Abstract
Two-component signal-transduction systems (TCSs) of bacteria are considered to form an intricate signal network to cope with various environmental stresses. One example of such a network in Escherichia coli is the signal transduction cascade from the EvgS/EvgA system to the PhoQ/PhoP system, where activation of the EvgS/EvgA system promotes expression of PhoP-activated genes. As a factor connecting this signal transduction cascade, we have identified a small inner membrane protein (65 aa), B1500. Expression of the b1500 gene is directly regulated by the EvgS/EvgA system, and b1500 expression from a heterologous promoter simultaneously activated the expression of mgtA and other PhoP regulon genes. This activation was PhoQ/PhoP-dependent and EvgS/EvgA-independent. Furthermore, deletion of b1500 from an EvgS-activated strain suppressed mgtA expression. B1500 is localized in the inner membrane, and bacterial two-hybrid data showed that B1500 formed a complex with the sensor PhoQ. These results indicate that the small membrane protein, B1500, connected the signal transduction between EvgS/EvgA and PhoQ/PhoP systems by directly interacting with PhoQ, thus activating the PhoQ/PhoP system.
Keywords: two-component signal transduction
The two-component signal transduction system (TCS) is the major system in bacteria for sensing environmental changes and transducing the information inside the cells to regulate gene expression (1). TCSs control the expression of genes for nutrient acquisition, virulence, antibiotic resistance, and numerous other pathways in diverse bacteria. TCS is composed of a membrane-bound sensor histidine kinase (HK) that perceives environmental stimuli and a cognate cytoplasmic response regulator (RR). The sensor HK monitors the environmental stimuli by autophosphorylating its conserved histidine residue, which in turn phosphorylates the conserved aspartate residue of its cognate response regulator. In most cases, RR binds to DNA regulatory sequences and affects transcription.
In Escherichia coli, 29 HKs, 32 RRs, and one histidine containing phosphotransfer (HPt) domain have been found by analyses of the E. coli K-12 genome (2). Oshima et al. proposed the existence of a network of functional interactions, such as cross-talks (transfer of phosphoryl groups from a sensory HK to a noncognate RR) and cascade signal transduction among multiple TCSs (3). Several examples of in vivo (4, 5, 6) and in vitro (7, 8) cross-talks, as well as signal transduction cascade between TCSs in E. coli (9–11), have been reported.
We reported signal transduction cascade between EvgS/EvgA and PhoQ/PhoP TCSs in E. coli. The EvgS/EvgA system confers acid resistance and multidrug resistance to E. coli, and it is very similar to the virulence-related BvgS/BvgA system in Bordetella pertussis (12, 13, 14), whereas the PhoQ/PhoP system responds to external Mg2+ and Ca2+ levels, regulating expression of the genes including Mg2+ transporters and LPS modification genes (15, 16). Cultivation of E. coli in neutral rich media results in activation of the PhoQ/PhoP system but not the EvgS/EvgA system. An addition of high concentration of Mg2+ turns off the PhoQ/PhoP system, presumably by activating the phosphatase activity of the sensor PhoQ (17). However, in the evgS1 mutant strain, which constitutively activates the EvgS/EvgA system, the PhoQ/PhoP system remains active even at high Mg2+ levels. This signal transduction between the two TCSs can also occur as a result of overproduction of the EvgA regulator, which rules out phosphotransfer between the activated sensor EvgS and the noncognate regulator PhoP. Moreover, enhanced transcription of the phoPQ genes did not further activate expression of the PhoQ/PhoP regulated genes, and no EvgA binding consensus sequence (12, 18) was found in the promoter regions of the PhoP regulon genes. Thus, activation of the EvgS/EvgA system may increase or at least maintain the level of phospho-PhoP by (i) inhibiting the phosphatase activity of PhoQ, (ii) activating the PhoQ kinase, or (iii) protecting the phospho-PhoP against dephosphorylation by PhoQ.
It has been reported that some small (<200-aa) proteins regulate TCSs to adapt cells for rapid environmental changes. In Salmonella enterica, PmrD, a cytoplasmic protein composed of 85 aa, connects the PhoQ/PhoP and PmrB/PmrA TCSs (19). PmrD binds to the phosphorylated form of the response regulator PmrA at the N-terminal domain, thus preventing both its intrinsic dephosphorylation and that promoted by its cognate sensor kinase PmrB (20). Another small protein in Salmonella and in E. coli is IraP (86 aa), which enhances RpoS stability by interacting with MviA (Salmonella, 21) or RssB (E. coli, 22). Moreover, CpxP, a periplasmic protein (166 aa) of E. coli, interacts with the sensor domain of a histidine kinase CpxA to inhibit the CpxA/CpxR pathway (23).
In the present study, we have identified a regulatory protein, B1500, whose expression was enhanced by the EvgS/EvgA system; moreover, this protein activated the PhoQ/PhoP system, thus connecting the two TCSs. B1500, composed of 65 aa, is localized in the inner membrane and forms a complex with the sensor PhoQ. Therefore, B1500 directly interacts with PhoQ to activate the PhoQ/PhoP system. To our knowledge, this is the first report of such a small membrane protein connecting two TCSs.
Results
Cloning and Sequencing of a Gene Involved in Signal Transduction Between EvgS/EvgA and PhoQ/PhoP Systems.
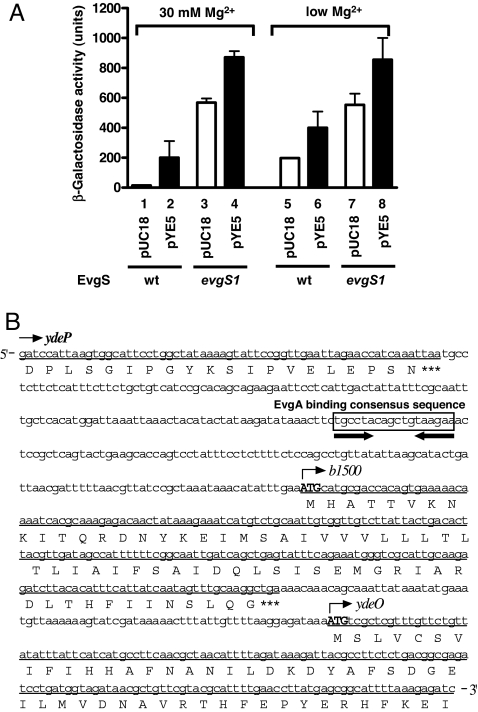
We set up a screening system to identify any additional factor(s) involved in signal transduction between the EvgS/EvgA and PhoQ/PhoP systems. Genomic DNA from MA204, a strain deleted for evgA, was partially digested with SauIIIAI and cloned into pUC18. This plasmid library was then transformed into MG1601, an E. coli strain with a lacZ gene under control of the mgtA promoter (a PhoP regulated gene) and selected for ampicillin and kanamycin resistance in the presence of X-gal and 100 mM MgCl2. At such a high concentration of Mg2+, the PhoQ/PhoP system is usually inactivated and mgtA expression is repressed. From this screening, we identified one positive plasmid, pYE5. As shown in Fig. 1A, MG1601/pYE5 grown at 30 mM Mg2+ concentration retained high mgtA transcriptional activity (column 2), whereas the control strain, MG1601/pUC18 at high Mg2+ concentration, showed suppressed expression (column 1). MG1601/pYE5 also showed higher mgtA transcriptional activity than MG1601/pUC18 at low Mg2+ concentration (columns 5 and 6).
Fig. 1.
Characterization of pYE5 plasmid obtained from shotgun screening. (A) Transcriptional activity of mgtA in MG1601/pUC18, MG1601/pYE5, MG1601evgS1/pUC18, and MG1601evgS1/pYE5 strains in the presence and absence of 30 mM MgCl2. (B) Nucleic acid sequence of the insert portion of pYE5 corresponds to AE000247 (complementary, 1581559–1582293). ORFs (underlined) included in the insert were the C-terminal part of ydeP, the complete b1500, and the N-terminal part of ydeO. Corresponding amino acids are shown under the line, and the stop codons are shown as ***. The EvgA binding consensus sequence (18) is boxed, and the inverted repeat is indicated by the bold arrows.
Sequencing of the insert region of pYE5 revealed the C-terminal portion of ydeP, the complete ORF of b1500, and the N-terminal portion of ydeO (Fig. 1B). All three of these genes have been reported to be directly regulated by EvgA, and b1500 composes an operon with ydeO (18). The gene ydeP codes for a putative oxidoreductase, and ydeO codes for a XylS/AraC-type regulatory protein (24). Overexpression of ydeP and ydeO confers acid resistance to E. coli (25), but the function of b1500 is unknown. In addition, an EvgA binding consensus sequence is present in the b1500/ydeO promoter (18) (Fig. 1B). Footprinting analysis showed that EvgA indeed bound directly to the promoter region of b1500 [supporting information (SI) Fig. 7]. Switching the host strain of MG1601/pYE5 to an EvgS-active mutant (MG1601evgS1) increased mgtA transcriptional activity at both low and high Mg2+ concentration (Fig. 1A, columns 4 and 8).
The b1500 Gene Is Involved in Signal Transduction Between EvgS/EvgA and PhoQ/PhoP.
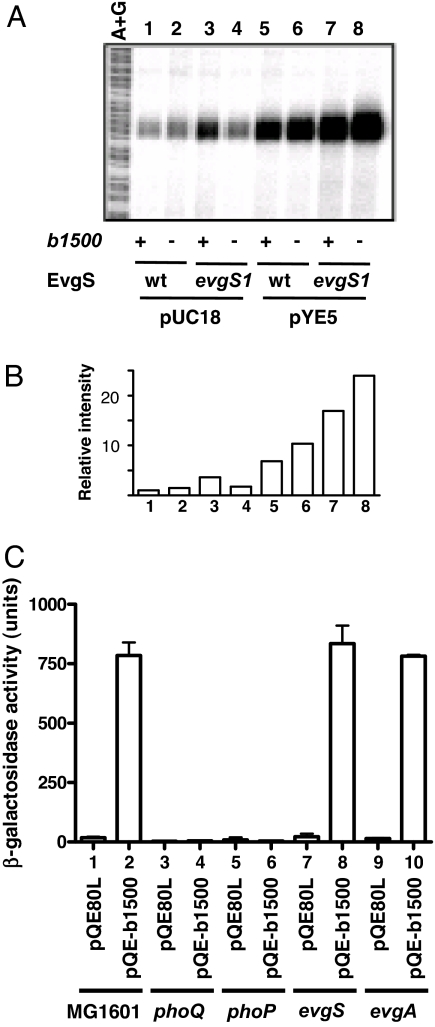
Because b1500 is the only gene in pYE5 that encodes a complete ORF, we hypothesized that b1500 may contribute to signal transduction between the EvgS/EvgA and PhoQ/PhoP systems. The wild-type strain and b1500-deleted strain showed repressed mgtA expression at high concentration of Mg2+ (Fig. 2 A and B, columns 1 and 2). When the EvgS/EvgA system was activated by the evgS1 mutation, mgtA expression was enhanced (Fig. 2 A and B, column 3) as reported (10). However, b1500-deleted evgS1 strain did not induce mgtA expression above the wild type level (Fig. 2 A and B, column 4). Furthermore, complementation of the MG1655evgS1 b1500 strain with pYE5 rescued the b1500 deletion (Fig. 2 A and B, column 8), whereas MG1655, MG1655 b1500, MG1655 evgS1 with pYE5 also enhanced mgtA expression (Fig. 2 A and B, columns 5, 6, and 7).
Fig. 2.
Functional analysis of b1500. (A) Deletion of b1500 suppresses mgtA transcription. S1 nuclease assay of mgtA was performed against the following strains grown in the presence of 30 mM MgCl2: lanes 1, MG1655/pUC18; lane 2, MG1655 b1500/pUC18; lane 3, MG1655 evgS1/pUC18; lane 4, MG1655 evgS1 b1500/pUC18; lane 5, MG1655/pYE5; lane 6, MG1655 b1500/pYE5; lane 7, MG1655 evgS1/pYE5; and lane 8, MG1655 evgS1 b1500/pYE5. A+G, Maxam–Gilbert sequencing ladder. (B) Radioactivity intensity of each lane in A relative to the radioactivity of lane 1. (C) Overexpression of b1500 promotes mgtA transcription in a PhoQ/PhoP-dependent and EvgS/EvgA-independent manner. Column 1, MG1601/pQE80L; column 2, MG1601/pQE-b1500; column 3, MG1607/pQE80L; column 4, MG1607/pQE-b1500; column 5, MG1622/pQE80L; column 6, MG1622/pQE-b1500; column 7, MG1601 evgS/pQE80L; column 8, MG1601 evgS/pQE-b1500; column 9, MG1601 evgA/pQE80L; and column 10, MG1601 evgA/pQE-b1500. Cells were grown in the presence of 30 mM MgCl2. For induction of b1500, IPTG, at a final concentration of 1 mM, was added to the culture at OD600 = 0.4.
Next, the b1500 ORF was expressed as a His-tag fusion protein in MG1601 and its phoQ-, phoP-, evgS-, and evgA-deleted strains. In the presence of 30 mM MgCl2, mgtA transcription was activated by expression of b1500 in MG1601 (Fig. 2C, column 2). In contrast, expression of b1500 had no effect in phoQ and phoP deleted strains (Fig. 2C, columns 4 and 6), which indicated that the transcriptional activation of mgtA by b1500 was PhoQ/PhoP-dependent. However, deletion of neither evgS nor evgA suppressed the activation of mgtA transcripton by b1500 (Fig. 2C, columns 8 and 10). Therefore, the transcriptional activation of mgtA was not due to the activation of the EvgS/EvgA system by b1500 but to a PhoQ/PhoP-dependent pathway.
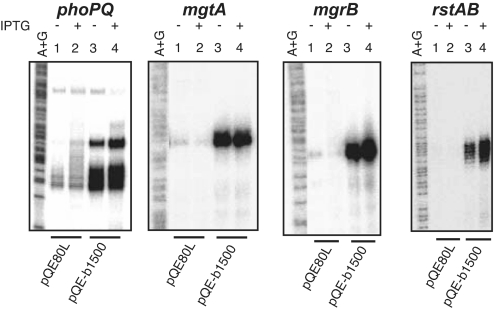
In our previous study (10), activation of the EvgS/EvgA system promoted transcriptional activity of PhoP regulated genes. Thus, we examined the effect of b1500 on transcriptional activities of four genes directly regulated by PhoP, namely, phoPQ, mgtA, mgrB, and rstAB (16) by S1 nuclease assay. As shown in Fig. 3, the presence of 30 mM MgCl2 repressed expression of these four genes (lanes 1 and 2). However, b1500 promoted expression of these four genes even in the presence of 30 mM MgCl2 (lanes 3 and 4). This indicated that b1500 promoted not only transcription of the mgtA gene, but also those of the other PhoP regulated genes, meaning that the PhoQ/PhoP system was activated. The genes, slyB, yrbL and hemL, which are also directly regulated by PhoP (16), were also activated by b1500 (SI Fig. 8). No apparent effect on transcription of the investigated promoters dependent on addition of isopropyl β-d-thiogalactopyranoside (IPTG) was observed, suggesting that only a small amount of B1500 was sufficient to activate the PhoQ/PhoP system.
Fig. 3.
Transcriptional activation of the PhoP regulon by b1500. S1 nuclease assay of phoPQ, mgtA, mgrB and rstAB was performed against cells grown in the presence of 30 mM MgCl2, with and without the addition of 1 mM IPTG at OD600 = 0.4. Lanes 1 and 2 are MG1601/pQE80L and Lanes 3 and 4 are MG1601/pQE-b1500.
The b1500 Gene Encodes a Small Membrane Protein.
The ORF, b1500, is only 198 bp long, and it is predicted to encode for a 65-aa protein. A BLASTP search of B1500 identified homologous ORFs in four Shigella species; Shigella flexneri, Shigella boydii, Shigella sonnei, and Shigella dysenteriae. Using MEMSTAT (26) and HMMTOP 2.0 (27) programs for topology predictions, we predicted that amino acids 19–40 may adopt a transmembrane domain structure (Fig. 4C).
Fig. 4.
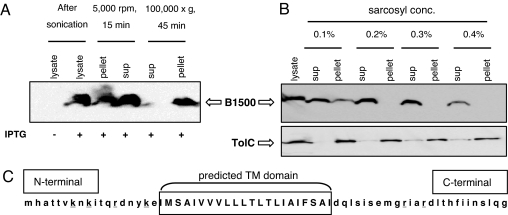
B1500 is localized in the inner membrane. (A) Western blot analysis against anti-His tag antibody for B1500 detection. MG1601/pQE-b1500 was grown in the presence of 30 mM MgCl2, and IPTG, at a final concentration of 1 mM, was added to the culture at OD600 = 0.4 for induction of B1500. Cells were lysed by sonication, followed by brief centrifugation and ultracentrifugation. (B) Sarcosyl treatment of the membrane fraction. Total membrane fraction after ultracentrifugation was treated with sarcosyl at different concentrations, followed by ultracentrifugation for the separation of inner and outer membrane. Anti-TolC antibody was used for the detection of TolC, an outer membrane protein control. (C) Predicted topology of B1500. Bold letters indicate the transmembrane domain predicted by the programs MEMSTAT and HMMTOP. Lysines and arginines are underlined.
To determine whether B1500 localized to the membrane, we overexpressed B1500 in MG1601, lysed the exponential phase cells by sonication, briefly centrifuged them to remove the cell debris and insoluble proteins, and ultracentrifuged the supernatant to obtain membrane fractions. To these fractions, Western blot analysis was performed by using an anti-His-tag antibody to detect the expressed His-tagged B1500 protein. As shown in Fig. 4A, B1500 was detected in the pellet after ultracentrifugation, indicating that it localized to the membrane. We next treated the membrane fraction with 0.1, 0.2, 0.3, and 0.4% sarcosyl and ultracentrifuged them to separate the inner (supernatant) and outer membrane (pellet). The majority of B1500 was detected from the inner membrane fraction, indicating that it localized to the inner membrane (Fig. 4B).
B1500 and PhoQ Interact in a Two-Hybrid Assay.
Given the fact that B1500 resided in the inner membrane, the most likely target of B1500 would be the sensor PhoQ. To investigate whether B1500 and PhoQ interact with each other, we used a bacterial two-hybrid system (28), which was recently applied to show direct interaction between membrane proteins of Bacillus subtilis (29). In this system, the proteins of interest are genetically fused to two fragments (T25 and T18) of the catalytic domain of B. pertussis adenylate cyclase and coexpressed in an E. coli deficient in endogenous adenylate cyclase. Interaction of the two hybrid proteins results in a functional complementation between the T25 and T18 fragments, which in turn relaxes catabolite repression and induces expression of lacZ among others. Hence, interaction of the two proteins can be studied by measuring the β-galactosidase activity.
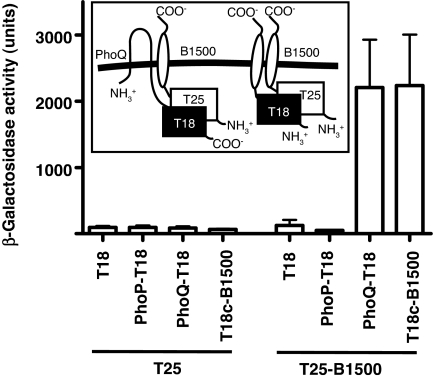
As demonstrated in Fig. 5, coexpression of T25-B1500 with PhoQ-T18 resulted in β-galactosidase activity that was 23-fold over the background (T25/T18), indicating that B1500 and PhoQ directly interacted with each other. Because the T25 fragment was fused to the N-terminal end of B1500, the N-terminal end of B1500 had to be exposed on the cytoplasmic side for the functional complementation between T25 and T18. Thus, according to the classification method suggested by S. J. Singer (30), B1500 is classified as a Type II membrane protein, because it spans the membrane once but has its N terminus on the cytoplasmic side of the cell and the C terminus on the exterior. This topology agrees with the “positive inside rule” (31), because four positively charged amino acids are clustered at the N-terminal end of B1500 (Fig. 4C). High β-galactosidase activity (Fig. 5) was also observed from a strain expressing T25-B1500 and T18c-B1500 (the T18 fragment was fused to the N-terminal end of B1500), suggesting that B1500 formed oligomers.
Fig. 5.
A bacterial two-hybrid assay reveals interaction between B1500 and PhoQ. β-Galactosidase activity was determined for adenylate cyclase-deficient E. coli strain, DHP1, harboring plasmids expressing B. pertussis adenylate cyclase fragment T18 and T25 either unfused or fused to b1500, phoP, and phoQ. β-Galactosidase activity above background levels indicates an interaction between the coexpressed hybrid constructs. Column 1, pT25/pT18; column 2, pT25/pT18-PhoP; column 3, pT25/pT18-PhoQ; column 4, pT25/pT18c-B1500; column 5, pT25-B1500/pT18; column 6, pT25-B1500/pT18-PhoP; column 7, pT25-B1500/pT18-PhoQ; and column 8, pT25-B1500/pT18c-B1500.
Discussion
The b1500 gene found by our genetic screening is located between the ydeP and ydeO genes (Fig. 1B). Nadler et al. (32) recently reported that YdeP and YdeO act conjointly to repress expression of the type III secretion system (TTSS) genes of enteropathogenic E. coli. When they overexpressed either ydeP or ydeO, TTSS protein production decreased. In contrast, overexpression of b1500 did not affect TTSS protein production. To date, no function has been assigned to b1500. In fact, an analysis performed in the 2005 E. coli annotation update (33) suggested that this gene did not exist in E. coli K-12, and therefore it was converted to an instance of the phantom genes class, implying that it is no longer considered an actual gene. However, in the present study, we have identified the gene, b1500, whose expression is directly induced by the EvgS/EvgA system, and expression of this gene from a heterologous promoter simultaneously activated the expression of PhoP regulon genes in a PhoQ/PhoP-dependent manner. These results suggest that b1500 is involved in signal transduction between EvgS/EvgA and PhoQ/PhoP TCSs, thus, associating a novel function with this small ORF.
As demonstrated in Fig. 4B, B1500 localized to the inner membrane. Membrane proteins such as NlpE (236 aa) and RcsF (134 aa), which are relatively small lipoproteins in the outer membrane of E. coli, activate the histidine kinases CpxA and RcsC, respectively (34, 35, 36). Although larger in size, YycH (458 aa) of B. subtilis is located external to the cell membrane and has one transmembrane domain at its N-terminal. Considering the location of this protein and the results of bacterial two-hybrid studies between YycH and the sensor YycG histidine kinase, YycH has been suggested to suppress the YycG/YycF system by direct interaction with the sensor domain of YycG (29, 37). Another example is that overproduction of DjlA (271 aa), an inner membrane protein belonging to the DnaJ family of E. coli, also activates the sensor RcsC (38). All of these membrane proteins interact with the histidine kinase sensor and not with the response regulator of TCSs. From our bacterial two-hybrid assay, we have found that B1500 also interacted with the sensor PhoQ to activate the PhoQ/PhoP system. Whether this activation results from inhibition of the phosphatase activity of PhoQ or from activation of the kinase activity of PhoQ remains to be investigated.
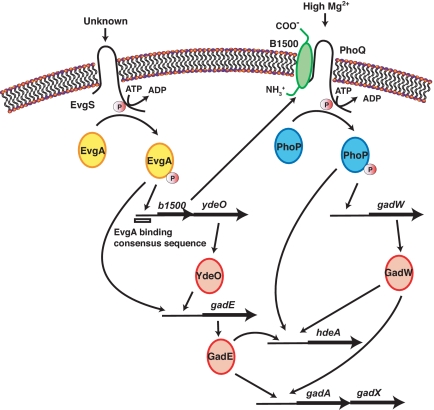
The EvgS/EvgA TCS is involved in acid resistance. Acid resistance in exponentially growing cells is induced by the overexpression of EvgA (18, 25, 39) and the activation of the sensor EvgS (unpublished results). The phosphorylated EvgA induces the b1500-ydeO operon, and the transcriptional regulator, YdeO, sequentially induces another transcriptional regulator, GadE. It has been reported that EvgA also directly induces GadE expression, forming a branched pathway of regulation (40). Subsequently, GadE directly up-regulates glutamate-dependent acid resistance genes, gadAX and gadBC, as well as hdeA, a periplasmic chaperone gene protecting the cell from organic acid stress, and hdeD, a putative membrane protein gene participating in acid resistance exhibited only at high cell densities (18, 40, 41, 42). These data indicate that the EvgS/EvgA system regulates the expression of acid resistance genes (Fig. 6).
Fig. 6.
A model of acid resistance controlled by EvgS/EvgA and PhoQ/PhoP TCSs connected by B1500.
Interestingly, the PhoQ/PhoP system also confers acid resistance. In a recent report, Zwir et al. (43) verified that PhoP played a critical role in the control of acid resistance: Transcription of hdeA and gadAX is induced at low Mg2+ in a PhoP- and YhiW (GadW)-dependent fashion (Fig. 6). These results indicate that both EvgS/EvgA and PhoQ/PhoP are involved in the expression of acid resistance genes. As shown by Masuda et al. (18), induction of the b1500-ydeO operon triggered acid resistance by YdeO, however, whether b1500 was involved in acid resistance had remained undetermined. In our recent study, deletion of b1500 from an EvgS-activated strain reduced the acid-resistant phenotype conferred by EvgS activation. Moreover, deletion of phoP or phoQ from an EvgS-activated strain also reduced such acid resistant phenotype (unpublished results). These results indicate that acid resistance induced by the activation of EvgS was strongly dependent on the PhoQ/PhoP system, suggesting a biological role for B1500.
In conclusion, the present study identified B1500 as a factor connecting the EvgS/EvgA and PhoQ/PhoP systems by directly interacting with PhoQ and increasing the level of PhoP phosphorylation. EvgS is reported to sense acidic pH in minimal media (40) and PhoQ of Salmonella also senses low pH by changing its conformation (44). By connecting the two systems, an intricate regulation network of acid resistance genes as shown in Fig. 6 is formed, which may help the cells for better adaptation to the acidic environment.
Materials and Methods
Bacterial Strains and Growth Conditions.
The E. coli strains used in this study are listed in Table 1. The phoP strain (MG1622) and phoQ strain (MG1607) were constructed with P1 transduction from the donors WP3022 and WQ3007 (16) to the recipient MG1601. Similarly, the evgA strain (MG1601 evgA) and evgS strain (MG1601 evgS) were constructed with P1 transduction from the donors MC204 and CS7 to the recipient MG1601. Deletion of b1500 was carried out by the method of Datsenko and Wanner (45). Primers b1500-H1 (5′-ACTGATTAACGATTTTTAACGTTATCCGCTAAATAAACATATTTGAAATGATTCCGGG GATCCGTCGACC-3′) and b1500-H2 (5′-ATTTCATATTTATAATTTGCTGTTTGTTTTCAGCC TTGCAAACTATTGATTGTAGGCTGGAGCTGCTTCG-3′) were used to delete the b1500 of strain BW25113. This BW25113 b1500 strain was used as the donor strain for P1 transduction to construct MG1655 b1500. The evgS1 mutants of MG1601, MG1655 and MG1655 b1500 were constructed by two consecutive P1 transductions with CS7 and KMR2001 as the donor strains.
Table 1.
Bacterial strains used in this study
| Strain | Description | Ref. or source |
|---|---|---|
| MG1655 | Wild-type E. coli | |
| MA204 | MC4100 evgA::kan | Laboratory stock |
| MC204 | MC4100 evgA::cat | Laboratory stock |
| CS7 | JC7623 evgS::cat | 12 |
| MG1601 | MC4100 mgtA:: placMu55 | 15 |
| MG1607 | MG1601 phoQ::cat | 15 |
| MG1622 | MG1601 phoP::cat | 15 |
| MG1601 evgS | MG1601 evgS::cat | This study |
| MG1601 evgA | MG1601 evgA::cat | This study |
| BW25113 b1500 | BW25113 b1500::kan | This study |
| MG1655 b1500 | MG1655 b1500::kan | This study |
| KMR2001 | KMY2001 fadL71::Tn10 | 12 |
| MG1601 evgS1 | MG1601 evgS1 fadL71::Tn10 | This study |
| MG1655 evgS1 | MG1655 evgS1 fadL71::Tn10 | This study |
| MG1655 evgS1 b1500 | MG1655 evgS1 fadL::Tn10 b1500::kan | This study |
| DHP1 | F-glnV44(AS) recA1 endA1 gyrA96 (Nalr) thi1 hsdR17 spoT1 rfbD1 cya- | 28 |
Bacteria were grown at 37° C in a Luria-Bertani (LB) medium [1% Bacto tryptone (Difco), 0.5% Bacto yeast extract (Difco), and 1% NaCl, pH 7.5]. MgCl2 was added to a final concentration of 30 mM when necessary. For induction of the T5 promoter of pQE-b1500, IPTG at a final concentration of 1 mM was added to the culture at OD600 of 0.4. Ampicillin, chloramphenicol, kanamycin, and tetracycline were used at 100 μg/ml, 25 μg/ml, 25 μg/ml, and 12.5 μg/ml, respectively.
Shotgun Screening.
A cloning vector, pUC18 (Nippon Gene), was digested with BamHI (Toyobo, Japan) and treated with alkaline phosphatase (calf intestine phosphatase; Nippon Gene). Genome DNA from MA204 (MC4100 evgA) was briefly digested with Sau3AI (Toyobo). The digested fragments larger than 1,000 bp were ligated to the BamHI-digested pUC18 and transformed to MG1601. Positive-blue colonies were selected on LB agar plates supplemented with 0.008% X-gal, 100 mM MgCl2, kanamycin, and ampicillin. Positive colonies were picked and amplified, and their plasmids were recovered. These plasmids were retransformed into MG1601 and spread onto the same supplemented LB agar plates for selection. The positive clones from this second selection were cultured in LB supplemented with 30 mM MgCl2, kanamycin, and ampicillin, and their β-galactosidase activity was assayed for the third selection. Plasmids were recovered from the final positive clones. Nucleic acid sequence of the insert region of these plasmids were performed by ABI3100 (Applied BioSystems) with pUC19F (5′-GGATGTGC TGCAAGGCGATTAAGTTGGGTA-3′) and pUC19R (5′-GTATGTTGTGTGGAATTGTGAGCG GATAAC-3′) primers, and a BLAST homology search of the acquired sequence was carried out.
Plasmid Construction.
The ORF of b1500 was amplified by PCR with pYE5 as the template and primers b1500FBamHI (5′-AAACATATCGGGATCCCGTGCGACCACAGT-3′) and b1500RHindIII (5′-TATTCCCAAGCTTGGGTTTGTTTTCAGCCT-3′). The PCR product was digested with BamHI and HindIII (Toyobo) and then ligated into BamHI and HindIII sites of pQE80L (Qiagen). The inserted region of the constructed plasmid was confirmed by sequencing, and the plasmid was named pQE-b1500. The amino acid sequence of the N-terminal region of the protein expressed from this plasmid is MRGSHHHHHHGSR. This sequence is fused with a B1500 fragment starting from the third amino acid (Ala) to the stop codon.
Bacterial two-hybrid vectors were created for PhoP, PhoQ, and B1500. The respective coding sequences were PCR-amplified introducing restriction sites, using the primers listed in SI Table 2. The PCR products were digested with respective restriction enzymes and cloned into plasmids, pT18, and pT25, resulting in pT18-PhoP, pT18-PhoQ, and pT25-B1500. The pT18 constructs expressed the respective genes fused to the N terminus of the B. pertussis adenylate cyclase gene fragment T18, and the pT25 constructs fused to the C terminus of fragment T25 (28). To fuse B1500 to the C terminus of fragment T18, the stop codon of T18 in pT18 was first deleted by ligating the PCR fragment amplified by primers T18F-KpnI and T18R-BamHI to pT18. This plasmid was named pT18c. A PCR-amplified fragment of b1500 digested with BamHI and NotI was cloned into the 3′ end of the T18 gene fragment of pT18c, resulting in pT18c-B1500.
β-Galactosidase Assay.
Cells grown to OD600 of 0.7–0.9 at 37° C in LB medium with appropriate supplements were subjected to β-galactosidase assay and expressed as Miller units (46). The data shown are means and standard deviations from three individual cultures.
RNA Preparation and S1 Nuclease Assay.
Total RNA was extracted with hot phenol from a mid-exponential-phase culture (OD600 0.7–0.9) at 60° C (47). S1 nuclease assay was carried out as described by Kato et al. (12). In brief, 32P-end-labeled probe was prepared by PCR, using the primers listed in SI Table 3, MC4100 genomic DNA as the template, and ExTaq DNA polymerase (Takara). A mixture of 32P-end-labeled probe and 100 μg of total RNA was incubated for 10 min at 75° C, then gradually cooled to 37° C and incubated overnight for hybridization, and finally subjected to S1 nuclease (Takara) digestion for 10 min at 37° C. Undigested RNA-probe DNA was extracted with phenol, precipitated with ethanol, and subjected to electrophoresis on a 6% (weight/volume) polyacrylamide sequencing gel containing 8 M urea. The radioactivity was measured by BAS1000 Mac (Fuji Film) and analyzed with MacBAS2.2 (Fuji Film).
Membrane Preparation and B1500 Detection.
Strain pQE-b1500/MG1601 was grown in an LB medium with appropriate supplements to a mid-exponential-phase (OD600 0.7–0.9). Cells were collected by centrifugation, and resuspended in lysis buffer [50 mM Tris·HCl (pH 8.0), 100 mM NaCl] with 1 mM PMSF. After sonication, the lysates were centrifuged for 10 min at 2,300 × g, 4° C, to remove cell debris and insoluble proteins. The supernatant was centrifuged again for 45 min at 100,000 × g, 4° C. The supernatant was collected as the cytoplasmic fraction, and the pellet was collected as the membrane fraction. For separation of the membrane to inner and outer membrane fractions, the membrane fraction was treated with 0.1, 0.2, 0.3, and 0.4% sarcosyl (N-lauroylsarcosine sodium salt solution; Fluka) for 1 h at room temperature followed by centrifugation for 45 min at 100,000 × g, 4° C. All of the samples were adjusted to equal final volume for comparison. Then, 10 μl of the samples were separated by SDS/PAGE, and the proteins were then transferred to a polyvinylidene difluoride membrane (Immobilon-P transfer membranes; Millipore) and probed with anti-His-tag antibody (GE Healthcare) or anti-TolC antibody (48). Detection was carried out with ECL Peroxidase-labeled anti-mouse antibody for anti-His tag antibody, ECL Peroxidase-labeled anti-rabbit antibody for anti-TolC antibody, and ECL Plus solution (ECL Plus Western blotting detection system; GE Healthcare). The results were analyzed by ImageQuant 400 (GE Healthcare).
Bacterial Two-Hybrid Assay.
The bacterial two-hybrid assay was performed essentially as described in ref. 28. Here, pT25-derived plasmids and pT18- or pT18c-derived plasmids were cotransformed into the adenylate cyclase-deficient E. coli strain, DHP1. Transformants were selected by using chloramphenicol and ampicillin. To quantify the interaction, β-galactosidase activity of mid-exponential-growth cells were measured.
Supplementary Material
Acknowledgments
We thank Drs. H. Aiba and K. Ho for valuable comments on the manuscript; Dr. D. Ladant for the generous gift of the plasmids pT25, pT18, and strain DHP1; and Dr. R. Misra for the anti-TolC antiserum. This work was supported by the Research and Development Program for New Bio-Industry Initiatives (2006–2010) of Bio-Oriented Technology Research Advancement Institution (BRAIN), Japan, and the “Academic Frontier” Project for Private Universities matching fund subsidy from the Ministry of Education, Culture, Sports, Science, and Technology (2004–2008), Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.A.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705768104/DC1.
References
- 1.Hoch JA, Silhavy TJ. Two-Component Signal Transduction. Washington, DC: ASM; 1995. [Google Scholar]
- 2.Mizuno T. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 3.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 4.Matsubara M, Mizuno T. Biosci Biotechnol Biochem. 1999;63:408–414. doi: 10.1271/bbb.63.408. [DOI] [PubMed] [Google Scholar]
- 5.Kim SK, Wilmes-Riesenberg MR, Wanner BL. Mol Microbiol. 1996;22:135–147. doi: 10.1111/j.1365-2958.1996.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 6.Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. Microbiology. 2002;148:69–78. doi: 10.1099/00221287-148-1-69. [DOI] [PubMed] [Google Scholar]
- 7.Yaku H, Kato M, Hakoshima T, Tsuzuki M, Mizuno T. FEBS Lett. 1997;408:337–340. doi: 10.1016/s0014-5793(97)00459-6. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 9.Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. J Bacteriol. 2003;185:5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eguchi Y, Okada T, Minagawa S, Oshima T, Mori H, Yamamoto K, Ishihama A, Utsumi R. J Bacteriol. 2004;186:3006–3014. doi: 10.1128/JB.186.10.3006-3014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eguchi Y, Utsumi R. Trends Biochem Sci. 2005;30:70–72. doi: 10.1016/j.tibs.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Kato A, Ohnishi H, Yamamoto K, Furuta E, Tanabe H, Utsumi R. Biosci Biotechnol Biochem. 2000;64:1203–1209. doi: 10.1271/bbb.64.1203. [DOI] [PubMed] [Google Scholar]
- 13.Utsumi R, Katayama S, Taniguchi M, Horie T, Ikeda M, Igaki S, Nakagawa H, Miwa A, Tanabe H, Noda M. Gene. 1994;140:73–77. doi: 10.1016/0378-1119(94)90733-1. [DOI] [PubMed] [Google Scholar]
- 14.Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. Microbiology. 2003;149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 15.Kato A, Tanabe H, Utsumi R. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castelli ME, Garcia Vescovi E, Soncini FC. J Biol Chem. 2000;275:22948–22954. doi: 10.1074/jbc.M909335199. [DOI] [PubMed] [Google Scholar]
- 18.Masuda N, Church GM. Mol Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 19.Kox LF, Wosten MM, Groisman EA. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato A, Groisman EA. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu X, Latif T, Bougdour A, Gottesman S, Groisman EA. Proc Natl Acad Sci USA. 2006;103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bougdour A, Wickner S, Gottesman S. Genes Dev. 2006;20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleischer R, Heermann R, Jung K, Hunkes S. J Biol Chem. 2007;282:8583–8593. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- 24.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda N, Church GM. J Bacteriol. 2002;184:6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones DT, Taylor WR, Thornton JM. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 27.Tusnády GE, Simon I. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 28.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szurmant H, Mohan MA, Imus PM, Hoch JA. J Bacteriol. 2007;189:3280–3289. doi: 10.1128/JB.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer SJ. Ann Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- 31.von Heijne G. J Mol Biol. 1986;192:287–290. doi: 10.1016/0022-2836(86)90365-7. [DOI] [PubMed] [Google Scholar]
- 32.Nadler C, Shifrin Y, Nov S, Kobi S, Rosenshine I. Infect Immun. 2006;74:839–849. doi: 10.1128/IAI.74.2.839-849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto K, Silhavy TJ. Proc Natl Acad Sci USA. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majdalani N, Heck M, Stout V, Gottesman S. J Bacteriol. 2005;187:6770–6778. doi: 10.1128/JB.187.19.6770-6778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castanie-Cornet M-P, Cam K, Jacq A. J Bacteriol. 2006;188:4264–4270. doi: 10.1128/JB.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szurmant H, Nelson K, Kim E-J, Perego M, Hoch JA. J Bacteriol. 2005;187:5419–5426. doi: 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke DJ, Holland IB, Jacq A. Mol Microbiol. 1997;25:933–944. doi: 10.1111/j.1365-2958.1997.mmi528.x. [DOI] [PubMed] [Google Scholar]
- 39.Nishino K, Yamaguchi A. J Bacteriol. 2001;183:1455–1458. doi: 10.1128/JB.183.4.1455-1458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma A, Masuda N, Foster JW. J Bacteriol. 2004;186:7378–7389. doi: 10.1128/JB.186.21.7378-7389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hommais F, Krin E, Coppée J-Y, Lacroix C, Yeramian E, Danchin A, Bertin P. Microbiology. 2004;150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 42.Mates AK, Sayed AK, Foster JW. J Bacteriol. 2007;189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Proc Natl Acad Sci USA. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prost LR, Daley ME, Sage VL, Bader MW, Moual HL, Klevit RE. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 47.Aiba H. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 48.Augustus AM, Celaya T, Husain F, Humbard M, Misra R. J Bacteriol. 2004;186:1851–1860. doi: 10.1128/JB.186.6.1851-1860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.