Abstract
The cell-mediated immune profile induced by a recombinant DNA vaccine was assessed in the simian/HIV (SHIV) and macaque model. The vaccine strategy included coimmunization of a DNA-based vaccine alone or in combination with an optimized plasmid encoding macaque IL-15 (pmacIL-15). We observed strong induction of vaccine-specific IFN-γ-producing CD8+ and CD4+ effector T cells in the vaccination groups. Animals were subsequently challenged with 89.6p. The vaccine groups were protected from ongoing infection, and the IL-15 covaccinated group showed a more rapidly controlled infection than the group treated with DNA vaccine alone. Lymphocytes isolated from the group covaccinated with pmacIL-15 had higher cellular proliferative responses than lymphocytes isolated from the macaques that received SHIV DNA alone. Vaccine antigen activation of lymphocytes was also studied for a series of immunological molecules. Although mRNA for IFN-γ was up-regulated after antigen stimulation, the inflammatory molecules IL-8 and MMP-9 were down-regulated. These observed immune profiles are potentially reflective of the ability of the different groups to control SHIV replication. This study demonstrates that an optimized IL-15 immune adjuvant delivered with a DNA vaccine can impact the cellular immune profile in nonhuman primates and lead to enhanced suppression of viral replication.
Keywords: HIV vaccine, immune response, cytokine adjuvant, T cell immunity
Cellular immune responses play an important role in the control of HIV-1 infection (1–6). Accordingly, vaccines that induce HIV-specific cellular immunity are being pursued (7–25). The exact nature of how the antigen-specific lymphocytes should be armed is still unknown. In the primate simian/HIV (SHIV) and simian immunodeficiency virus (SIV)-challenge models, induction of a high level of CD8 T cell immunity is associated with reduced viremia and protection of CD4 cells (19–25). A number of studies in primates as well as HIV-infected humans have indicated that suppressed viral replication and long-term nonprogression are associated with higher and more complex cellular immune responses. However, to date there has been only a limited understanding of the immune responses in vaccinated animals, which maintain control of viral replication.
DNA vaccines are attractive candidates for vaccine development because of their safety and relative ease of production (26–29). Studies have experimented with engineering DNA vaccines to include their own immune expansion signals (30–38). In mouse studies, one of the most potent drivers of cytotoxic T lymphocyte (CTL) immunity was IL-15 (35). IL-15 has more recently gained a great deal of attention in both mouse and macaque models because of its role in enhancing and prolonging antigen-specific CD8+ memory T cell survival. IL-15 was first identified by Waldmann and colleagues (39), and by Grabstein and associates (40) and is now known to play an important role in stimulation, proliferation, and survival of a memory CD8+ T cells (41–47). Zhang and coworkers (45) demonstrated in an in vivo mouse model that IL-15 provides effective and discriminating stimulation of the memory phenotype, CD44hi CD8+, T cells. Ku et al. (42) reported the division of memory CD8+ T cells is stimulated by IL-15. The effects of IL-15 on CD8+ T cells have led to the use of IL-15 with vaccines. Kutzler et al. (35) found that IL-15 significantly enhanced the cytotoxic response when a plasmid encoding IL-15 was coadministered with an HIV-1 DNA vaccine and could improve protection in an influenza mouse challenge model. Moore et al. (46) further demonstrated that IL-15 could expand the innate and adaptive immune responses by coadministering the FL gene. Oh et al. (47) evaluated vaccines expressing either IL-2 or IL-15 in combination with HIV-1 gp160 antigen. Both cytokines enhanced long-lasting CD4+ cellular immunity. However, IL-15 also supported long lasting CD8+ T cell-mediated immunity.
In this study we demonstrate that the cytokine vaccine adjuvant pmacIL-15 induced a unique immunological profile in vivo that significantly impacted subsequent viral challenge outcome. Interestingly, the increase in IFN-γ was not, by itself, an adequate predictor of the vaccine's ability to control viral challenge. Importantly, infection versus vaccination produced unique immune phenotypes.
Results
IFN-γ Response to SIV239 Gag After Immunization.
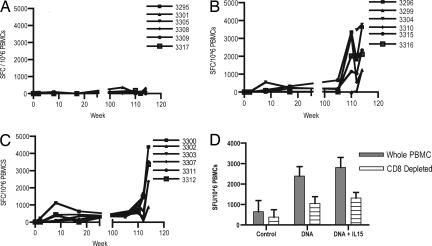
Three groups of macaques each were immunized by intramuscular injection (Table 1). We assessed the induction of an antigen-specific immune response to SIV gag in all macaques by an IFN-γ ELISA-linked immunospot (ELISpot) assay. After one immunization, one of six in the group that received pmacIL-15 (Fig. 1C) responded. After the second immunization, there was an increase in SIVgag cellular responses, three of six macaques responded in Group 2, which had an average of 135 spot-forming cells (SFC) per 106 peripheral blood mononuclear cells (PBMCs). An enhanced gag-specific immune response in Group 3 was also observed; six of six macaques had an average of 442 SFC/106 PBMCs (Fig. 1C). After the third immunization, there appeared to be further boosting in group 2, with little effect in group 3 animals.
Table 1.
Immunization schedule
| Immunization | Group 1 | Group 2 | Group 3 |
|---|---|---|---|
| Week 0 | Control | pSIVgag | pSIVgag + pmacIL-15 |
| Week 5 | Control | pSIVgag | pSIVgag + pmacIL-15 |
| Week 12 | Control | pSIVgag | pSIVgag + pmacIL-15 |
| Week 104 | Control | pSIVgag, pSIVpol, pHIVenv | pSIVgag pSIVpol, pHIVenv, pmacIL-15 |
| Week 108 | Control | pSIVgag, pSIVpol, pHIVenv | pSIVgag pSIVpol, pHIVenv, pmacIL-15 |
| Week 112 | Control | pSIVgag, pSIVpol, pHIVenv | pSIVgag pSIVpol, pHIVenv, pmacIL-15 |
Fig. 1.
IFN-γ-producing cells after the primary and secondary sets of immunization. Samples were taken 2 weeks after each injection and assessed for an SIVgag antigen-specific response by ELISpot. (A–C) The number of cells able to secrete IFN-g after SIVgag in vitro stimulation of PBMCs isolated from naïve macaques (A) pCSIVgag (B), pCSIVgag, and pmacIL15 immunized macaques (C) is presented as SFC per 1 million PBMCs. (D) After the final immunization, we evaluated the contribution of the CD8 T cells to the observed population of cells from macaques secreting IFN-γ.
Macaques were immunized three times at weeks 104, 108, and 112 with the DNA vaccine that encoded SIV gag. In addition, at these time points, SIVpol and HIV-1env plasmids were incorporated. We continued to use the ELISpot assay to monitor the number of SIVgag-specific IFN-γ-secreting effector cells (Fig. 1). At the time of the fourth injection, the average number of effector cells in the group immunized with plasmid vaccine alone was 225 SFC per 106 PBMCs. Animals that were coinjected with pmacIL-15 generated 355 SFC per 106 PBMCs.
In a dramatic fashion, the rest period appeared to substantially improve the vaccine-induced responses, by almost 10-fold. The number of effector cells in Group 2 increased to 2,460 SFC per 106 PBMCs after injection 4. Group 3, which received a coinjection of pmacIL-15, also had an increased number of effector cells (2,235 SFC per 106 PBMCs). After injections 5 and 6, the number of effectors able to secrete IFN-γ in groups 2 (2,389 SFC per 106 PBMCs) and 3 (2,389 SFC per 106 PBMCs) did not increase significantly. These data support that substantial immune maturation and expansion of vaccine-induced T cells takes place during the extended rest period.
We evaluated the contribution of CD8+ and CD4+ T cells to the observed population of cells secreting IFN-γ (Fig. 1D). One month after final immunization, isolated PBMCs and PBMCs depleted of CD8+ T cells were analyzed for the ability to secrete IFN-γ after stimulation with the gag peptide. Depletion of CD8+ T cells led to the assessment of the number of CD4+ T cells able to secrete IFN-γ. The animals that were vaccinated with DNA vaccine alone demonstrated a total number of effector cells of 2,389 SFC per 106 PBMCs with 1,057 CD4+ SFC per 106 PBMCs. The animals that were coimmunized with pmacIL-15 had a total of 2,808 SFC per 106 PBMCs and 1,316 CD4+ SFC per 106 PBMCs. Overall, the data demonstrate that IFN-γ results were similar between the vaccine groups at the time of viral challenge.
Control of Viral Replication.
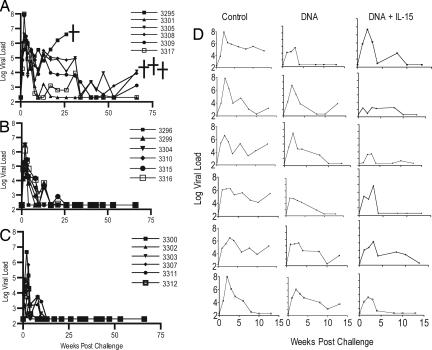
All animals were challenged with 300 monkey infectious doses (MID) SHIV89.6p by the i.v. route 11 weeks after the final injection. The average viral loads in the control group at week 2 after challenge or at the peak viral load was 7 logs (Fig. 2A). One macaque was killed at week 25, and three were killed at week 60 because of AIDS-like symptoms. Animals that received vaccine DNA alone controlled viral load by week 26 (Fig. 2B). All animals in the group that received DNA vaccine with pmacIL-15 controlled viral load by week 12 (Fig. 2C). The peak viral loads of group 2 was 5.6 logs. The viral loads for these animals was significantly lower by week 10 when compared with the control group (P = 0.020). The average peak viral load for animals that received pmacIL-15 was 3.8 log, which was significantly lower than the animals in the control group (P = 0.001). The viral loads in this group remained significantly lower than the control group at all time points (P < 0.05). In addition, the group of animals that received DNA plus pmacIL-15 exhibited a significantly lower viral load at weeks 2, 4, and 6 as compared with the group of animals that received DNA alone (P < 0.05).
Fig. 2.
Viral load after challenge of cynomologous macaques with 100 MID of SHIV89.6p. (A–C) Viral load is presented for control (A), SIV DNA (B), SIV DNA + pmacIL15-immunized (C) macaques. The assay has a threshold sensitivity of 200 RNA copies per milliliter of plasma with interassay variations averaging 0.5 log10. (D) The viral loads for the first 15 weeks for each individual macaque. Several animals were killed because of AIDS-like syndrome.
Lymph node biopsies were taken 57 weeks after challenge (Table 2.) The tissue samples demonstrated that, of the five animals that remained alive in the control group, two had virus-positive axillary and inguinal lymph nodes. Three of the six animals that received SHIV DNA had either a positive axillary or inguinal lymph node. Only one of six animals in the group that received IL-15 was demonstrated to be positive for virus in a single lymph node sample.
Table 2.
Viral replication in lymph nodes
| Control |
DNA |
DNA plus pmacIL-15 |
||||||
|---|---|---|---|---|---|---|---|---|
| Animal | Ax | Ing | Animal | Ax | Ing | Animal | Ax | Ing |
| 3295 | t | t | 3296 | + | − | 3300 | + | − |
| 3301 | − | − | 3299 | − | + | 3302 | − | − |
| 3305 | + | + | 3304 | − | + | 3303 | − | − |
| 3308 | + | + | 3310 | − | − | 3307 | − | − |
| 3309 | − | − | 3315 | − | − | 3311 | − | − |
| 3317 | NT | − | 3316 | − | − | 3312 | NT | − |
t, terminated prior to biopsy; Ax, axilary lymph node; Ing, Inguinal lymph node; +, positive; −, negative; NT, no tissue.
IFN-γ-Producing Cells After Challenge.
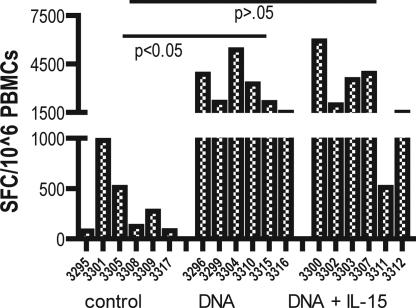
Samples were obtained 12 weeks after challenge and assessed for cells able to secrete IFN-γ after stimulation with SIVgag (Fig. 3). The cellular immune responses in the control group at no time point reached the same level of responses generated in the vaccine groups (Fig. 3). However, animal 3301, in the control group, was able to exhibit some control of viral replication. This animal reached a level of 1,000 SFC per 106 PBMCs. The group that was immunized with DNA vaccine alone had a significantly higher level of IFN-γ compared with the control group (P < 0.05). Interestingly, the low secondary response of animal 3311 resulted in the lack of a significant difference between the control group and the animals that received DNA combined with IL-15. We next sought to examine more markers to gain additional insight into the basis for immune control by these vaccines.
Fig. 3.
IFN-γ-producing cells after IV challenge with SHIV89.6p virus. Samples were taken 12 weeks after challenge. PBMCs were isolated by a standard Percoll separation technique and assessed for a gag antigen-specific response by ELISpot. The data represent the number of cells able to secrete IFN-γ after SIVgag in vitro stimulation of PBMCs isolated from vaccine-naïve macaques, pCSIVgag-vaccinated, and pCSIVgag- and pmacIL15-immunized macaques.
Proliferation of T Cells by DNA and IL-15 Coinjected Groups.
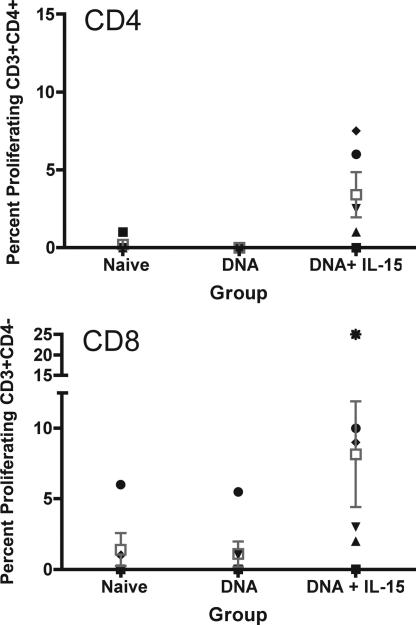
Lymphocytes that are fully differentiated are capable of proliferating after in vitro antigen stimulation. We investigated the ability of the vaccine-specific CD8+ and CD4+ effector cells to proliferate antigen stimulation. PBMCs were isolated from all macaques 2 weeks after the final immunization. Cells were incubated with CFSE, washed, and stimulated with SIVgag antigen for 5 days. The data obtained demonstrated little proliferative capacity for the lymphocytes isolated from macaques immunized with DNA vaccine. The proliferative responses were dramatically improved in pmacIL-15 coimmunized animal groups (Fig. 4). An average of 3% gag-specific CD4+ T cell and 8% of the CD8+ cells were proliferating in the pmacIL-15 covaccinated animals.
Fig. 4.
T cell proliferative responses to SIVgag. PBMCs were stained with CFSE and stimulated with SIVgag peptides for 5 days. Standard surface staining protocol was followed for CD3/CD4-positive cells.
Inflammatory Panel of Cytokines.
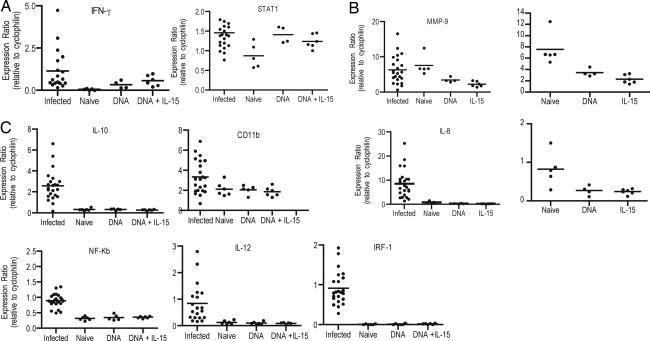
To further understand the immunological profiles induced by these DNA vaccines, PBMCs taken after the final vaccination were stimulated in vitro for 6–12 h with SIVgag antigen. After in vitro stimulation, RNA was isolated. In addition to our naïve controls and the two vaccine groups, PBMCs from SIV-infected macaques were isolated and stimulated in vitro in an analogous manner to the vaccine group (Fig. 5). Antigen-specific expression levels of a number of genes were altered as a result of SIV DNA vaccination. The genes for IFN-γ, STAT1 (Fig. 5A) are clearly up-regulated in PBMCs isolated from vaccinated macaques and stimulated with SIVgag. There was no expression of IFN-γ after antigen stimulation of PBMCs in the control group.
Fig. 5.
Gene expression after SIV gag stimulation, quantitative RT-PCR. Five million PBMCs were taken from the macaques 4 weeks after the sixth and final immunization stimulated in vitro with SIVgag peptides, and mRNA was extracted. Data are presented for effector function (IFN-g and STAT1) (A), inflammatory response (MMP-9 and IL-8) (B), and to assess other immunological markers of T cell activation (C). Data presented are mRNA levels compared with control mRNA of cyclophilin.
We further observed that MMP9 and IL-8 (Fig. 5B) were down-modulated in antigen-specific cells in the vaccine groups as compared with naïve cells. However, MMP-9 and IL-8 gene expression is high in SIV-infected virally suppressed macaques when the PBMCs were stimulated with SIVgag antigen. Furthermore, IL-8 gene expression in PBMCs of infected macaques was higher than PBMCs isolated from naïve macaques with SIVgag antigen.
Fig. 5C illustrates that although several genes do not vary between naïve and vaccinated animals, there is a clear increase in specific immune-related gene expression in SIV-infected macaques. Specifically, gene expression for IL-10, CD11b, NFκB, IL-12, and IRF-1 are increased above background levels or those observed in naïve animals. NFκB and IRF-1 are particularly interesting because HIV requires these transcription factors for its efficient intracellular replication. Antigen-specific cells that are not up-regulated in NFκB and IRF-1 expression may reduce the number of target cells for HIV-1 replication. These data suggest that an effective T cell vaccine should drive immune expansion in a manner that does not provide activation signals needed for efficient pathogen replication.
Discussion
In this study, we demonstrate significant protection against SHIV89.6p replication and pathogenesis in macaques coimmunized with SHIV DNA and a plasmid IL-15 adjuvant. Coimmunization with pmacIL-15 improved the ability to rapidly suppress viral replication and quickly control the infection. The group that was vaccinated with DNA alone also was able to control viral replication; however, the viral peak was several logs higher, and control of viral replication in all animals took two times longer, week 25 as compared with week 12 in the pIL-15 vaccine group. The SHIV89.6p challenge model can lead to a rapid and dramatic loss of CD4 lymphocytes as we observed in our control animals. Therefore, this model does not adequately represent what is seen in humans. Yet, despite this discrepancy, the model does allow us to associate the level of a lentiviral viral suppression as well as protection of CD4 lymphocytes with the observed immune response. Over the course of immunization, we noted that IL-15 plasmid adjuvant did not appear to dramatically increase the magnitude of IFN-γ-producing cells. However, the pIL-15 vaccine animals did exhibit higher proliferative responses, with a higher ratio of CD8+ T cell proliferation compared with CD4+ T cell proliferation.
During the differentiation of effector T cells into memory T cells, the proliferative potential in response to antigenic stimulation increases substantially (48–50). The ability to vigorously proliferate upon infection results in the generation of a large pool of secondary effector T cells and is believed to be central to providing optimal protective immunity. Oh et al. (51) proposed that IL-15 induced CD8 cells with higher avidity and suggested that these cells ultimately had higher levels of proliferation. In addition, Younes et al. (52) demonstrated that HIV-1-specific CD8+ T cells in acute HIV-1 infection exhibit strong ex vivo proliferative capacities, whereas this effector function is rapidly lost in the presence of ongoing viral replication (52). Importantly, Kutzler et al. (35) reported that IL-15 could drive CD4 independent maturation of effector CD8 T cells.
We assessed a series of genes associated with a cellular immune response. There was a measurable difference in only a small subset of genes between cells isolated from control animals and vaccinated animals. The level of IFN-γ gene expression increased when cells were stimulated with SIVgag peptides concurrently with increased gene expression of STAT1, a signal transducer activated by IFN. However, what was most interesting was the down-modulation for genes encoding IL-8 and matrix metalloproteinase 9 (MMP-9), two molecules associated with the proinflammatory state and the establishment of chronic infection. IL-8 is a chemokine produced during the inflammatory response to signal neutrophils. MMP-9 has also been associated with chronic inflammatory autoimmune diseases (53–56). We observed a decreased level of MMP-9 gene expression when cells from vaccinated macaques were stimulated with antigen as compared with naïve animals stimulated with SIVgag antigen. Inflammatory molecules do not appear to be associated with a vaccine response that efficiently controlled this infection challenge.
This study demonstrates that DNA vaccination plus pmacIL-15 can contribute to enhanced immune profiles and ultimately control SHIV89.6P challenge. The mechanism of control of viral replication has not completely been elucidated. Although it is likely that the number of effector cells able to secrete IFN-γ is most likely not the sole correlate of protection, the use of ELISpot analysis gives us the ability to make some comparison between studies where vaccinated macaques have demonstrated lower viral peaks and delayed viral replication after SHIV challenge. A study by Chong et al., (57) with DNA vaccines and IL-15/IL-12 adjuvants demonstrated that macaques immunized with SIVgag alone and challenged with 300 MID of SHIV89.6p had partial suppression of viral replication. The monkeys achieved a level of 1,000–5,000 IFN-γ-secreting cells. Many of the responses dropped to 50% by 4 weeks after the final injection, but the best controllers had the higher numbers of IFN-γ-producing cells. McKenna et al. (58), using an attenuated rabies vector that expressed HIVenv and SIVgag, induced a level of IFN-γ-secreting effector cells of 120–300 SFC per 106 PBMCs. Viral replication after challenge of 50 MID of SHIV89.6p was suppressed 3 logs. Demberg et al. (59), using adenovirus, observed higher levels (1,000–2,500) IFN-γ-secreting cells. And, when challenged with 30 MID of SHIV89.6P, viral loads were significantly lower than controls at set point, but complete control was not obtained. Studies by Thorner et al. (60) are using heterologous adenovirus immunizations to further increase the level of cellular immune responses. Finally, Robinson et al. (61) have incorporated GM-CSF into their DNA/MVA vaccine regimen. The GM-CSF enhanced antibodies to SHIV89.6p, but complete viral suppression was not observed.
Our data obtained from CFSE proliferation study demonstrated an average of 3% gag-specific CD4+ T cells, and 8% of the CD8+ cells were proliferating in the pmacIL-15 covaccinated animals. A higher proliferative capacity is indicative of higher precursor frequency. Further examination of such defined and adjuvanted DNA vaccines in challenge settings appear to be a useful tool for probing correlates of pathogen immunity and may provide interesting immune phenotypes for clinical study.
Materials and Methods
Animals.
Macaques were housed at the Southern Research Institute (SRI) in Frederick, MD. These facilities are accredited by the American Association for the Accreditation of Laboratory Animal Care International and meet National Institutes of Health standards as set forth in the Guidelines for Care and Use of Laboratory Animals. The University of Pennsylvania Institutional Animal Care and Use Committee (IACUC) reviewed and approved all procedures carried out by SRI.
DNA Plasmids.
The pCSIVgag plasmid expresses a 37-kDa fragment of the SIV core protein. This rev-independent expression vector and pCSIVpol and pCHIVenv have been optimized for high-level expression as described (62). Macaque IL-15 was cloned and expression analysis completed (sequence from GenBank, accession number U19843) as described by Kutzler (M.A.K., unpublished work).
Immunization Schedule and Sample Collection.
Plasmids were manufactured and purified by Puresyn (Malvern, PA) and formulated in 0.15 M citrate solution and 0.25% bupivicaine at a pH of 6.5. The immunization schedule is outlined in Table 1. Groups of six cynomologous macaques were immunized three times intramuscularly with buffer, 2 mg of pSIVgag DNA, or 2 mg of pSIVgag DNA coinjected with 2 mg of pmacIL-15. The macaques were then rested 84 weeks before performing the second set of immunizations. The second series of immunizations included an increase in dose to 3 mg of pCSIVgag and pmacIL-15 and incorporated 3 mg of pSIVpol and pHIVenv.
Peptides.
Peptides corresponding to the entire coding region of SIVmac239 gag protein were obtained from the AIDS Reagent Reference Repository (National Institutes of Health). These 15 oligomers overlapping by 11 aa were resuspended in DMSO at a final concentration of ≈100 mg/ml and mixed as pools for ELISpot analysis.
Ifn-γ ELISpot Assay.
EliSpot using IFN-γ reagents purchased from MabTech (Nacka Strand, Sweden) and nitrocellulose plates from Millipore (Billerica, MA) as performed previously (63). A positive response is defined as >50 SFC per 1 million PBMCs and two times above background. A second set of PBMCs was depleted of CD8+ lymphocytes with α-CD8 depletion beads according to the manufacturer's protocol (Dynal, Carlsbad, CA) before plating cells in triplicate with peptides.
CFSE Staining for T Cell Proliferation.
PBMCs were incubated with CFSE (5 μM) and incubated for 8 min at 37°C. The cells were washed and incubated with antigens (SIVp27/gag peptide mix) at a concentration of 5 μg/ml for 5days at 37°C in 96-well plates. Cultures without gag peptide was used to determine the background proliferative response. Standard surface-staining protocol was followed for CD4+ cells by using α-human CD4-PE (BD–Pharmingen, San Diego, CA) monoclonal antibody. The frequency of CD4+ T cells was determined by gating on CD4+ T cells. The data were analyzed by using the FlowJo program (Ashland, OR).
Gene Expression Analysis.
Five million PBMCs were stimulated for 6–12 h. PBMCs from infected macaques served as reference samples. The infected animals were treated with ART to suppress active viral replication, thus reducing viral pathogenesis. mRNA was isolated by using the RNA-BEE RNA isolation kit (TEL-TEST, Friendswood, TX).The gene-expression patterns of multiple genes were monitored with the VEM-XP multiplex assay (Althea Technologies, Inc., San Diego, CA) and the GenomeLab GeXP Analysis System Multiplex RT-PCR assay (Beckman, Fullerton, CA). For each reaction, 3 μl of RNA were mixed with 1.5 μl of 10× DNase buffer (Ambion, Austin, TX) as described earlier (64).
Infection.
Primates were challenged with 300 MID by the i.v. route with SHIV89.6P (kindly provided by Norman Letvin, Harvard University, Cambridge, MA) 11 weeks after the final boost.
SHIV Viral RNA Quantitation.
SHIV viral RNA was quantitated by using a procedure described by Silvera et al. (65). The assay has a threshold sensitivity of 200 RNA copies per milliliter of plasma with interassay variations averaging 0.5 log10.
Lymph Node Biopsies and in Situ Hybridization.
Formalin-fixed, paraffin-embedded lymph node biopsies were stained for SIV RNA by using a method previously described by Hirsch et al. (66). The sections were reacted with NBT/BCIP (Vector Laboratories, Orton Southgate, U.K.) for 10 h, rinsed with distilled water, counterstained with nuclear fast red (Sigma, St. Louis, MO), and examined with a Zeiss Z1 microscope (Thornwood, NY).
Acknowledgments
This research was supported in part by National Institutes of Health (NIH) Grants N01-AI-50010, P01-A1–071739, and R01-A1-071186 and the National Institutes of Health Intramural Research Program.
Abbreviations
- SHIV
simian/human immunodeficiency virus
- SIV
simian imunodeficiency virus
- PBMC
peripheral blood mononuclear cell
- ELISpot
ELISA-linked immunospot
- SFC
spot-forming cells
- MID
monkey infectious doses.
Footnotes
The authors declare no conflict of interest.
References
- 1.Cao H, Kanki P, Sankale JL, Dieng-Sarr A, Mazzara GP, Kalams SA, Korber B, Mboup S, Walker BD. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinaldo C, Huang XL, Fan ZF, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, et al. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 5.Rowland-Jones SL, Dong T, Dorrell L, Ogg G, Hansasuta P, Krausa P, Kimani J, Sabally S, Ariyoshi K, Oyugi J, et al. Immunol Lett. 1999;66:9–14. doi: 10.1016/s0165-2478(98)00179-5. [DOI] [PubMed] [Google Scholar]
- 6.Rowland-Jones SL, Nixon DF, Aldhous MC, Gotch F, Ariyoshi K, Hallam N, Kroll JS, Froebel K, McMichael A. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 7.Barouch DH, Pau MG, Custers JH, Koudstaal W, Kostense S, Havenga MJ, Truitt DM, Sumida SM, Kishko MG, Arthur JC, et al. J Immunol. 2004;172:6290–6297. doi: 10.4049/jimmunol.172.10.6290. [DOI] [PubMed] [Google Scholar]
- 8.Evans DT, Bricker JE, Desrosiers RC. J Virol. 2004;78:11715–11725. doi: 10.1128/JVI.78.21.11715-11725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fultz PN, Stallworth J, Porter D, Novak M, Anderson MJ, Morrow CD. Virology. 2003;315:425–437. doi: 10.1016/s0042-6822(03)00546-4. [DOI] [PubMed] [Google Scholar]
- 10.Koopman G, Mortier D, Hofman S, Niphuis H, Fagrouch Z, Norley S, Sutter G, Liljestrom P, Heeney JL. J Gen Virol. 2004;85:2915–2924. doi: 10.1099/vir.0.80226-0. [DOI] [PubMed] [Google Scholar]
- 11.Letvin NL, Huang Y, Chakrabarti BK, Xu L, Seaman MS, Beaudry K, Korioth-Schmitz B, Yu F, Rohne D, Martin KL, et al. J Virol. 2004;78:7490–7497. doi: 10.1128/JVI.78.14.7490-7497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makitalo B, Lundholm P, Hinkula J, Nilsson C, Karlen K, Morner A, Sutter G, Erfle V, Heeney JL, et al. J Gen Virol. 2004;85:2407–2419. doi: 10.1099/vir.0.79869-0. [DOI] [PubMed] [Google Scholar]
- 13.Matano T, Kobayashi M, Igarashi H, Takeda A, Nakamura H, Kano M, Sugimoto C, Mori K, Iida A, et al. J Exp Med. 2004;199:1709–1718. doi: 10.1084/jem.20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nacsa J, Radaelli A, Edghill-Smith Y, Venzon D, Tsai WP, Morghen CG, Panicali D, Tartaglia J, Franchini G. Vaccine. 2004;22:597–606. doi: 10.1016/j.vaccine.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Paterson Y, Johnson RS. Exp Rev Vaccines. 2004;3:S119–S134. doi: 10.1586/14760584.3.4.s119. [DOI] [PubMed] [Google Scholar]
- 16.Wang SW, Bertley FM, Kozlowski PA, Herrmann L, Manson K, Mazzara G, Piatak M, Johnson RP, Carville A, et al. AIDS Res Hum Retroviruses. 2004;20:846–859. doi: 10.1089/0889222041725253. [DOI] [PubMed] [Google Scholar]
- 17.Johnson PR, Schnepp BC, Connell MJ, Rohne D, Robinson S, Krivulka GR, Lord CI, Zinn R, Montefiori DC, Letvin NL, et al. J Virol. 2005;79:955–965. doi: 10.1128/JVI.79.2.955-965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan MA, Chong SY, Rose NF, Megati S, Lopez KJ, Schadeck EB, Johnson JE, Masood A, Piacente P, Druilhet RE, et al. AIDS Res Hum Retroviruses. 2004;20:989–1004. doi: 10.1089/aid.2004.20.989. [DOI] [PubMed] [Google Scholar]
- 19.Boyer JD, Cohen AD, Vogt S, Schumann K, Nath B, Ahn L, Lacy K, Bagarazzi ML, Higgins TJ, Baine Y, et al. J Infect Dis. 2000;181:476–483. doi: 10.1086/315229. [DOI] [PubMed] [Google Scholar]
- 20.Boyer JD, Chattergoon M, Ugen K, Shah A, Bennett M, Cohen A, Nyland S, Lacy K, Bagarazzi M, Higgins T, et al. Clin Immunol. 1999;90:100–107. doi: 10.1006/clim.1998.4616. [DOI] [PubMed] [Google Scholar]
- 21.MacGregor RR, Boyer JD, Ugen KE, Lacy KE, Bagarazzi ML, Chattergoon M, Baine Y, Higgins T, Ciccarelli R, Coney L, et al. J Infect Dis. 1998;178:92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 22.Calarota S, Bratt G, Nordlund S, Hinkula J, Leandersson A-C, Sandström E, Wahren B. Lancet. 1998;351:1320–1325. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- 23.Letvin NL, Mascola JR, Sun Y, Gorgone DA, Yang Z, Chakrabarti B, Rao SS, Schimitz, Montefiori DC, Barker BR, et al. Nature. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, Davies ME, McDermott AB, O'Connor DH, Fridman A, et al. J Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mwau M, Cebere I, Sutton J, Chikoti P, Winstone N, Wee EG, Beattie T, Chen YH, Dorrell L, McShane H, et al. J Gen Virol. 2004;85:911–919. doi: 10.1099/vir.0.19701-0. [DOI] [PubMed] [Google Scholar]
- 26.Shedlock DJ, Weiner DB. J Leukoc Biol. 2000;68:793–806. [PubMed] [Google Scholar]
- 27.Barouch DH, Craiu A, Kuroda MJ, Schmitz JE, Zheng XX, Santra S, Frost JD, Krivulka GR, Lifton MA, Crabbs CL, et al. Proc Natl Acad Sci USA. 2000;97:4192–4197. doi: 10.1073/pnas.050417697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, Bilska M, Craiu A, Zheng XX, Krivulka GR, et al. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 29.Kim JJ, Ayyavoo V, Bagarazz ML, Chattergoon M, Boyer JD, Wang B, Weiner DB. Vaccine. 1997;15:879–883. doi: 10.1016/s0264-410x(96)00260-5. [DOI] [PubMed] [Google Scholar]
- 30.Kim JJ, Bagarazzi ML, Trivedi N, Hu Y, Kazahaya K, Wilson DM, Ciccarelli R, Chattergoon MA, Dang K, Mahalingam S, et al. Nat Biotech. 1997;15:641–646. doi: 10.1038/nbt0797-641. [DOI] [PubMed] [Google Scholar]
- 31.Kim JJ, Nottingham LK, Sin JI, Tsai A, Morrison L, Oh J, Dang K, Hu Y, Kazahaya K, Bennett M, et al. J Clin Invest. 1998;102:1112–1124. doi: 10.1172/JCI3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JJ, Trivedi NN, Nottingham LK, Morrison L, Tsai A, Hu Y, Mahalingam S, Dang K, Ahn L, Doyle NK, et al. Eur J Immunol. 1998;28:1089–1103. doi: 10.1002/(SICI)1521-4141(199803)28:03<1089::AID-IMMU1089>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 33.Sin JI, Kim JJ, Zhang D, Weiner DB. Hum Gene Ther. 2001;12:1091–1102. doi: 10.1089/104303401750214302. [DOI] [PubMed] [Google Scholar]
- 34.Kim JJ, Ayyavoo V, Bagarazzi ML, Chattergoon MA, Dang K, Wang B, Boyer JD, Weiner DB. J Immunol. 1997;158:816–826. [PubMed] [Google Scholar]
- 35.Kutzler M, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY, Ramanathan MP, Parkinson R, Kudchodkar S, Tamura Y. J Immunol. 2005;175:112–123. doi: 10.4049/jimmunol.175.1.112. [DOI] [PubMed] [Google Scholar]
- 36.Sin JI, Kim JJ, Arnold RL, Shroff KE, McCallus D, Pachuk C, McElhiney SP, Wolf MW, Pompa-de Bruin SJ, Higgins TJ, et al. J Immunol. 1999;162:2912–2921. [PubMed] [Google Scholar]
- 37.Boyer JD, Robinson TM, Kutzler MA, Parkinson R, Calarota S, Sidhu M, Lewis M, Pavlakis GN, Felber BK, Weiner DB. J Med Primatol. 2005;34:262–270. doi: 10.1111/j.1600-0684.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- 38.Schadeck EB, Sidhu M, Egan MA, Chong SY, Piacente P, Masood A, Garcia-Hand D, Cappello S, Roopchand V, Megati S, et al. Vaccine. 2006;24:4677–4687. doi: 10.1016/j.vaccine.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Waldmann T, Tagaya Y, Bamford R. Int Rev Immunol. 1998;16:205–226. doi: 10.3109/08830189809042995. [DOI] [PubMed] [Google Scholar]
- 40.Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, et al. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- 41.Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, Ahmed R. J Exp Med. 2002;195:1541–1548. doi: 10.1084/jem.20020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. Science. 2000;288:675–678. doi: 10.1126/science.288.5466.675. [DOI] [PubMed] [Google Scholar]
- 43.Sprent J. Microbes Infect. 2003;5:227–231. doi: 10.1016/s1286-4579(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 44.Waldmann T. Arthritis Res. 2002;4:S161–S167. doi: 10.1186/ar584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 46.Moore AC, Kong WP, Chakrabarti BK, Nabel GJ. J Virol. 2002;76:243–250. doi: 10.1128/JVI.76.1.243-250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh S, Berzofsky JA, Burke DS, Waldmann TA, Perera LP. Proc Natl Acad Sci USA. 2003;100:3392–3397. doi: 10.1073/pnas.0630592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaech SM, Hemby S, Kersh E, Ahmed R. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 49.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Proc Natl Acad Sci USA. 2004;101:16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Nat Immun. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 51.Oh S, Perera LP, Burke DS, Waldmann TA, Berzofsky JA. Proc Natl Acad Sci USA. 2004;101:15154–15159. doi: 10.1073/pnas.0406649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Younes S, Bade Y, Dumont AR, Boulassel M, Grossman Z, Routy J, Sekaly R. J Exp Med. 2003;198:1909–1922. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahrens D, Koch AE, Pope RM, Stein-Picarella M, Niedbala MJ. Arthritis Rheum. 1996;39:1576–1587. doi: 10.1002/art.1780390919. [DOI] [PubMed] [Google Scholar]
- 54.Konttinen YT, Halinen S, Hanemaaijer R, Sorsa T, Hietanen J, Ceponis A, Xu JW, Manthorpe R, Whittington J, Larsson A, et al. Matrix Biol. 1998;17:335–347. doi: 10.1016/s0945-053x(98)90086-5. [DOI] [PubMed] [Google Scholar]
- 55.El-Shabrawi YG, Christen WG, Foster SC. Curr Eye Res. 2000;20:211–214. [PubMed] [Google Scholar]
- 56.Faber-Elmann A, Sthoeger Z, Tcherniack A, Dayan M, Mozes E. Clin Exp Immunol. 2002;127:393–398. doi: 10.1046/j.1365-2249.2002.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chong SY, Egan MA, Kutzler MA, Megati S, Masood A, Roopchard V, Garcia-Hand D, Montefiori DC, Quiroz J, Rosati M, et al. Vaccine. 2007;25:4967–4982. doi: 10.1016/j.vaccine.2006.11.070. [DOI] [PubMed] [Google Scholar]
- 58.McKenna PM, Koser ML, Carlson KR, Montefiori DC, Letvin NL, Papaneri AB, Pomerantz RJ, Dietzschold B, Silvera P, McGettigan JP, et al. J Inf Dis. 2007;195:980–988. doi: 10.1086/512243. [DOI] [PubMed] [Google Scholar]
- 59.Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, Lee EM, Pal R, Venzon D, Grant R, et al. J Virol. 2007;81:3414–3427. doi: 10.1128/JVI.02453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thorner AR, Lemckert AA, Goudsmit J, Lynch DM, Ewald BA, Denholtz M, Havenga MJ, Barouch DH. J Virol. 2006;80:12009–12016. doi: 10.1128/JVI.01749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson HL, Montefiori DC, Villinger F, Robinson JE, Sharma S, Wyatt LS, Earl PL, McClure HM, Moss B, Amara RR. Virology. 2006;352:285–294. doi: 10.1016/j.virol.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 62.Nappi F, Scjmeider R, Zolotukhin A, Smulevitch S, Michalowski D, Bear J, Felber BK, Pavlakis GN. J Virol. 2001;10:4558–4569. doi: 10.1128/JVI.75.10.4558-4569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyer JD, Kumar S, Robinson T, Parkinson R, Wu L, Lewis M, Weiner DB. J Med Primatol. 2006;35:202–209. doi: 10.1111/j.1600-0684.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 64.Chen Q, Vansant G, Oades K, Pickering M, Wei JS, Song YK, Montforte J, Khan J. J Med Diag. 2007;9:80–88. doi: 10.2353/jmoldx.2007.060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silvera P, Richardson MW, Greenhouse J, Yalley-Ogunro J, Shaw N, Mirchandani J, Kamel Khalili K, Zagury JF, Lewis MG, Rappaport J. J Virol. 2002;76:3800–3809. doi: 10.1128/JVI.76.8.3800-3809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins WR, Montefiori DC. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]