Abstract
Phylogenetic analysis of the cation/proton antiporter superfamily has uncovered a previously unknown clade of genes in metazoan genomes, including two previously uncharacterized human isoforms, NHA1 and NHA2, found in tandem on human chromosome 4. The NHA (sodium hydrogen antiporter) family members share significant sequence similarity with Escherichia coli NhaA, including a conserved double aspartate motif in predicted transmembrane 5. We show that HsNHA2 (Homo sapiens NHA2) resides on the plasma membrane and, in polarized MDCK cells, localizes to the apical domain. Analysis of mouse tissues indicates that NHA2 is ubiquitous. When expressed in the yeast Saccharomyces cerevisiae lacking endogenous cation/proton antiporters and pumps, HsNHA2 can confer tolerance to Li+ and Na+ ions but not to K+. HsNHA2 transformants accumulated less Li+ than the salt-sensitive host; however, mutagenic replacement of the conserved aspartates abolished all observed phenotypes. Functional complementation by HsNHA2 was insensitive to amiloride, a characteristic inhibitor of plasma membrane sodium hydrogen exchanger isoforms, but was inhibited by phloretin. These are hallmarks of sodium–lithium countertransport activity, a highly heritable trait correlating with hypertension. Our findings raise the possibility that NHA genes may contribute to sodium–lithium countertransport activity and salt homeostasis in humans.
Keywords: sodium–lithium countertransport, yeast expression, red blood cell, pancreas
The regulation of salt, pH, and volume is prerequisite to all forms of life, and central to these homeostatic mechanisms is the transmembrane exchange of H+ for cations (Na+ or K+). In bacteria, an array of Na+/H+ antiporters convert the proton motive force (established by the respiratory chain or the F1Fo-ATPase) into sodium gradients that drive other energy-requiring processes (solute transport or flagellar motors), transduce environmental signals into cell responses, and even function in drug efflux (TetL and MdfA) (1–3). In plants, newly discovered members of the NHE (sodium hydrogen exchanger) family of Na+/H+ exchangers sequester Na+ and K+ within vacuoles as a principal means of salt tolerance, regulate vacuolar pH to control flower color, and are essential for development (4, 5). In metazoans, the role of NHE in cytosolic and compartmental pH regulation has been implicated in the control of cell cycle and cell proliferation, vesicle trafficking, and compartmental biogenesis (6–8). Specific isoforms of NHE localize distinctly to basolateral or apical membranes to control transepithelial sodium fluxes (4, 9). In mammals, NHE dysfunction leads to a host of pathophysiological conditions that include hypertension, epilepsy, postischemic myocardial arrhythmias, gastric and kidney disease, diarrhea, and glaucoma.
The last decade witnessed an explosion in the number of genomic sequences deposited in databases worldwide, and these are now awaiting functional analysis. Automated annotation programs have identified >550 sequence entries as putative Na+/H+ exchangers. In an effort to understand the evolutionary origins and distribution of Na+/H+ exchangers, we undertook a comprehensive phylogenetic analysis of the superfamily of monovalent cation/proton antiporters (CPA) that have in common a transmembrane organization of 12 predicted hydropathic helices with detectable sequence similarity (10). This superfamily has two main subdivisions, named CPA1 and CPA2, according to the nomenclature of Transport Classification Database established by Milton Saier (http://www.tcdb.org/). Members of the CPA1 group include the well characterized NHE family of electroneutral Na+(K+)/H+ exchangers represented by nine paralogs in human (NHE1–9). In contrast, we found that virtually all eukaryotic members of the CPA2 group were previously unknown or poorly characterized. Among these was a new family of related genes in animals that we named NHA on the basis of their similarity to fungal NHA genes and bacterial NhaA genes (10). There are two paralogs, NHA1 and NHA2, in all completely sequenced metazoan genomes, including nematodes, fly, puffer fish, mouse, and human.
The identification of an entire family of phylogenetically distinct antiporters that are conserved from bacteria to humans opens up a new line of investigation. In this study, we describe Homo sapiens NHA2 (HsNHA2) as a prototypic metazoan example of the NHA family. Sequence similarity with Escherichia coli NhaA (EcNhaA) served to guide functional characterization by heterologous expression in yeast. Patterns of tissue distribution, chromosomal location, and inhibitor sensitivity point to this gene as a likely candidate for the sodium–lithium countertransport (SLC) activity reported in red blood cells (11), lymphoblasts, and fibroblasts (12–14), and suggest an important potential physiological role in hypertension.
Results
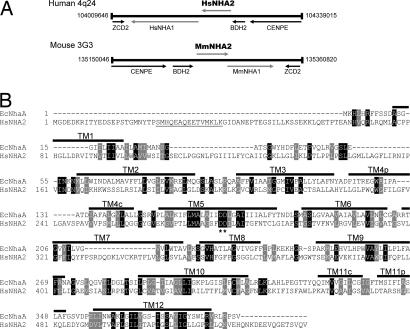
The presence of two paralogous NHA genes in the metazoan genomes of nematodes, insects, fish, and mammals is indicative of an early gene duplication event. Indeed, NHA1 and NHA2 appear in tandem on human chromosome 4, and the syntenic mouse chromosome 3 (Fig. 1A), and share amino acid identity of 55% over 515 aa [69% similarity; supporting information (SI) Fig. 5]. An alignment of human NHA2 sequence with that of EcNhaA reveals a significant conservation of residues (18% identity and 33% similarity over 388 aa) extending over 12 predicted transmembrane domains (Fig. 1B). Of note, two aspartic acid residues in TM5 of EcNhaA that have previously been shown to be critical for ion transport (15) are conserved in the metazoan NHA. Other residues of interest include H356 (TM8) and R432 (TM10) in NHA2, which may be equivalent to functionally important residues H225 and K300, respectively, in EcNhaA (16). Compared with the related NHE family of CPA1 antiporters that have long, hydrophilic C-terminal domains (150–300 aa) (10), the NHA proteins are distinguished by much shorter C-terminal tails following predicted TM12.
Fig. 1.
Gene location and sequence similarity of NHA. (A) Tandem arrangement of NHA1 and NHA2 on human chromosome 4 and the syntenic mouse chromosome 3 showing relative locations of genes. (B) Sequence alignment of human NHA2 (HsNHA2) with EcNhaA, showing locations of transmembrane domains (TM1–TM12), based on the structure of EcNhaA (22). Asterisks mark the conserved aspartate motif in TM5. Black and gray boxes highlight sequences of identity and similarity, respectively. The N-terminal peptide of HsNHA2 used for antibody generation is underlined.
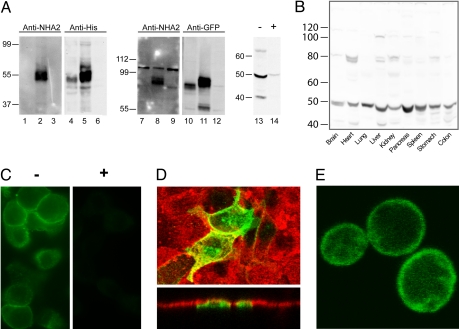
Antibodies raised against a unique N-terminal peptide of HsNHA2 (Fig. 1B, underlined) identified a band of the expected size in membranes from yeast heterologously expressing His9-tagged HsNHA2 (55 kDa), or GFP-HsNHA2 (90 kDa), whereas no cross-reactivity was seen in membranes from yeast transformed with His9-HsNHA1 (Fig. 2A). The partial conservation of residues (7 of 15) within the antigenic peptide between mouse and human NHA2 allowed us to test expression in mouse: A strongly reacting band of ≈50 kDa was observed in pancreatic lysates, and preincubation of antibody with the antigenic peptide greatly attenuated the band intensity (Fig. 2A, lanes 13–14). Expression of NHA2 appeared to be ubiquitous in all mouse tissues examined (Fig. 2B), with high levels in pancreas. Indirect immunofluorescence of pancreatic β cells derived from rat indicated plasma membrane distribution of the endogenous protein (Fig. 2C). In the polarized, kidney-derived MDCK cell line transfected with GFP-HsNHA2, confocal microscopy showed that fluorescence was restricted to the apical domain, as seen by colocalization with fluorescent lectin (Rhodamine-wheat germ agglutinin) (Fig. 2D). Similarly, yeast cells expressing GFP-HsNHA2 showed predominant plasma membrane localization, albeit with some retention within intracellular compartments (Fig. 2E).
Fig. 2.
Distribution and subcellular localization of NHA2. (A) Western blot of membranes isolated from yeast AB11c expressing HsNHA1 (lanes 1, 4, 7, and 10), HsNHA2 (lanes 2, 5, 8, and 11), or neither (lanes 3, 6, 9, and 12). Antibodies raised against an N-terminal peptide of HsNHA2 recognizes a ≈55 kDa polypeptide in yeast membranes expressing HsNHA2 only (lanes 2 and 8) and not HsNHA1 (lanes 1 and 7). Control antibodies against epitope tags recognize both NHA proteins (anti-His, lanes 4 and 5; anti-GFP, lanes 10 and 11). Mouse pancreatic lysates were treated with anti-NHA2 antibody after preincubation in the absence (−, lane 13) or presence (+, lane 14) of antigenic peptide for 1 h. (B) Western blot, generated by using anti-NHA2 antibody, of lysates (100 μg) from the indicated mouse tissues. A prominent band of ≈50 kDa was observed in all tissues. (C) Immunofluorescence micrograph of rat pancreatic β cells INS-1 (832/13) generated by using anti-NHA2 antibody (−, Left) and peptide-blocked anti-NHA2 antibody (+, Right). (D Upper) Confocal fluorescence image of polarized MDCK cells treated with Rhodamine-labeled wheat germ agglutinin (red) and transfected with GFP-HsNHA2 (green). (Lower) Two cells expressing GFP fluorescence show apical localization in the z plane. (E) Confocal fluorescence image of B31 yeast expressing GFP-HsNHA2 at the cell surface.
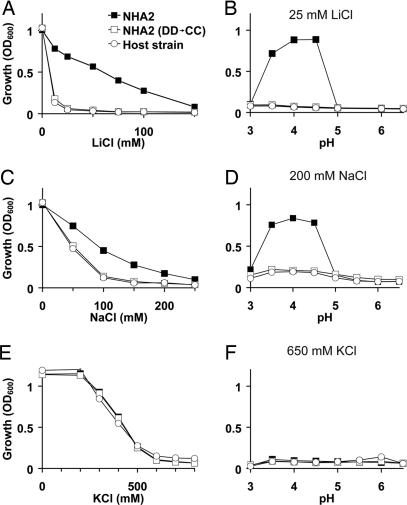
A useful strategy to obtain functional insight on a new gene is by phenotype complementation of the orthologous gene knockouts in yeast. We evaluated the ability of heterologously expressed HsNHA2 to rescue the salt-sensitive growth phenotype of the yeast strain AB11c (ena1-4Δnhx1Δnha1Δ) lacking three major salt-handling mechanisms. As expected, the host strain showed dose-dependent growth sensitivity to cationic salts of Li+, Na+, and K+. We show that HsNHA2 conferred tolerance to Li+ and Na+ but not to K+ (Fig. 3A, C, and E). Mutagenic replacement of the two aspartate residues, D278 and D279, of HsNHA2 that align with residues D163 and D164 of EcNhaA as shown in Fig. 1B, abolished the ability of HsNHA2 to confer salt-tolerant growth (double mutant DD→CC) (Fig. 3), consistent with a conserved critical role of these acidic residues in mediating cation transport. The complementation efficacy of HsNHA2 in yeast, for both LiCl and NaCl sensitivity, was optimal between pH 3.5 and 4.5 (Fig. 3 B and D), whereas there was no complementation of KCl sensitivity at all tested pH (Fig. 3F). In contrast, heterologous expression of mammalian NHE6 and NHE9 could confer robust growth at 1 M KCl in yeast strains lacking endogenous antiporters (17).
Fig. 3.
Functional complementation of salt-sensitive yeast phenotypes by heterologous expression of HsNHA2. The salt-sensitive yeast strain AB11c (Host strain) was transformed with His9-tagged HsNHA2 (NHA2) or the corresponding mutant with both Asp-278 and Asp-279 replaced by Cys [NHA2 (DD→CC)]. Yeast were grown in APG pH 4 medium supplemented with LiCl (A and B), NaCl (C and D), or KCl (E and F), as indicated, and growth was determined by optical density of the culture at 600 nm (OD600) after 20 h at 30°C. APG medium was supplemented with salt, as indicated; pH was adjusted by using phosphoric acid or Tris, as appropriate (B, D, and F); and cultures were grown to saturation. Data are the average of triplicate determinations.
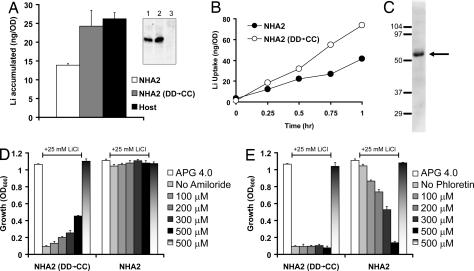
Because HsNHA2 was expressed on yeast plasma membrane, it seemed likely that salt tolerance was achieved by cation extrusion. Consistent with this hypothesis, HsNHA2 transformed yeast grown overnight in nontoxic levels of LiCl (2 mM) accumulated less Li+ relative to the DD→CC mutant or host strain (Fig. 4A). Similarly, addition of toxic levels of LiCl (25 mM) to exponentially growing cells resulted in lower rate of Li+ uptake in HsNHA2 transformants (Fig. 4B). We confirmed that expression levels of mutant were similar to that of wild-type HsNHA2 in yeast membranes (Fig. 4A Inset). Given the ubiquitous tissue distribution of NHA2 and the robust Li+ transport and tolerance phenotypes, we searched the literature for previous reports consistent with a physiological role for this transporter. We noted that SLC activity, postulated to be a variation of Na+(Li+)/H+ antiport, has long been linked to essential hypertension although the gene(s) responsible for this activity have not yet been identified (11, 14, 18). Defining characteristics of SLC activity are the absence of inhibition by amiloride, a diagnostic inhibitor of plasma membrane NHE isoforms, and inhibition by phloretin (19). We show that HsNHA2-mediated tolerance to LiCl (25 mM) was not inhibited by amiloride added to the growth medium (up to 500 μM) (Fig. 4D). Unexpectedly, salt-tolerant growth of the DD→CC mutant increased in the presence of amiloride; this may be due to weak base effects of the drug or to blocking of a sodium uptake channel. However, phloretin elicited a dose-dependent inhibition of the Li+-tolerant phenotype in HsNHA2 transformed cells but not in the corresponding DD→CC mutant (Fig. 4E). Neither amiloride nor phloretin was toxic to yeast growth in the absence of LiCl. Because phloretin is a relatively nonspecific inhibitor of glucose uniporters and other transporters, we also tested the effect of quinidine, previously reported to inhibit SLC activity (19). Like phloretin, quinidine inhibited the ability of NHA2 to confer salt-tolerant growth of yeast, without accompanying toxicity (SI Fig. 6). SLC activity appears to be ubiquitous and has been extensively characterized in erythrocytes (11, 20, 21). Consistent with NHA2 as a potential candidate for hypertension-linked SLC function, we show endogenous expression of NHA2 in red blood cells (Fig. 4C).
Fig. 4.
NHA2 has hallmarks of SLC activity. (A) Li+ accumulation was measured in yeast strain AB11c (Host) expressing His9-HsNHA2 (NHA2) or the corresponding mutant [NHA2 (DD→CC)] after overnight incubation in low levels of LiCl (2 mM). (Inset) Both WT (lane 1) and mutant NHA2 (lane 2) are expressed at similar levels in host strain AB11c (lane 3). (B) Li+ uptake was monitored in yeast strains expressing His9-HsNHA2 (NHA2) or mutant [NHA2 (DD→CC)]. Cultures were incubated in APG pH 4 medium supplemented with 25 mM LiCl for up to 1 h, as indicated. (C) Western blot of human erythrocyte lysate with anti-NHA2 antibody, showing a prominent band at ≈52 kDa (arrow). Amiloride (D) or phloretin (E) was added to yeast cultures, at the concentrations indicated. Yeast strains expressing NHA2 or DD→CC mutant were grown in APG pH 4 medium in the absence or presence of 25 mM LiCl as indicated by the line above the graphs. Dose-dependent inhibition of salt-tolerant growth (NHA2 transformed cells) was seen with phloretin (E) but not amiloride (D). Neither drug inhibited growth in the absence of LiCl (500 μM; last bar).
Discussion
A surprising and particularly useful outcome of our phylogenetic analysis was the inclusion of the well studied E. coli NhaA antiporters in the CPA2 subgroup (11). Many years of intensive biochemical and mutagenic study of EcNhaA recently culminated in the solution of a crystal structure of this membrane protein at 3.45 Å resolution by Padan and colleagues (22), the first and only structure of a cation/proton antiporter. Analogous to bacterial NhaA (23, 24), yeast strains expressing HsNHA2 were tolerant to Li+ and Na+ toxicity but not to K+. The similarity in sequence with bacterial NhaA suggests that the newly identified mammalian NHA (and other members of the CPA2 group) may have transport characteristics distinct from the ubiquitous NHE antiporters of the CPA1 subgroup, and they may therefore play a novel role in cell physiology. Unlike electroneutral NHE that mediate a 1:1 exchange of cations for protons (25), NhaA antiporters are electrogenic, with a H+/Na+ stoichiometry of 2 (26). Although we have not yet examined electrogenicity of transport by HsNHA2, there have been reports of electrogenic transport by fungal NHA antiporters (27, 28). Both NHE and NhaA show steep pH-dependent activation; however, their specific responses are quite distinct. Although the NHE are activated by cytosolic acidification (29), bacterial NhaA are essentially shut off below pH 6.5, with the Vmax for transport increasing by three orders of magnitude to maximal activity at pH 8.5 (30). These characteristics have a profound influence on the physiological role of the antiporter, allowing the bacterium to maintain pH homeostasis and survive in hypersaline and hyperalkaline environments. When expressed in yeast, HsNHA2 also has a very narrow pH range to complement salt-sensitive phenotypes (Fig. 3), although the acidic pH optimum is likely a reflection of the H+ motive force set up by the plasma membrane H+-ATPase, PMA1. Given the plasma membrane localization and Na+ selectivity of HsNHA2, we expect that the inwardly directed sodium gradient would provide the corresponding driving force for antiporter activity in mammalian cells. Electrogenic (nH+/Na+) exchange in this scenario would be expected to hyperpolarize the plasma membrane. Testing this prediction awaits the establishment of a functional assay of NHA2 in mammalian cells.
The ubiquitous tissue distribution and plasma membrane localization of NHA2 prompted us to search the literature for reports of Na+ and Li+ antiporter activity that might point to a physiological role. Increased SLC is a highly heritable trait and well known marker of essential hypertension commonly monitored in erythrocytes, although the gene(s) responsible have been sought after for more than two decades (11). SLC is considered to represent an alternative mode of Na+(Li+)/H+ antiport, and the ubiquitous NHE1 antiporter was initially an attractive candidate for this activity. Indeed, one study demonstrated that an alternatively spliced variant of NHE1, lacking the first three predicted transmembrane domains and the putative amiloride binding site, could mediate amiloride-insensitive SLC, although this truncated form of the transporter is devoid of Na+/H+ exchange function and may not have a physiological role (31). However, a comprehensive genetic analysis appears to have ruled out linkage between the NHE1 gene locus and SLC activity, pointing instead to the possibility of an unknown gene mediating SLC (32). Given the expression of HsNHA2 in red blood cells (Fig. 4C), we considered the possibility that this previously uncharacterized antiporter contributed to SLC activity. We showed that inhibitor characteristics of NHA2 expressed in yeast matched those of SLC activity, including insensitivity to amiloride (a hallmark of plasma membrane NHE), and sensitivity to phloretin and quinidine. In addition to genetic predisposition to hypertension and cardiovascular disease, elevated SLC has also been reported in subjects with diabetic and IgA nephropathy (33, 34). There is evidence that renal Li+ clearance, which is inversely related with proximal tubule Na+ reabsorbtion, is reduced in patients with high SLC (35). In this regard, the apical localization of NHA2 in the kidney-derived MDCK cell line is consistent with a role in Na+ reabsorption. Finally, we note that the gene loci of HsNHA1 and HsNHA2 (4q24) lie within a chromosomal region associated with hypertension in numerous linkage studies, including a sib-pair study linking SLC activity to MN blood group (4q28-q31) on chromosome 4 (36). In particular, one study in baboon mapped quantitative trait loci associated with SLC activity to chromosome 5, which is syntenic to human chromosome 4, with a maximum multipoint LOD score of 9.3 near microsatellite marker D4S1645 (37). Taken together, these findings raise the possibility that the previously uncharacterized NHA antiporters contribute to SLC activity and are candidate genes for essential hypertension.
Materials and Methods
Chemicals.
Amiloride hydrochloride hydrate (catalog no. A7410–5G), phloretin (catalog no. P7912–250MG), and quinidine (catalog no. Q3625–5G) were all purchased from Sigma.
Plasmids, Site-Directed Mutagenesis, Yeast Strains, and Growth Media.
Human NHA1 (HsNHA1) cDNA clone (accession number BM908201.1) and human NHA2 (HsNHA2) cDNA clone (accession number NM_178833) were purchased from Open Biosystems. The gene was amplified by PCR to include an N-terminal MluI site immediately before codon 1 (ATG) and a NotI site following the termination codon, and subsequently cloned into the equivalent sites of plasmid pSM1052 (a gift from Susan Michaelis, The Johns Hopkins University, Baltimore). This placed the ORF behind an N-terminal His9 or GFP epitope tag and under control of the constitutive PGK1 promoter in the yeast expression vector, as described earlier for yeast PMR1 (38, 39). Two conserved aspartic acid residues (D278 and D279) in HsNHA2 were mutated into cysteine (DD→CC) by using mutagenic primers in conjunction with the PCR. HsNHA2 was also subcloned into mammalian expression vector pEGFPC2 (Clontech) by using existing EcoRI and engineered NotI restriction sites.
Saccharomyces cerevisiae strains AB11c or B31W lacking endogenous Na+ pumps and antiporters (40) were used as host for heterologous expression of HsNHA2. Yeast cultures were grown at 30°C in APG, a synthetic minimal medium with minimal salt (10 mM arginine, 8 mM phosphoric acid, 2% glucose, 2 mM MgSO4, 1 mM KCl, 0.2 mM CaCl2, and trace minerals and vitamins) (41). Salt-sensitive growth was monitored by inoculating 0.2 ml of APG medium in a 96-well microplate with 4 μl of a saturated seed culture. After incubation at 30°C for 20–72 h, cultures were gently resuspended, and the OD600 was recorded on a FLUOStar Optima plate reader (BMG Labtechnologies).
Antibody Generation and Purification.
Polyclonal antibodies were raised in rabbit against a 15-aa peptide of HsNHA2 (24SMHQEAQEETVMKLK38C). HsNHA2 peptide (1 mg/ml) was immobilized on SulfoLink Coupling Gel (Pierce) through the C-terminal cysteine according to manufacturer's instructions. Peptide-specific antibodies were purified from antisera as described, concentrated by Centricon filtration (YM30; Millipore), and stored in PBS supplemented with 0.02% NaN3 at 4°C.
SDS/PAGE and Biochemical Techniques.
Total yeast lysates were prepared by using glass bead methods (42) from equal numbers of cells (4 OD600 units). Total lysates from mouse tissues were extracted by using N+ buffer (60 mM Hepes, pH 7.4, 150 mM NaCl, 3 mM KCl, 5 mM Na3EDTA, 3 mM EGTA, and 1% Triton X-100). Samples (50 μg) were subjected to SDS/PAGE and Western blotting. HsNHA2 was detected on a Western blot by anti-His6 antibody (1:5,000 dilution; BD Biosciences), anti-GFP antibody (1:2,000; Abcam), and anti-HsNHA2 antibody (1:2,000 dilution), as described in the figure legends. Where specified, purified anti-HsNHA2 antibody (1:2,000) was preincubated with antigenic peptide (5 μg/μl) at 25°C for 1 h. Horseradish peroxidase-coupled anti-rabbit (1:5,000; GE Healthcare) or anti-mouse secondary antibody (1:10,000; Amersham Biosciences) was used in conjunction with ECL reagents (Amersham Biosciences) to visualize protein bands.
Confocal Microscopy.
Confocal images were taken on a Zeiss LSM410 laser confocal microscope equipped with a Zeiss ×100 oil immersion lens. Live yeast cells harboring GFP-HsNHA2 were grown overnight and then visualized and imaged directly under microscope. MDCK cells were cultured in RPMI medium (Mediatech) containing 10% FBS (Invitrogen). Cells were grown on glass slides and transiently transfected with pEGFP-HsNHA2 by using LipofectAmine 2000 (Invitrogen) according to the manufacturer's instructions. One day after transfection, cells were placed at 4°C for 20 min and then treated with Rhodamine-conjugated wheat germ agglutinin (WGA) for 20 min to label apical membrane. After brief wash with PBS buffer, cells were visualized and imaged under microscope.
Immunofluorescence.
Rat pancreatic β cell line INS-1 (832/13) (43) was kindly provided by Christopher Newgard of Duke University (Durham, NC). The cells were cultured in RPMI 1640 medium with 10% FCS, 10 mM Hepes, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.05 mM 2-mercaptoethanol. β cells were fixed in 4% paraformaldehyde for 20 min, washed three times in PBS, and blocked with BSA (10% for 60 min). The coverslips were incubated with affinity-purified anti-HsNHA2 antibody (1:1,000 dilution) for 1 h. Where specified, purified anti-HsNHA2 antibody (1:1,000) was preincubated with antigenic peptide (5 μg/μl) at 25°C for 1 h. Secondary antibodies used were anti-rabbit Alexa Fluor 488 (1:1,000; Molecular Probes). Cells were imaged on a fluorescence microscope.
Li+ Uptake in Yeast.
For measurement of overnight Li+ accumulation, yeast strains were seeded in 24-well plates in APG pH 4.0 medium with or without 2 mM LiCl. After growth for 20 h at 30°C, yeast cells were harvested by centrifugation. For measurement of time-dependent Li+ uptake, LiCl was at a final concentration of 25 mM, and 1-ml yeast samples were harvested at the indicated time points. Harvested samples were washed three times with water and then resuspended in 1,050 μl of water. Fifty microliters of the sample was used for measurement of cell density (OD600). The remainder (1,000 μl) was centrifuged, and the pellet was dissolved in 25 μl of 1 M nitric acid. After overnight incubation at 37°C, 25 μl of 1 M NaOH was added to neutralize nitric acid. Then 950 μl of water was added to make final volume of 1 ml of solution. The diluted samples were analyzed for Li+ concentration by using a PerkinElmer Life Sciences AAnalyst 600 graphite furnace atomic absorption spectrophotometer.
Supplementary Material
Acknowledgments
We thank Michael Poli for technical assistance, Dr. Hana Sychrova for providing yeast strains, and Dr. Olga Kovbasnjuk and the Hopkins Digestive Diseases Basic Research Development Core Center (R24 DK64388) for help with confocal microscope. This work was supported by a grant from the National Institutes of Health to R.R. (DK54214) and a grant from the Binational Science Foundation jointly awarded to R.R. and Prof. Etana Padan (BSF2005013).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707120104/DC1.
References
- 1.Krulwich TA, Jin J, Guffanti AA, Bechhofer H. J Mol Microbiol Biotechnol. 2001;3:237–246. [PubMed] [Google Scholar]
- 2.Krulwich TA, Lewinson O, Padan E, Bibi E. Nat Rev Microbiol. 2005;3:566–572. doi: 10.1038/nrmicro1181. [DOI] [PubMed] [Google Scholar]
- 3.Lewinson O, Padan E, Bibi E. Proc Natl Acad Sci USA. 2004;101:14073–14078. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhter S, Nath SK, Tse CM, Williams J, Zasloff M, Donowitz M. Am J Physiol Cell Physiol. 1999;276:C136–C144. doi: 10.1152/ajpcell.1999.276.1.C136. [DOI] [PubMed] [Google Scholar]
- 5.Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S. Nature. 2000;407:581. doi: 10.1038/35036683. [DOI] [PubMed] [Google Scholar]
- 6.Putney LK, Barber DL. J Biol Chem. 2003;278:44645–44649. doi: 10.1074/jbc.M308099200. [DOI] [PubMed] [Google Scholar]
- 7.Brett CL, Tukaye DN, Mukherjee S, Rao R. Mol Biol Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denker SP, Barber DL. J Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gekle M, Mildenberger S, Sauvant C, Bednarczyk D, Wright SH, Dantzler WH. Am J Physiol. 1999;277:F251–F256. doi: 10.1152/ajprenal.1999.277.2.F251. [DOI] [PubMed] [Google Scholar]
- 10.Brett CL, Donowitz M, Rao R. Am J Physiol Cell Physiol. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 11.Canessa M, Adragna N, Solomon HS, Connolly TM, Tosteson DC. N Engl J Med. 1980;302:772–776. doi: 10.1056/NEJM198004033021403. [DOI] [PubMed] [Google Scholar]
- 12.Morrison AC, Boerwinkle E, Turner ST, Ferrell RE. Am J Hypertens. 2005;18:653–656. doi: 10.1016/j.amjhyper.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Schork NJ, Gardner JP, Zhang L, Fallin D, Thiel B, Jakubowski H, Aviv A. Hypertension. 2002;40:619–628. doi: 10.1161/01.hyp.0000037131.41957.a8. [DOI] [PubMed] [Google Scholar]
- 14.Zerbini G, Podesta F, Meregalli G, Deferrari G, Pontremoli R. J Hypertens. 2001;19:1263–1269. doi: 10.1097/00004872-200107000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Inoue H, Noumi T, Tsuchiya T, Kanazawa H. FEBS Lett. 1995;363:264–268. doi: 10.1016/0014-5793(95)00331-3. [DOI] [PubMed] [Google Scholar]
- 16.Padan E, Venturi M, Gerchman Y, Dover N. Biochim Biophys Acta. 2001;1505:144–157. doi: 10.1016/s0005-2728(00)00284-x. [DOI] [PubMed] [Google Scholar]
- 17.Hill JK, Brett CL, Chyou A, Kallay LM, Sakaguchi M, Rao R, Gillespie PG. J Neurosci. 2006;26:9944–9955. doi: 10.1523/JNEUROSCI.2990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerbini G, Gabellini D, Ruggieri D, Maestroni A. J Am Soc Nephrol. 2004;15(Suppl 1):S81–S84. doi: 10.1097/01.asn.0000093371.65105.8f. [DOI] [PubMed] [Google Scholar]
- 19.Pandey GN, Sarkadi B, Haas M, Gunn RB, Davis JM, Tosteson DC. J Gen Physiol. 1978;72:233–247. doi: 10.1085/jgp.72.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strazzullo P, Siani A, Cappuccio FP, Trevisan M, Ragone E, Russo L, Iacone R, Farinaro E. Hypertension. 1998;31:1284–1289. doi: 10.1161/01.hyp.31.6.1284. [DOI] [PubMed] [Google Scholar]
- 21.Romero JR, Rivera A, Monari A, Ceolotto G, Semplicini A, Conlin PR. J Hum Hypertens. 2002;16:353–358. doi: 10.1038/sj.jhh.1001387. [DOI] [PubMed] [Google Scholar]
- 22.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 23.Padan E, Schuldiner S. J Exp Biol. 1994;196:443–456. doi: 10.1242/jeb.196.1.443. [DOI] [PubMed] [Google Scholar]
- 24.Ros R, Montesinos C, Rimon A, Padan E, Serrano R. J Bacteriol. 1998;180:3131–3136. doi: 10.1128/jb.180.12.3131-3136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demaurex N, Orlowski J, Brisseau G, Woodside M, Grinstein S. J Gen Physiol. 1995;106:85–111. doi: 10.1085/jgp.106.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taglicht D, Padan E, Schuldiner S. J Biol Chem. 1993;268:5382–5387. [PubMed] [Google Scholar]
- 27.Ohgaki R, Nakamura N, Mitsui K, Kanazawa H. Biochim Biophys Acta. 2005;1712:185–196. doi: 10.1016/j.bbamem.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Kinclova-Zimmermannova O, Gaskova D, Sychrova H. FEMS Yeast Res. 2006;6:792–800. doi: 10.1111/j.1567-1364.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 29.Aronson PS, Nee J, Suhm MA. Nature. 1982;299:161–163. doi: 10.1038/299161a0. [DOI] [PubMed] [Google Scholar]
- 30.Taglicht D, Padan E, Schuldiner S. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 31.Zerbini G, Maestroni A, Breviario D, Mangili R, Casari G. Diabetes. 2003;52:1511–1518. doi: 10.2337/diabetes.52.6.1511. [DOI] [PubMed] [Google Scholar]
- 32.Lifton RP, Hunt SC, Williams RR, Pouyssegur J, Lalouel JM. Hypertension. 1991;17:8–14. doi: 10.1161/01.hyp.17.1.8. [DOI] [PubMed] [Google Scholar]
- 33.Mangili R, Bending JJ, Scott G, Li LK, Gupta A, Viberti G. N Engl J Med. 1988;318:146–150. doi: 10.1056/NEJM198801213180304. [DOI] [PubMed] [Google Scholar]
- 34.Boero R, Fabbri A, Degli Esposti E, Guarena C, Forneris G, Lucatello A, Sturani A, Quarello F, Fusaroli M, Piccoli G. Am J Kidney Dis. 1993;21:61–65. doi: 10.1016/0272-6386(93)70096-h. [DOI] [PubMed] [Google Scholar]
- 35.Weder AB. N Engl J Med. 1986;314:198–201. doi: 10.1056/NEJM198601233140402. [DOI] [PubMed] [Google Scholar]
- 36.Weder AB, Schork NJ, Julius S. Hypertension. 1991;17:977–981. doi: 10.1161/01.hyp.17.6.977. [DOI] [PubMed] [Google Scholar]
- 37.Kammerer CM, Cox LA, Mahaney MC, Rogers J, Shade RE. Hypertension. 2001;37:398–402. doi: 10.1161/01.hyp.37.2.398. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Marchi V, Wang R, Rao R. Biochemistry. 1999;38:14534–14541. doi: 10.1021/bi9911233. [DOI] [PubMed] [Google Scholar]
- 39.Mandal D, Rulli SJ, Rao R. J Biol Chem. 2003;278:35292–35298. doi: 10.1074/jbc.M306166200. [DOI] [PubMed] [Google Scholar]
- 40.Maresova L, Sychrova H. Mol Microbiol. 2005;55:588–600. doi: 10.1111/j.1365-2958.2004.04410.x. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Navarro A, Ramos J. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorin A, Rosas G, Rao R. J Biol Chem. 1997;272:9895–9901. doi: 10.1074/jbc.272.15.9895. [DOI] [PubMed] [Google Scholar]
- 43.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.