Abstract
Colon cancer has been viewed as the result of progressive accumulation of genetic and epigenetic abnormalities. However, this view does not fully reflect the molecular heterogeneity of the disease. We have analyzed both genetic (mutations of BRAF, KRAS, and p53 and microsatellite instability) and epigenetic alterations (DNA methylation of 27 CpG island promoter regions) in 97 primary colorectal cancer patients. Two clustering analyses on the basis of either epigenetic profiling or a combination of genetic and epigenetic profiling were performed to identify subclasses with distinct molecular signatures. Unsupervised hierarchical clustering of the DNA methylation data identified three distinct groups of colon cancers named CpG island methylator phenotype (CIMP) 1, CIMP2, and CIMP negative. Genetically, these three groups correspond to very distinct profiles. CIMP1 are characterized by MSI (80%) and BRAF mutations (53%) and rare KRAS and p53 mutations (16% and 11%, respectively). CIMP2 is associated with 92% KRAS mutations and rare MSI, BRAF, or p53 mutations (0, 4, and 31% respectively). CIMP-negative cases have a high rate of p53 mutations (71%) and lower rates of MSI (12%) or mutations of BRAF (2%) or KRAS (33%). Clustering based on both genetic and epigenetic parameters also identifies three distinct (and homogeneous) groups that largely overlap with the previous classification. The three groups are independent of age, gender, or stage, but CIMP1 and 2 are more common in proximal tumors. Together, our integrated genetic and epigenetic analysis reveals that colon cancers correspond to three molecularly distinct subclasses of disease.
Keywords: classification, DNA methylation, genetic alterations
Colorectal cancer (CRC) is the second and fourth most common cancer in men and women, respectively (1). Approximately 70% of colorectal cancers are sporadic, with no inherited predisposition. A stepwise progression model involving two distinct genetic pathways has been proposed to explain the etiology of colon cancer from benign neoplasm to adenocarcinoma (2). One class of genetic alterations involves mutations of oncogenes and tumor-suppressor genes that directly control cell birth and death, such as APC, KRAS, and p53. Another involves mutations of DNA mismatch repair genes.
In addition to these genetic alterations, cancer initiation and promotion can occur by epigenetic mechanisms (3). CpG methylation is the best characterized epigenetic change in the mammalian genome. Whereas CpG dinucleotides are underrepresented in the mammalian genome, approximately half of all human genes contain a CpG-rich region called a “CpG island” in the 5′ area, often encompassing the promoter and transcription start site of the associated gene (4, 5). Gene silencing by hypermethylation of CpG islands (including tumor-suppressor genes) is a common event in tumors. Further, hypermethylation of specific genes such as ERα, MYOD1, and N33 occurs in the normal colon tissue of aging individuals (6, 7), and hypermethylation of the secreted frizzled-related gene family (SFRPs) is detectable in aberrant crypt foci (8). The early occurrence of epigenetic alterations led to a hypothesis that they allow for the subsequent accumulation of both genetic and epigenetic alterations that promote tumor development and progression.
Importantly, certain individuals appear predisposed to aberrant promoter hypermethylation, including at several tumor-suppressor genes (9). This phenomenon, termed CpG island methylator phenotype (CIMP), provides an alternative pathway to promote colon cancer (10). Several independent studies have linked CIMP to distinct genetic and clinical features, including high rates of BRAF and KRAS mutation, low rates of p53 mutations, specific histology (mucinous, poorly differentiated), familial occurrence, and distinct clinical outcome (11). However, the current view of the formation of colon cancer does not fully reflect the molecular heterogeneity of the disease. Here, we analyzed both genetic and epigenetic alterations in primary colorectal cancers and found that, molecularly, colon cancer consists of three distinct subclasses, each of which is fairly homogeneous.
Results
Clinical Variables and Epigenetic and Genetic Alterations.
We analyzed colorectal cancers from 97 individual CRC patients selected solely based on tissue availability. Clinical characteristics of the patients are summarized in Table 1. DNA isolated from grossly microdissected cancers was analyzed to determine the methylation status of 27 promoter-associated CpG islands selected based on prior studies. For each gene, the average methylation level measured quantitatively and the frequency of positive cases (with methylation level greater than >15%) are shown in supporting information (SI) Table 6. In an initial analysis, we selected 20 cases to compare methylation analysis for the same genes by different methods [methylated CpG island amplification (MCA) (12), combined of bisulfite restriction enzyme amplification (COBRA) (13), or bisulfite pyrosequencing (14)] and found excellent correlation in methylation between the methods (similar results were observed for 92% cases, using MCA or pyrosequencing methods, and 95% cases using COBRA or pyrosequencing methods). Therefore, we combined all results together for further analysis. Methylation frequencies for the 27 genes we examined ranged from 5.2 to 98.9%. Five genes, ERα, MyoD1, N33, HPP1, and SFRP1, were hypermethylated in >80% of cancer cases, suggestive of age-related methylation (6, 15). Indeed, when we examined the methylation of these genes in normal-appearing mucosa from the same patients, we found substantial methylation in normal colon and significant correlation between patient age and methylation of each gene (R = 0.36, P = 0.0005 for ERα; R = 0.42, P < 0.001 for MyoD1; R = 0.45, P < 0.0001 for N33; R = 0.45, P < 0.0001 for SFRP1; and R = 0.33, P = 0.002 for HPP1; see SI Fig. 6). This was not found for any of the other genes examined. Therefore, as previously proposed, we called these five genes Type-A genes for age-related and all other genes Type-C genes for tumor-specific.
Table 1.
Clinical characteristics of 97 CRC patients
| Characteristic | n = 97 |
|---|---|
| Age | |
| Median age in years, range | 68 years (25–98 years) |
| Missing data | 2 |
| Gender | |
| Female | 29 (30%) |
| Male | 66 (68%) |
| Missing data | 2 (2%) |
| Location | |
| Proximal | 38 (39%) |
| Distal | 44 (45%) |
| Missing data | 15 (16%) |
| Stage | |
| I or II | 43 (44%) |
| III or IV | 37 (38%) |
| Missing data | 17 (18%) |
We next determined the status of BRAF mutation (using pyrosequencing), KRAS mutation (using mutant allele specific amplification), p53 mutation (using single-strand conformational polymorphism and sequencing), and microsatellite instability (using the classical panel) in these same cases. BRAF mutation was observed in 11 of 87 cancers (12.6%); KRAS mutation was found in 43 of 94 cancers (45.7%); and 44 of 93 patients (47.3%) had p53 mutation. Of the 97 tumors evaluated for microsatellite instability, 22 (22.7%) had high levels of microsatellite instability (MSI-H).
CIMP Affects Most Genes.
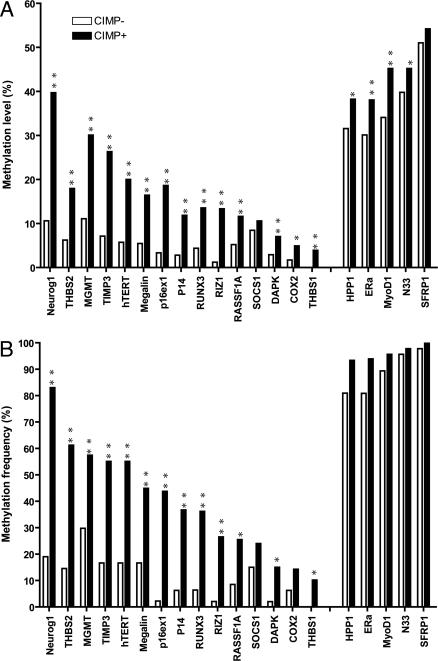
It was shown that methylation clusters in specific colorectal cancer subsets termed CIMP, and CIMP was originally defined based on seven cancer-specific MINT markers with hypermethylation at 2 or more loci (9). Using the original definition, 49 cases studied here were defined as CIMP-positive (51%) and 48 cases were CIMP negative. We compared the average methylation measured quantitatively at the additional 20 genes between these two groups and found that all genes except SFRP1 and SOCS1 showed significantly higher methylation density in the CIMP-positive group (Fig. 1A). When we analyzed the frequency of methylation-positive cases (methylation density >15%), we found all 15 Type-C genes except SOCS1 showed significantly higher frequency of methylation in the CIMP-positive group; the 5 Type-A genes showed no difference between these two groups (Fig. 1B).
Fig. 1.
Comparison of methylation level and frequency for 20 genes between CIMP-positive and negative groups. (A) Comparison of the mean methylation level of each gene between CIMP-positive group and CIMP-negative group. All genes except SFRP1 and SOCS1 showed significantly higher methylation level in the CIMP-positive group. (B) Comparison of methylation frequency of positive cases for each gene between CIMP-positive and -negative groups. Methylation-positive case is defined by methylation level >15%. All Type-C genes except SOCS1 showed significantly higher frequency of methylation in CIMP-positive group, whereas 5 Type-A genes (on the Right) showed no difference between these two groups. **, P < 0.001; *, P < 0.05
Three Distinct Clusters of Colon Cancers.
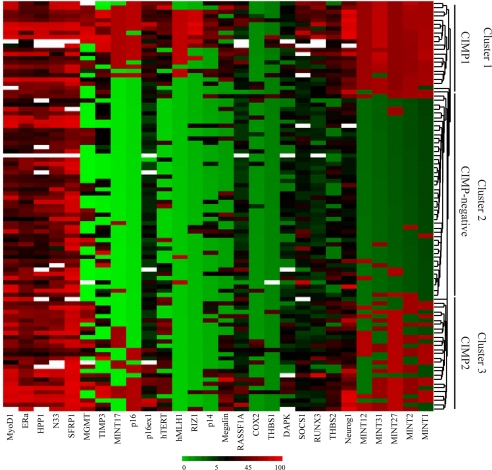
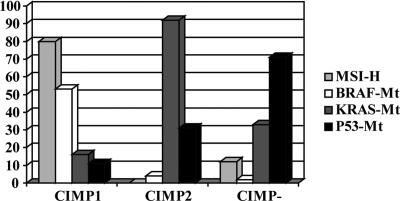
To explore the underlying patterns of gene-methylation changes, we performed unsupervised hierarchical clustering analysis, using the methylation of 27 genes as a continuous variable within primary CRC patients. Three separate clusters were identified by this analysis, one of which corresponded very closely to the previous CIMP-negative group (middle cluster in Fig. 2) showing low or less methylation for all genes we examined. Surprisingly, CIMP-positive cases fit into two subgroups: CIMP1 (cluster 1 in Fig. 2) and CIMP2 (cluster 3 in Fig. 2). When we compared the genetic alterations within these three clusters, each of them corresponded to very distinct genetic profiles (Fig. 3). CIMP1 cases showed a significantly higher frequency of MSI and BRAF mutations (80% and 53%, respectively) but few KRAS and p53 mutations (16% and 11%, respectively). Conversely, CIMP2 was associated with a high frequency of KRAS mutations (92%), but MSI and BRAF mutation occurred rarely (0% and 4% respectively) with a low rate of p53 mutation (31%). CIMP-negative cases had a higher rate of p53 mutation (71%) and lower rates of MSI (12%) and mutations of BRAF (2%) and KRAS (33%). Thus, each of MSI, BRAF, KRAS, and p53 alterations were unevenly distributed within the three groups (Fig. 3), and all of the P values were statistically significant (<0.0001 by Fisher's exact test).
Fig. 2.
Unsupervised hierarchical clustering analysis on the basis of 27 methylation markers. Three separate clusters were generated by this analysis with one cluster corresponding very closely to the previous CIMP-negative group (middle cluster), and CIMP-positive cases were separated into two subgroups, CIMP1 (cluster 1) and CIMP 2 (cluster 3).
Fig. 3.
Comparison of the genetic alterations among the three clusters. Each cluster corresponds to very distinct genetic profiles. CIMP1 is characterized by high frequency of MSI (80%) and BRAF mutations (53%), CIMP2 is characterized by a higher rate of KRAS mutations (92%), and CIMP negative is characterized by high frequency of p53 mutations (71%).
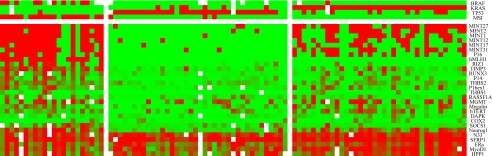
Based on the hierarchical clustering results, we used both genetic and epigenetic information to perform K-means clustering, which identifies the most homogeneous clusters. The three groups classified from this analysis (Fig. 4) were largely overlapping with the previous classification, with only 17 (18%) cases being reclassified. By K-Means clustering, 22 cases were classified as CIMP1 (23%), 37 cases (38%) were classified as CIMP2, and 38 cases (39%) were classified as CIMP negative.
Fig. 4.
K-means clustering analysis on the basis of both genetic and epigenetic markers. K-means clustering including genetic information yielded very homogenous groups. Twenty-two cases were classified as CIMP1 (23%), 37 cases (38%) were classified as CIMP2, and 38 cases (39%) were classified as CIMP negative. This clustering largely overlaps with the previous hierarchical clustering with only 17 cases (18%) reclassified.
To assess the reliability and reproducibility of the classification, first we performed bootstrap analysis (resampling with replacement method) (16) to determine the level of confidence of the clustering. As shown in SI Fig. 7, we observed three main blocks robustly clustered in bootstrap datasets, suggesting that each of these three classes is fairly stable. Interestingly, the middle cluster (CIMP2) shows more heterogeneity than the other two clusters. We also compared the current classification with our classification in ref. 9 in 49 CRC patients. We found that 44 cases (90%) remained in the same groups, with only 5 cases being reclassified (Table 2). These results show that these three newly identified clusters largely overlap with the previous classification. Together, our results suggest that combined genetic and epigenetic characteristics subclassify colorectal cancer into three distinct groups.
Table 2.
Comparison between previous and current classifications based on 49 CRC patients
| Group | Previous study* |
Current study |
||
|---|---|---|---|---|
| Old categories | Cases, no. | New categories | No. of cases, % agreement | |
| 1 | MSI+/CIMP+ | 12 | CIMP1 | 12 (100%) |
| 2 | MSI−/CIMP+ | 16 | CIMP2 | 14 (88%) |
| CIMP1 | 2 | |||
| 3 | MSI+/CIMP− | 4 | CIMP-negative | 3 (75%) |
| CIMP1 | 1 | |||
| 4 | MSI−/CIMP− | 17 | CIMP-negative | 15 (88%) |
| CIMP2 | 2 | |||
We next analyzed whether the different CRC subclasses identified correlated to distinct clinical characteristics. Among the three groups, there was no significant difference in age, gender, or stage (Table 3). However, a significantly higher incidence of proximal colon cancer was found in both CIMP1 and CIMP2 groups (63% of proximal tumors in CIMP 1 and 60% in CIMP 2) compared with the CIMP-negative group (24% of proximal tumors, P = 0.004 by Fisher's exact test).
Table 3.
Patient clinical characteristics in each cluster
| Characteristic | CIMP1 (n = 22) | CIMP2 (n = 37) | CIMP negative (n = 38) | P |
|---|---|---|---|---|
| Age | ||||
| Median, years | 68 | 69 | 67 | 0.90 |
| Range, years | 25–88 | 26–85 | 29–98 | |
| Missing data (N) | 1 | 1 | 0 | |
| Gender (N) | ||||
| Female | 6 | 14 | 9 | 0.40 |
| Male | 15 | 22 | 29 | |
| Missing data | 1 | 1 | 0 | |
| Location (n) | ||||
| Proximal | 12 | 18 | 8 | 0.004 |
| Distal | 7 | 12 | 25 | |
| Missing data | 3 | 7 | 5 | |
| Stage (n) | ||||
| 1 or 2 | 8 | 18 | 17 | 0.84 |
| 3 or 4 | 6 | 14 | 17 | |
| Missing data | 8 | 5 | 4 |
Optimal Markers to Predict the Three Groups.
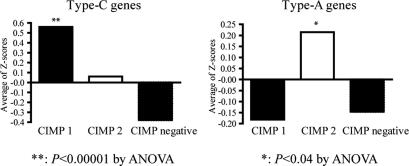
We further examined in detail the epigenetic signatures among three groups of CRC identified (CIMP1, CIMP2, and CIMP negative). By Kruskal–Wallis tests, we found that all Type-C genes (except for COX2, DAPK, and RASSF1A) showed significant differences among these groups (see SI Table 7 for details). The three genes showing no difference had very low levels of methylation overall. For Type-A genes, only MYOD1 showed a statistically significant difference among the groups. However, there was a nonsignificant trend for increased methylation of ERα, HPP1, N33, and SFRP1 in CIMP2 compared with the other groups. Next, we used Z-score method to assign equal weight for methylation of each gene by substituting all raw methylation values in each data set with their respective Z-scores (see SI Materials and Methods for details), and assigned methylation scores for each patient based on the average Z-scores of either Type-A genes or Type-C genes. As shown in Fig. 5, the methylation score for Type-C genes was significantly higher in CIMP1, followed by CIMP2, and CIMP-negative cases were the lowest (0.56, 0.06 and −0.38, respectively, P < 0.001). Interestingly, the methylation score for Type-A genes was significantly higher among CIMP2 (0.21) compared with CIMP1 (−0.18) and CIMP-negative (−0.15) individuals (P < 0.04).
Fig. 5.
Comparison of methylation for Type-C genes and Type-A genes among the three clusters. A Z-score method was used to standardize the methylation level of each gene and each patient was assigned methylation scores based on the average Z-scores of either Type-C genes or Type-A genes. (Left) For Type-C genes, the average methylation Z-score was significantly higher in CIMP1 compared with other two groups (P < 0.001). (Right) For Type-A genes, the average methylation Z-score was significantly higher in CIMP2 group (P = 0.04).
To determine which individual genetic or epigenetic alteration can best predict these three groups clinically, we calculated the sensitivity, specificity, positive and negative predictive values and κ coefficient value (assessment for reliability) for each marker. Table 4 shows the top 10 single markers for predicting each group. Based on κ coefficient, the best single marker to predict CIMP1 group is hMLH1 methylation, whereas KRAS mutation is the best predictive marker for CIMP2 group, and p53 mutation is the best predictive marker for CIMP-negative group. As expected, the two genetic markers MSI-H and BRAF mutation were also among the best predictors for CIMP1, with a high degree of accuracy determined by sensitivity and predictive values. Several methylation markers are also on the top of the list for predicting each cluster; hMLH1, TIMP3, and MINT17 methylation were most closely linked to CIMP1, methylation of MINT2 and MINT27 were associated with CIMP2, and lack of methylation of MINT1, MINT2, MINT27, and MINT31 predicted CIMP negativity.
Table 4.
Predictive values of each marker to identify three clusters
| Marker | κ coefficient | Specificity, % | Sensitivity, % | PPV, % | NPV, % |
|---|---|---|---|---|---|
| CIMP1 | |||||
| Genetic | |||||
| BRAF-MT | 0.60 | 99 | 53 | 91 | 88 |
| MSI-H | 0.82 | 96 | 86 | 86 | 96 |
| Epigenetic | |||||
| hMLH1-M | 0.91 | 100 | 86 | 100 | 96 |
| TIMP3-M | 0.62 | 84 | 86 | 61 | 95 |
| MINT17-M | 0.52 | 84 | 73 | 57 | 91 |
| MINT1-M | 0.49 | 75 | 86 | 50 | 95 |
| RIZ1-M | 0.48 | 93 | 50 | 69 | 86 |
| SOCS1-M | 0.47 | 86 | 63 | 55 | 90 |
| MINT12-M | 0.44 | 76 | 77 | 49 | 92 |
| RUNX3-M | 0.43 | 85 | 60 | 52 | 89 |
| P16-M | 0.41 | 81 | 64 | 50 | 88 |
| MINT31-M | 0.38 | 71 | 77 | 44 | 91 |
| P14-M | 0.36 | 83 | 55 | 48 | 86 |
| CIMP2 | |||||
| Genetic | |||||
| KRAS-MT | 0.85 | 88 | 100 | 84 | 100 |
| P53-WT | 0.30 | 60 | 72 | 53 | 77 |
| MSS/MSI-L | 0.31 | 37 | 100 | 49 | 100 |
| Epigenetic | |||||
| MINT27-M | 0.39 | 63 | 78 | 57 | 83 |
| MINT2-M | 0.30 | 68 | 62 | 55 | 75 |
| Neurog1-M | 0.30 | 71 | 59 | 56 | 73 |
| MINT31-M | 0.22 | 68 | 54 | 51 | 71 |
| Megalin-M | 0.21 | 72 | 49 | 51 | 69 |
| MINT1-M | 0.15 | 67 | 49 | 47 | 68 |
| hMLH1-UM | 0.26 | 32 | 100 | 47 | 100 |
| CIMP-negative | |||||
| Genetic | |||||
| P53-MT | 0.70 | 81 | 92 | 75 | 94 |
| KRAS-WT | 0.41 | 63 | 81 | 59 | 84 |
| Epigenetic | |||||
| MINT27-UM | 0.71 | 81 | 92 | 76 | 94 |
| MINT2-UM | 0.58 | 68 | 95 | 65 | 95 |
| MINT31-UM | 0.53 | 63 | 95 | 62 | 95 |
| MINT1-UM | 0.55 | 63 | 97 | 63 | 97 |
| MINT12-UM | 0.46 | 56 | 95 | 58 | 94 |
| Neurog1-UM | 0.45 | 60 | 89 | 59 | 90 |
| P16-UM | 0.41 | 47 | 100 | 55 | 100 |
PPV, positive predictive value; NPV: negative predictive value; MT, mutation; M, methylated; UM, unmethylated.
To explore whether a combination of markers could provide greater accuracy than individual markers in predicting subtypes of CRC, we selected the top five predictive markers based on predictive values and analyzed them together. For the CIMP1 group, a combination analysis of five markers (BRAF mutation and methylation of hMLH1, TIMP3, MINT1, and RIZ1) indicates that having three positive markers results an excellent positive predictive value and negative predictive value (94% and 94% respectively, Table 5). For the CIMP2 group, no combination performs better than KRAS mutation alone. In CIMP-negative group, p53 mutation and lack of methylation at MINT27, MINT2, MINT31, and MINT1 are the top five best markers, and a combination of any three markers gave 73% positive predictive value and 100% negative predictive value (Table 5). The performance of these markers in classifying CRC should, however, be validated in independent studies.
Table 5.
Predictive values for combination markers to identify each of the three clusters
| Positive, no. | κ coefficient | Specificity, % | Sensitivity, % | PPV, % | NPV, % |
|---|---|---|---|---|---|
| CIMP 1 (BRAF-Mutation, hMLH1-Meth, Timp3-Meth, MINT1-Meth, and RIZ1-Meth) | |||||
| 2 of 5 | 0.80 | 95 | 86 | 83 | 96 |
| 3 of 5 | 0.81 | 99 | 77 | 94 | 94 |
| CIMP2 (KRAS-Mutation, MINT27-Meth, MINT2-Meth, MINT31-Meth, and Megalin-Meth) | |||||
| 2 of 5 | 0.40 | 60 | 84 | 56 | 86 |
| 3 of 5 | 0.30 | 45 | 89 | 50 | 87 |
| CIMP Negative (p53-Mutation, MINT27-Unmeth, MINT2-Unmeth, MINT31-Unmeth, MINT1-Unmeth) | |||||
| 2 of 5 | 0.55 | 61 | 100 | 62 | 100 |
| 3 of 5 | 0.72 | 76 | 100 | 73 | 100 |
Discussion
In this study, we show that primary colorectal cancers cluster into three distinct subclasses based on epigenetic and genetic profiles: CIMP1, intense methylation of multiple genes and MSI and BRAF mutations; CIMP2, methylation of a limited group of genes, increased methylation level for age-related genes, and mutation in KRAS; and CIMP negative, rare methylation with p53 mutation. These three groups are relatively homogeneous on a molecular level and likely representative of three different subclasses of disease.
These data suggest that colon cancer can be divided into substantially distinct groups in a way similar to breast cancers, where hormone status and HER2 amplification define distinct groups (17), and to leukemias, where specific chromosomal changes define very different diseases (18). The three colorectal cancer groups also differ clinically; CIMP1 and CIMP2 are more often proximal; CIMP1 has a good prognosis because it consists mostly of MSI-H cancers (19, 20), whereas CIMP2 has a poor prognosis (21). Moreover, they may have distinct precancerous lesions such as HPP/serrated adenomas for CIMP1 (22, 23), and villous adenomas for CIMP2 (24). It is unclear whether these three groups reflect initiations of cancer in distinct precancerous cells (as hypothesized for breast cancer), or reflect entirely different diseases (with a different cause/epidemiology) that affect the same precancerous cells. Nevertheless, they are sufficiently distinct to merit consideration in clinical trials and clinical management of colorectal cancer.
The mechanistic basis of these two CIMP in colon cancer remain unknown. One possibility is that genetic events that activate methylases or inactivate methylation-protection factors explain CIMP1, where increased methylation degree and frequency is observed for multiple CpG islands including a number of tumor-suppressor genes, such as hMLH1, p16, p14, etc. Another possibility is environmental exposure-related CIMP etiology, possibly explaining CIMP2, in which methylation spreading could be a molecular signature of environmental exposure by targeting age-related genes (25). In this case, increased methylation may not be directly linked to the methylation machinery, but to a constitutional predisposition to environment-DNA interactions, such as chronic inflammation or an exaggerated response to tissue injury (25, 26).
Our data also confirm that CIMP affects many genes, not just a subset of genes, and show that there are two distinct CIMPs with potentially different causes. The optimal markers for CIMP remain unclear. A recent article by the Laird group (27) used a panel of five-markers by MethyLight method, and concluded that a new panel of genes outperforms the classic panel. However, this study possibly focused mainly on the CIMP1 group and largely underestimated the CIMP2 group. In our study, we also included three of the five genes (Neurog1, RUNX3, and SOCS1) from the previous report. All three markers performed well to identify CIMP1 confirming the previous study, but they did not perform well for identifying the CIMP2 group. Among all of the methylation markers we analyzed, the original markers (all MINT markers) still show the best predictive values, and the combination of them could best define CIMP2. However, in this study, genetic markers performed equally well or better than epigenetic markers in some cases, highlighting the importance of integrated genetic and epigenetic analysis to resolve the heterogeneity in cancers.
In summary, by integrating genetic and epigenetic analysis, we show that colon cancers correspond to three molecularly distinct subclasses of disease. Further studies will be needed to quantify the prognostic utility of our findings. It will also be important to study the epidemiology and clinical courses of these three subclasses of colon cancers. We suggest that molecular classification of all cancers by combined genetic and epigenetic analyses will improve our understanding of the diseases and the selection of optimal therapy.
Materials and Methods
Further details of tissue samples, DNA methylation analysis, mutation analysis, and statistical analysis used in this study are described in SI Materials and Methods.
Tissue Samples.
We collected samples of primary colorectal tumors and adjacent normal-appearing tissues from 97 patients selected solely on the basis of availability.
DNA Methylation Analysis.
We used different methods (MCA, COBRA, MSP, and bisulfite-pyrosequencing) to study the methylation status of 27 promoter region CpG islands (see also details in SI Table 8).
Mutation Analysis.
Mutations of KRAS and p53 were determined by mutant allele specific PCR for KRAS codons 12 or 13 and single-strand conformational polymorphism and sequencing for p53 (10, 28). BRAF mutations at exon 11 and 15 were determined by the pyrosequencing method.
Statistical Analysis.
Correlation between methylation and clinical variables were analyzed by Fisher's exact test for categorical variables and Spearman correlation analysis for continuous variables. Unsupervised hierarchical clustering and K-means clustering analyses were used to identify potential distinct subgroups among CRC patients based on either epigenetic or combined of genetic and epigenetic profiling. Bootstrapping cluster analysis (16) was performed to assess the reliability of clustering results. The difference of molecular and clinical variables among each cluster was analyzed by the Kruskal–Wallis test. Sensitivity, specificity, positive and negative predictive values, and κ coefficient values were calculated to determine the sensitivity and specificity of either single molecular marker or combination of markers to predict each subgroup of CRC patients.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA098006 and CA105346. J.-P.J.I. is an American Cancer Society Clinical Research professor supported by a generous gift from the F. M. Kirby Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704652104/DC1.
References
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Bock C, Walter J, Paulsen M, Lengauer T. PLoS Comput Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones PA, Takai D. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 6.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JPJ. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 7.Issa JPJ, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Bin Y, Akiyama Y, van Engeland M, et al. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 9.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyota M, Ohe-Toyota M, Ahuja N, Issa JPJ. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa JP. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 12.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JPJ. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 13.Xiong ZG, Laird PW. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colella S, Shen L, Baggerly KA, Issa JPJ, Krahe R. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 15.Issa JP. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 16.Kerr MK, Churchill GA. Proc Natl Acad Sci USA. 2001;98:8961–8965. doi: 10.1073/pnas.161273698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M. Clin Trans Oncol. 2006;8:7–14. doi: 10.1007/s12094-006-0089-6. [DOI] [PubMed] [Google Scholar]
- 18.Mrozek K, Heerema NA, Bloomfield CD. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 19.Issa JPJ. Clin Cancer Res. 2003;9:2879–2881. [PubMed] [Google Scholar]
- 20.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 21.Shen L, Catalano PJ, Benson A., III, O'Dwyer P, Hamilton SR, Issa JPJ. Clin Cancer Res. 2007;13:6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iino H, Jass JR, Simms LA, Young J, Leggett B, Ajioka Y, Watanabe H. J Clin Pathol. 1999;52:5–9. doi: 10.1136/jcp.52.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minoo P, Baker K, Goswami R, Chong G, Foulkes WD, Ruszkiewicz AR, Barker M, Buchanan D, Young J, Jass JR. Gut. 2006;55:1467–1474. doi: 10.1136/gut.2005.082859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chirieac LR, Shen LL, Catalano PJ, Issa JP, Hamilton SR. Am J Surg Pathol. 2005;29:429–436. doi: 10.1097/01.pas.0000155144.53047.7d. [DOI] [PubMed] [Google Scholar]
- 25.Issa JPJ, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 26.Issa JP, Shen L, Toyota M. Gastroenterology. 2005;129:1121–1124. doi: 10.1053/j.gastro.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 27.Weisenberger DJ, Siegmund D, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 28.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, et al. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.