2004;20:563–575. doi: 10.1016/s1074-7613(04)00110-4. [DOI] [PubMed] [Google Scholar]
- 13.Perez OD, Nolan GP. Immunol Rev. 2006;210:208–228. doi: 10.1111/j.0105-2896.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 14.Farber DL, Acuto O, Bottomly K. Eur J Immunol. 1997;27:2094–2101. doi: 10.1002/eji.1830270838. [DOI] [PubMed] [Google Scholar]
- 15.Farber DL, Luqman M, Acuto O, Bottomly K. Immunity. 1995;2:249–259. doi: 10.1016/1074-7613(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 16.Hall SR, Heffernan BM, Thompson NT, Rowan WC. Eur J Immunol. 1999;29:2098–2106. doi: 10.1002/(SICI)1521-4141(199907)29:07<2098::AID-IMMU2098>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 17.Hussain SF, Anderson CF, Farber DL. J Immunol. 2002;168:1557–1565. doi: 10.4049/jimmunol.168.4.1557. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan S, Warke VG, Nambiar MP, Wong HK, Tsokos GC, Farber DL. Blood. 2001;97:3851–3859. doi: 10.1182/blood.v97.12.3851. [DOI] [PubMed] [Google Scholar]
- 19.Merkenschlager M, Terry L, Edwards R, Beverley PC. Eur J Immunol. 1988;18:1653–1661. doi: 10.1002/eji.1830181102. [DOI] [PubMed] [Google Scholar]
- 20.Young JL, Ramage JM, Gaston JS, Beverley PC. Eur J Immunol. 1997;27:2383–2390. doi: 10.1002/eji.1830270937. [DOI] [PubMed] [Google Scholar]
- 21.Amyes E, McMichael AJ, Callan MF. J Immunol. 2005;175:5765–5773. doi: 10.4049/jimmunol.175.9.5765. [DOI] [PubMed] [Google Scholar]
- 22.Brod SA, Rudd CE, Purvee M, Hafler DA. J Exp Med. 1989;170:2147–2152. doi: 10.1084/jem.170.6.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anjos SM, Shao W, Marchand L, Polychronakos C. Genes Immun. 2005;6:305–311. doi: 10.1038/sj.gene.6364211. [DOI] [PubMed] [Google Scholar]
- 24.Novak TJ, Farber D, Leitenberg D, Hong SC, Johnson P, Bottomly K. Immunity. 1994;1:109–119. doi: 10.1016/1074-7613(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 25.Chandok MR, Farber DL. Semin Immunol. 2004;16:285–293. doi: 10.1016/j.smim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Kong G, Dalton M, Wardenburg JB, Straus D, Kurosaki T, Chan AC. Mol Cell Biol. 1996;16:5026–5035. doi: 10.1128/mcb.16.9.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnan A, Di Bartolo V, Mura AM, Boyer C, Richelme M, Lin YL, Roure A, Gillet A, Arrieumerlou C, Acuto O, et al. J Exp Med. 2001;194:491–505. doi: 10.1084/jem.194.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q, Weiss A. Mol Cell Biol. 1996;16:6765–6774. doi: 10.1128/mcb.16.12.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KM, Chuang E, Griffin M, Khattri R, Hong DK, Zhang W, Straus D, Samelson LE, Thompson CB, Bluestone JA. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 30.Jago CB, Yates J, Camara NO, Lechler RI, Lombardi G. Clin Exp Immunol. 2004;136:463–471. doi: 10.1111/j.1365-2249.2004.02478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovett-Racke AE, Trotter JL, Lauber J, Perrin PJ, June CH, Racke MK. J Clin Invest. 1998;101:725–730. doi: 10.1172/JCI1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholz C, Patton KT, Anderson DE, Freeman GJ, Hafler DA. J Immunol. 1998;160:1532–1538. [PubMed] [Google Scholar]
- 33.Viglietta V, Kent SC, Orban T, Hafler DA. J Clin Invest. 2002;109:895–903. doi: 10.1172/JCI14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greve B, Vijayakrishnan L, Kubal A, Sobel RA, Peterson LB, Wicker LS, Kuchroo VK. J Immunol. 2004;173:157–163. doi: 10.4049/jimmunol.173.1.157. [DOI] [PubMed] [Google Scholar]
- 35.Liossis SN, Ding XZ, Dennis GJ, Tsokos GC. J Clin Invest. 1998;101:1448–1457. doi: 10.1172/JCI1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang M, Setoyama Y, Tsuzaka K, Yoshimoto K, Amano K, Abe T, Takeuchi T. Clin Exp Immunol. 2002;129:160–168. doi: 10.1046/j.1365-2249.2002.01833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi T, Tsuzaka K, Pang M, Amano K, Koide J, Abe T. Int Immunol. 1998;10:911–921. doi: 10.1093/intimm/10.7.911. [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury B, Krishnan S, Tsokos CG, Robertson JW, Fisher CU, Nambiar MP, Tsokos GC. J Immunol. 2006;177:8248–8257. doi: 10.4049/jimmunol.177.11.8248. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury B, Tsokos CG, Krishnan S, Robertson J, Fisher CU, Warke RG, Warke VG, Nambiar MP, Tsokos GC. J Biol Chem. 2005;280:18959–18966. doi: 10.1074/jbc.M501048200. [DOI] [PubMed] [Google Scholar]
- 40.Perez OD, Nolan GP. Nat Biotechnol. 2002;20:155–162. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 41.Krutzik PO, Hale MB, Nolan GP. J Immunol. 2005;175:2366–2373. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- 42.Irish JM, Kotecha N, Nolan GP. Nat Rev Cancer. 2006;6:146–155. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]