Abstract
Herpes simplex virus entry into cells requires four glycoproteins, gB, gD, gH, and gL. Binding of gD to one of its receptors triggers steps requiring the core fusion proteins, gB and the gH/gL heterodimer. There is evidence that gH/gL initiates hemifusion of cells, but whether this complex interacts physically with gB to cause complete fusion is unknown. We used bimolecular complementation (BiMC) of enhanced yellow fluorescent protein (EYFP) to detect glycoprotein interactions during cell–cell fusion. The N- or C-terminal half of EYFP was fused to the C terminus of gD, gB, and gH to form six chimeric proteins (Dn, Dc, Bn, Bc, Hn, and Hc). BiMC was detected by confocal microscopy. Receptor-bearing (C10) cells cotransfected with Dn and Bc or Dn, Hc, and untagged gL exhibited EYFP fluorescence, indicative of interactions between gD and gB and between gD and gH/gL. EYFP complementation did not occur in cells transfected with gL, Bc, and Hn. However, when gD was coexpressed with these other three proteins, cell–cell fusion occurred and the syncytia exhibited bright EYFP fluorescence. To separate glycoprotein expression from fusion, we transfected C10 cells with gL, Bc, and Hn for 20 h and then added soluble gD to trigger fusion. We detected fluorescent syncytia within 10 min, and both their number and size increased with exposure time to gD. Thus, when gD binds its receptor, the core fusion machinery is triggered to form a multiprotein complex as a step in fusion and possibly virus entry.
Keywords: BiMC, fusion, HSV, interaction, EYFP
Membrane fusion allows exchange of materials between cellular compartments enclosed by lipid membranes (1). Similarly, entry of enveloped viruses into cells allows the viral contents to be exposed by fusion of its envelope with a target cell membrane. Fusion requires disruption of both layers of the two membranes. For most enveloped viruses, a single surface glycoprotein undergoes conformational changes that bring the bilayer of the virus in proximity with that of the host cell and fusion ensues.
In contrast, herpesvirus entry requires three virion glycoproteins, gB and a gH/gL heterodimer, that function as the core fusion machinery. Some herpesviruses require additional proteins; e.g., alpha herpesviruses initiate fusion by binding of glycoprotein gD to a cell receptor (HVEM or nectin-1) (2). A conformational change then exposes the normally hidden receptor binding residues of gD. This change and/or the exposed residues trigger gB and gH/gL to effect virus–cell and cell–cell fusion (3, 4).
Based on crystal structure, gB has features that are typical of viral fusion proteins, and its domain structure is similar to that of the postfusion form of glycoprotein G of vesicular stomatitis virus (VSV) (5, 6). G is both the receptor binding and fusion protein of VSV, suggesting that gB is also a fusogen. However, herpes simplex virus (HSV) gB does not function as a fusion protein in the absence of gH/gL (3, 7, 8). Although the structure of gH/gL is unknown, indirect evidence suggests that this complex also has fusogenic properties. For example, synthetic peptides corresponding to several regions of HSV gH associate with membranes (9). Fusion-compromised HSV gH mutants have been isolated, emphasizing the importance of gH for fusion (10–12).
Influenza HA and Env A of avian sarcoma-leukemia virus (ASLV) can trigger hemifusion, whereby the altered outer leaflet of the target membrane allows mixing of the lipid layers but not of the contents of the virus (or effector cell) and target cell (13, 14). Full fusion allows content mixing, and for these viruses, this is accomplished by the same protein that causes hemifusion. For HSV, gH/gL can effect hemifusion in the absence of gB. However, this process still requires gD and its receptor. Additionally, content mixing requires all four glycoproteins (8), emphasizing the complexity of fusion induced by HSV glycoproteins. Although the hemifusion study clearly shows a role for both gH/gL and gB in fusion, a major issue is whether the process of cell–cell fusion (and virus entry) requires direct protein–protein interactions among gD, gB, and/or gH/gL. To address this issue, we used bimolecular complementation (BiMC) (15, 16). The idea is to split the gene for a reporter protein with an observable function into two inactive pieces, each of which is separately fused to the gene for the protein of interest. When the chimeric proteins are coexpressed, the two halves of the split reporter come close enough to restore activity (complementation), provided that the proteins of interest interact with each other. We used complementation of two halves of a variant of enhanced yellow fluorescent protein (EYFP), called Venus, to provide visual results in cells (17, 18). Venus was engineered to overcome the need to incubate cells at low temperature for long periods of time to enhance development of the inherent fluorescence, properties of its parent form. EYFP complementation is essentially irreversible, thereby stabilizing weak or transient interactions between the proteins of interest (15, 16).
We created six fusion proteins, each consisting of one Venus fragment (Venus N or Venus C) linked to the C terminus of either gD, gB, or gH (termed Dn, Dc, Bn, Bc, Hn, and Hc) and examined interactions of various combinations expressed in transfected cells by confocal microscopy. We found that chimeric gD interacted in receptor-bearing cells with chimeric gB in the absence of gH/gL; it also interacted with chimeric gH/gL in the absence of gB. In contrast, chimeric forms of gB and gH/gL did not interact with each other in the absence of gD. However, when cells were cotransfected with plasmids for gD and gL, along with chimeric forms of gB and gH/gL, syncytia formed and BiMC occurred, indicating that gB and gH/gL formed a complex within each syncytium. Does this complex form in cells during glycoprotein synthesis or does it happen once the proteins are on the cell surface and gD contacts a receptor on an adjacent cell? To answer this question, we transfected receptor-bearing C10 cells with chimeric forms of gB and gH along with untagged gL and then added exogenous gD306t to trigger fusion. Fluorescent syncytia were seen as rapidly as 10 min after gD addition, indicating that binding of gD to its receptor concurrently triggers both fusion and the interaction between gB and gH/gL. We suggest that this interaction is a necessary event in the pathway to cell–cell fusion.
Results
Analysis of EYFP-Tagged gD, gB, and gH.
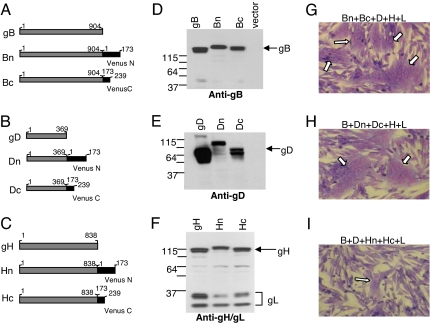
DNA encoding the N- and C-terminal fragments of the Venus form of the EYFP gene (18) were ligated to the 3′ ends of ORFs for full-length gD, gB, and gH in the pCAGGS plasmid (19). This placed the tags on the C terminus of each glycoprotein (Fig. 1 A–C). We left gL untagged. To test the properties of the chimeras, we transfected receptor-bearing B78H1-nectin-1 (C10) cells (20) with individual ones or pairs of plasmids. When gH was involved, cells were also cotransfected with gL, because it is required for proper folding and transport of gH (21). We examined cell lysates by SDS/PAGE and Western blotting (Fig. 1 D–F). All constructs were expressed, and their electrophoretic mobilities were consistent with the expected sizes of the chimeras. The presence of “mature” gL (37 K band in Fig. 1F) indicated proper formation and processing of each gH/gL complex (10, 22). To verify that each protein retained its function in cell–cell fusion, we transfected C10 cells with plasmids for each construct, along with the other three entry glycoproteins, and monitored fusion by formation of multinucleated cells (syncytia). Each of the chimeric proteins was functional [Fig. 1 G–I and supporting information (SI) Fig. 6]. We did similar experiments with pairwise combinations of the chimeras (along with the appropriate untagged glycoproteins). All pairs were functional except when chimeras of gD and gH were used (SI Fig. 6). We confirmed the results by a quantitative luciferase fusion assay (19) (not shown).
Fig. 1.
Construction and analysis of EYFP constructs. (A–C) Glycoprotein-EYFP chimeras used in this study. (D and E) Western blot analysis. C10 cells were transfected with EYFP-tagged or WT HSV proteins. Cell lysates were analyzed by using R68 anti-gB (D), R8 anti-gD (E), or R137 anti-gH/gL (F) antibodies. (G–I) Giemsa staining of syncytia. C10 cells were transfected with plasmids expressing gB, gD, gH, and gL; in each case, a pair of plasmids for the EYPF-tagged version were used in place of the WT version: G, Bn-Bc; H, Dn-Dc; I, Hn-Hc. Arrows denote syncytia.
Homologous Interactions of gB, gD, and gH/gL.
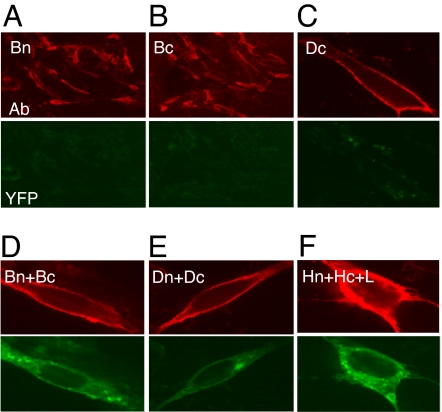
Because the gB ectodomain crystallizes as a trimer (6), we hypothesized that the cytoplasmic tails of this protein are in close proximity and would drive BiMC. To test this, we transfected C10 cells with plasmids for Bn, Bc, or Dc (Fig. 2 A–C) and incubated the cells for 20 h. Fixed cells were stained with monoclonal antibodies (mAbs) to gB or gD to detect surface protein expression (red channel) and also examined for EYFP fluorescence (green channel). As expected, cells that expressed only one chimera were positive for gB but exhibited no EYFP fluorescence and only minimal autofluorescence (Fig. 2 A–C). However, cells that were cotransfected with Bn+Bc exhibited bright EYFP fluorescence (Fig. 2D), indicating that the C termini of gB trimers are close together and allow BiMC. EYFP was seen both internally and on the cell surface (Fig. 2D). Similar results were obtained when we cotransfected cells with Dn+Dc (Fig. 2E), suggesting that gD is oligomeric on transfected cells. These results are consistent with those from cross-linking studies showing that virion gD is dimeric (23). Interestingly, we also detected gH/gL oligomers (Fig. 2F), a new finding not expected from biochemical experiments with the isolated gH/gL heterodimer (24). Our results suggest that gH/gL-gH/gL oligomers are expressed on the cell surface as well as inside the cells.
Fig. 2.
Glycoprotein homooligomerization. C10 cells were transfected, fixed, and stained with the appropriate antibodies (red): anti-gB (A, B, and D), anti-gD (C and E), or anti-gH (F). Fluorescent images were captured at ×20 (A and B) or ×100 (C–F) magnification. The same camera settings were used for green and red fluorescence.
Because we obtained BiMC for each glycoprotein, we wanted to be certain that fluorescence was due to the presence of a natural oligomer and not to an artifact—e.g., overexpression that might drive two independent proteins together. Therefore, we did two types of experiments. First, we transfected cells with decreasing amounts of DNA and observed a concurrent decrease in the number of cells expressing the particular glycoprotein (red) and the number of cells exhibiting EYFP fluorescence (green) (shown for gD in SI Fig. 7A). In all cases where we observed green cells, we also detected the particular glycoprotein. Cells that appear less bright for EYFP than their neighbors were also less bright for antibody staining. As a second demonstration of specificity, we carried out “cold” competition experiments. This classic biochemical approach uses an unlabeled competitor to compete for binding with the labeled versions of the same molecule. Here, we cotransfected C10 cells with plasmids for both chimeras along with increasing amounts of plasmid DNA for untagged gB, gD, or gH as appropriate (SI Fig. 7B). Using gD as an example, if the interaction is specific, then untagged gD will interact with either of the tagged forms of gD and fluorescence should decrease as a function of competitor concentration. In fact, this is what we observed for each homooligomeric interaction (Fig. 2) and all heterooligomeric interactions involving gD. As a final control, we cotransfected C10 cells with Dn plus a control glycoprotein [A33 from vaccinia virus (25)] tagged with the N-terminal Venus fragment (described in SI Fig. 7C). Although we detected abundant gD (red), we observed no green fluorescence due to EYFP complementation. Similar results were found for gB and gH (not shown). Thus, the untagged glycoprotein specifically competed with one or the other chimeric partner and led to a decrease in BiMC. We conclude that formation of homooligomers of gB and gD leads to EYFP complementation. Likewise, formation of higher-order oligomers of gH/gL also leads to BiMC.
Heterologous Interactions Occur Among the Entry Glycoproteins.
Because our major interest is in learning whether heterologous interactions occur among gB, gD, and gH/gL in the context of virus entry and cell fusion, we next asked whether we could detect them using BiMC. We first tested heterologous combinations under conditions where no cell–cell fusion could occur—i.e., by omitting one essential glycoprotein or by using B78H1 cells lacking a gD receptor (20). We then did similar experiments but under conditions where we expected cell–cell fusion (syncytium formation) to occur—i.e., with all four glycoproteins present plus a functional gD receptor.
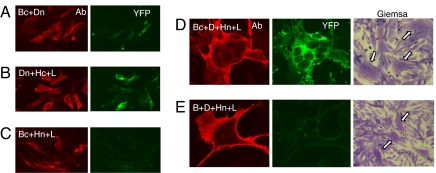
When cells were transfected with Bc+Dn, many of the gB-expressing cells exhibited EYFP fluorescence indicative of a gB/gD interaction (Fig. 3A). The same was true when the tags were reversed (i.e., Bn+Dc; not shown). EYFP fluorescence was diminished by cotransfection with increasing amounts of DNA for untagged gD or gB, indicative of specificity (not shown). Likewise, EYFP fluorescence was detected in cells transfected with plasmids for Dn+Hc+gL (Fig. 3B). Importantly, no EYFP was detected in cells cotransfected with plasmids for Bc+Hn+gL (Fig. 3C) or with plasmids expressing the gB and gH chimeras with the reverse tags (not shown). These results suggest that gD interacts individually with both gB and gH but that gB and gH do not interact with each other in the absence of gD. Perhaps interactions between gD and other entry glycoproteins normally occur transiently even in the absence of receptor or they differ from interactions that occur when gD engages receptor.
Fig. 3.
BiMC detects heterologous interactions between glycoproteins. C10 cells were transfected with the indicated DNAs for 20 h, fixed, and stained with anti-gD mAbs (A, B, D, and E) or fixed, permeabilized, and stained with anti-gB mAbs (C). All were analyzed by immunofluorescent assay for protein (red) and YFP (green). Confocal images were captured at ×40 (A–C) or ×100 (D and E) magnification. All images were captured by using the same camera setting. In a parallel experiment (D and E), cells were stained with Giemsa and examined at ×20 magnification. Arrows indicate syncytia.
Because Venus-tagged gD can interact with tagged gB and tagged gH and form stable complexes before fusion, we could not follow what happened to these complexes during fusion. Moreover, these interactions between gD and the other glycoproteins occurred in the parental B78 cells that lack a gD receptor (not shown). However, the lack of an interaction between gB and gH/gL (in the absence of gD) was important because we could then determine whether they associate with each other under conditions that permit fusion. Therefore, we cotransfected C10 cells with Bc+Hn+gL+gD for 20 h to allow for protein expression and cell–cell fusion (Fig. 3D). As expected, syncytia formed (Fig. 3D, Giemsa stain) and the fused cells expressed gD (red). The syncytia also exhibited bright EYFP fluorescence. To control for autofluorescence in syncytia, we transfected cells with gB+gD+Hn+gL, leaving only one EYFP partner (Hn) in the transfection mix (Fig. 3E). Here, the syncytia exhibited dull or no green fluorescence. Because gH is functional only when paired with gL (21), we assume that the interaction is between Bc and an intact Hn/gL complex. We conclude that although gB and gH/gL do not interact before fusion, they do interact at some point during syncytium formation. Moreover, this interaction did not occur in parental cells lacking a gD receptor (not shown).
Fusion Can Be Triggered with Soluble gD (gD306t).
Because C10 cells transfected with Bc+gD+Hn+gL were expressing all four glycoproteins and undergoing fusion during the 20-h incubation, we could not tell whether the gB and gH association occurred during fusion or was a consequence of it. Therefore, we explored ways to separate protein expression from fusion. Earlier studies showed that gD306t complements the infectivity of a virus lacking the gD gene (26). We asked whether we could use gD306t to trigger fusion of cells that were already expressing gB, gH, and gL.
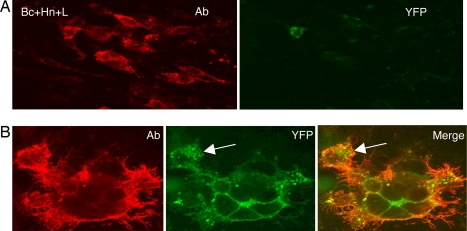
We cotransfected C10 cells with wild-type forms of gB, gH, and gL for 20 h and then added gD306t for an additional 4 h. We detected syncytia, although there were fewer (≈50% as many) than when the plasmid for full-length gD was included in the transfection mix (not shown). However, these results encouraged us to try it with chimeric gB and gH. Coverslips were seeded with 104 C10 cells and transfected for 20 h with Bc+Hn plus untagged gL. As expected, no syncytia formed because no gD was present (Fig. 4A). When we added gD306t and incubated the cells for 4 h, we detected green syncytia (Fig. 4B). Both gB protein and EYFP fluorescence were seen on the outside edges of the syncytia. The gB/gH/gL complex was also concentrated in the compressed cytoplasm that surrounded the nuclei, thereby defining the individual cells within the syncytia. Although the protein stain did not reveal these cells, we believe that this is a matter of antibody access to all parts of this particular syncytium. When we permeabilize the cells, both the protein stain and the green fluorescence were observed throughout the syncytia (SI Fig. 8). When we costained for both proteins, gH colocalized with gB (not shown). Occasionally, we observed a green fluorescent cell that was incompletely fused to the rest of the syncytium (arrows in Fig. 4), as though it was in the process of fusing. These observations suggested that we could incubate the transfected cells with soluble gD for shorter times to follow both gB/gH association and fusion as they developed. To some extent, how rapidly we could detect BiMC depended on the rapid development of Venus EYFP fluorescence (17, 18).
Fig. 4.
Synchronization of fusion with soluble gD. (A) C10 cells were transfected with Bc+Hn+gL plasmids for 20 h, fixed, permeabilized, stained with anti-gB mAbs (red), and also examined for YFP in the green channel. (B) Same as in A, except that at 20 h posttransfection, 250 μg/ml gD306t was added and cells were incubated for an additional 4 h before fixation. Images were captured at ×40 (A) or ×100 (B) magnification. The same camera setting was used to capture red, green, and merged images. In the merged image, the green fluorescence is very bright (yellow) or less bright (orange). An arrow indicates an incompletely fused cell.
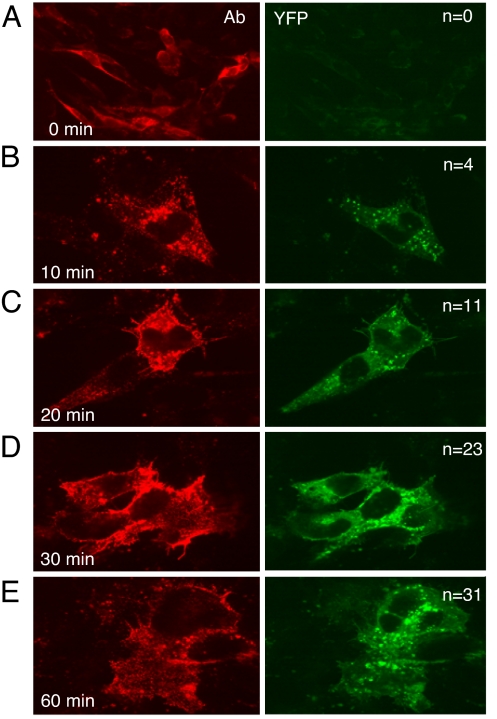
Cells were transfected as before with gL, Bc, and Hn for 20 h. As expected, no syncytia were observed (Fig. 5A). We then added exogenous gD306t for different times posttransfection. Surprisingly, we detected fusion (cells with two to three nuclei per cell) within 10 min after gD306t addition (Fig. 5B). Although only a small number of cells formed syncytia over the 1-h time course, both the total number of syncytia on each coverslip (n in Fig. 5) and the total number of nuclei per syncytium increased over time (Fig. 5 C–E). In fact, we observed EYFP fluorescence in every developing syncytium, indicating that Bc and Hn always interacted concurrently with fusion. The interaction did not occur when gD was absent, nor did it occur in B78H1 cells lacking a gD receptor (not shown). Also, no gB/gH/gL interaction and no fusion was triggered by gD(Δ222–224)306t (not shown), a form of gD that is unable to bind gD receptors. This control was important in light of our observation that gD can interact with gB and gH/gL in the absence of a receptor. Thus, the interaction between gB and gH/gL occurred only when gD was able to interact with its receptor, nectin-1.
Fig. 5.
Time course of fusion triggered by gD306t. C10 cells were transfected with Bc+Hn+gL plasmids for 20 h. Soluble gD306t was added for 0 (A), 10 (B), 20 (C), 30 (D), or 60 min (E). Cells were fixed, permeabilized (A), and stained with anti-gB mAbs (red). EYFP was examined in green channel. Confocal images were captured at ×40 (A) or ×100 (B–E) magnification. The numbers of syncytia per coverslip (one experiment) are indicated by n.
These observations, together with the rapidity of the interaction after gD addition, support the idea that the interaction between gB and gH/gL occurs concurrently with fusion. Moreover, this event occurs on the cell surface in response to gD binding to its receptor.
Discussion
In many viruses, envelope glycoproteins are in a multimeric state, which might be essential for optimal structure of the virion, and HSV is no exception in this regard. All of the glycoproteins that decorate the HSV virion surface are homooligomers (e.g., gB) or partners in a heterooligomeric complex with another glycoprotein (e.g., gH/gL). Data from the crystal structure show that gB is a trimer (6). Likewise, gH must form a noncovalent complex with gL on cell surfaces and in the virion envelope to be functional (21, 22, 24). Chemical cross-linking studies identified several interactions between glycoproteins on the virion (23). However, it has been argued that these interactions between the essential glycoproteins do not indicate formation of functional complexes in the virion envelope (27). Because these associations change during virus entry (28), it is possible that a fusion complex forms when HSV contacts a cell. Recent electron microscopy studies suggest that the glycoproteins coalesce at one end of the envelope when the virus contacts a cell (K. Grunewald, personal communication). Another method that has been used to identify members of a fusion complex is coimmunoprecipitation (29). Although an interaction between gD and gH/gL was detected, how it related to fusion was not clear.
We used BiMC to investigate protein–protein interactions in vivo. Most BiMC studies have focused on soluble proteins, although several have examined interactions of integral membrane proteins (30–32). One problem that is mentioned in these reports is where to place the tag. Although the tag could potentially hinder the overall folding of the protein or its ability to carry out its normal function, we found that this was not a problem when the proteins were expressed singly or in homooligomeric combinations. BiMC due to homooligomerization of gB was expected based on crystal structure of the soluble protein (6). We considered it likely that gD also dimerizes, based on cross-linking studies (23, 27). However, BiMC of gH/gL to form higher-order oligomers is a new finding. Because BiMC stabilizes transient or weak interactions, these higher-order oligomers may not be normally stable enough to detect either by immunoprecipitation or employing a truncated form of gH bound to gL. In any case, we propose that gH/gL may form homooligomers that partner with gB trimers during fusion.
The heterologous combinations that we have examined provide important information regarding the interactions between HSV glycoproteins both before and during fusion. Despite abundant data about herpesvirus entry, most of what we know concerns the initial binding steps. We know that HSV initially attaches to cells by binding of gC and gB to cell-surface glycosaminoglycans (33). The next step, which is essential, involves binding of gD to its receptor along with associated conformational changes that launch the fusion cascade. Fusion requires both gB and gH/gL. If any of the four proteins is missing, virus entry does not occur, nor does cell–cell fusion (34–36). According to a recent study (8), binding of gD to its receptor is followed by hemifusion carried out by gH/gL, and full fusion is completed by the combination of gH/gL and gB. Our BiMC data suggest that gD triggers gH/gL and gB to interact with each other concurrently with fusion. Is the interaction a consequence of fusion, or is it a step in the pathway to fusion? Although a definitive answer is lacking, we noted that sometimes a cell that was adjacent to a syncytium was incompletely fused to the rest of the multinucleated giant cell. These incompletely fused cells exhibited EYFP fluorescence due to the interaction of Bc with Hn, suggesting that the gB/gH/gL interaction is a step in fusion and not a consequence of it.
Do other interactions take place as well? We found that gB and gD can form a stable complex that does not interfere with their ability to function in fusion. In contrast, when a complex between gD and gH/gL is stabilized by BiMC, no fusion occurs (SI Fig. 6). We suggest that gD and gH/gL may normally interact in a transient manner, and that when a receptor is present, hemifusion occurs. However, if this interaction is stabilized, as is the case with the chimeric forms, hemifusion might occur, but full fusion would be blocked because no gB/gH/gL interaction could occur. This model is consistent with the observations made by Subramanian and Geraghty (8). Future studies of hemifusion using the tagged forms of gB and gH may resolve this issue.
The BiMC approach opens up other avenues for future study. For example, we should be able to follow both fusion and gB–gH/gL interactions in real time and examine whether various gD mutants trigger the gB–gH/gL interaction without triggering cell–cell fusion. Such studies should provide more details of the mechanism by which the quartet of HSV glycoproteins cooperate to cause both cell–cell fusion and virus entry.
Materials and Methods
Construction of Plasmids Expressing HSV Glycoproteins Tagged with EYFP Fragments.
All chimeric constructs were cloned into the pCAGGS expression vector (19). EYFP sequences and full-length glycoproteins gB, gD, and gH (KOS strain) were PCR-amplified. The glycoproteins primers excluded the natural translation stop codons to allow in frame ligation with N (Yn) or C-terminal (Yc) portions of the EYFP ORF. The template for EYFP sequence was plasmid pCS2 (18). For fusion constructs carrying the Yn, DNA for residues 1–173 was amplified. A translation stop codon was incorporated after codon 173. For fusion constructs involving the Yc, DNA for EYFP 173–239 was amplified. The following primers were used. gD fwd: 5′-CGCGAATTCATGGGGGGGGCTGCCGCCAGGTT. gD rev: 5′-CCGCTAGGTACCGTAAAACAAGGGCTGGTGCGA. gB fwd: 5′-CGCGAATTCATGCACCAGGGCGCCCCCTCGT. gB rev: 5′-CCGCTAGGTACCCAGGTCGTCCTCGTCGGCGTCA. gH fwd: 5′-GCGGAGCTCATGGGGAATGGTTTATGGTT. gH rev: 5′-CCGCTAGCATGCTTCGCGTCTCCAAAAAAA. EYFP fwd Kpn: 5′-CCGGGTACCATGGTGAGCAAGGGCGAGGAGCTGTT. EYFP fwd 173 Kpn: 5′-CCGGGTACCGACGGCGGCGTGCAGCTCGCCGACCACTA. EYFP fwd Sph: 5′-CCGGCATGCATGGTGAGCAAGGGCGAGGAGCTGTT. EYFP fwd 173 Sph: 5′-CCGGCATGCGACGGCGGCGTGCAGCTCGCCGACCACTA. EYFP 173 rev Xho: 5′-GCCTCGAGGTCCTCGATGTTGTGGCGGATCTT. EYFP rev Xho: 5′-CGCCTCGAGTTACTTGTACAGCTCGTCCATGCCGAGA.
Cells.
Mouse melanoma cells (B78H1) expressing nectin-1 (C10) were grown in 10% FBS-DMEM containing 500 μg/ml G418 (20). The parental cell line B78H1 was propagated in the absence of G418. CHO-K1 cells (a gift from P. G. Spear, Northwestern University Feinberg School of Medicine, Chicago) were grown in Ham's F12 medium containing 10% FBS.
Antibodies.
Polyclonal antibodies used in this study were as follows. Rabbit (R) serum R8 was raised against HSV-2 gD and cross-reacts with HSV-1 gD (37). R137 was prepared against purified HSV-1 gHt-gL (24). R68 was prepared against purified and denatured HSV-1 gB (37). SS55 and A22 mAbs were raised against wild-type gB (38). MC5, MC14, and MC23 mAbs were raised against HSV-2-infected Vero cells (J.C.W., R.J.E., and G.H.C., unpublished data).
Fusion Assay.
C10 cells were transfected with 1 μg of total DNA encoding for the indicated glycoproteins. At 24 h posttransfection, cells were fixed, stained with Giemsa (GIBCO/BRL), and scored for syncytium formation.
Fluorescence Assay for BiMC.
B78H1 or B78H1-C10 cells were seeded on glass coverslips and transfected with the indicated plasmids by using GenePorter (Gene Therapy Systems). For any given construct, ≈50% of the cells were transfected by using 125 ng of plasmid DNA, based on antibody staining of cells that were fixed and permeabilized (with 0.1% Triton X-100) to reveal total protein and costaining of nuclei with DAPI (not shown). To synchronize fusion, 250 μg/ml soluble gD was added 20 h after transfection, and the cells were incubated for the indicated times. Cells were fixed with 3% PFA and quenched with 50 mM NH4Cl. When indicated, cells were fixed and then permeabilized. Cells were treated with 10% goat serum and labeled with the appropriate antibodies. For gB, we used mAbs A22+SS55 (38); for gD, we used mAbs MC5+MC14+MC23 (39); and for gH, we used polyclonal R137 (24). Coverslips were washed with PBS, incubated with the appropriate Alexa Fluor 594-conjugated goat anti-IgG (Invitrogen), and mounted in ProLong Gold Antifade reagent (Invitrogen). For confocal microscopy, we used a Nikon TE2000-U inverted microscope coupled to a PerkinElmer confocal imaging system. A two-line argon krypton laser emitting at 488 and 568 nm was used to excite the fluorescence of Alexa Fluor 594.
Supplementary Material
Acknowledgments
We thank Huan Lou and Manuel Ponce de Leon for excellent technical assistance, Florent Bender and Claude Krummenacher for many helpful discussions, and A. Miyawaki (Brain Science Institute, Saitama, Japan) for the Venus constructs. This work was supported by National Institute of Allergy and Infectious Diseases Grants AI-076231 (formerly NS-036731) (to R.J.E.) and AI-18289 (to G.H.C.). B.R. was supported in part by the Merck Summer Research Fellowship Program at the School of Veterinary Medicine, University of Pennsylvania.
Note Added in Proof.
A similar approach was just reported by Avitabile et al. (42).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707452104/DC1.
References
- 1.Wu SR, Haag L, Hammar L, Wu B, Garoff H, Xing L, Murata K, Cheng RH. J Biol Chem. 2007;282:6752–6762. doi: 10.1074/jbc.M609125200. [DOI] [PubMed] [Google Scholar]
- 2.Spear PG, Eisenberg RJ, Cohen GH. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 3.Krummenacher C, Carfi A, Eisenberg RJ, Cohen GH. In: Viral Entry into Host Cells. Pohlmann S, Simmons G, editors. Austin, TX: Landes Bioscience; 2007. in press. [Google Scholar]
- 4.Spear PG, Manoj S, Yoon M, Jogger CR, Zago A, Myscofski D. Virology. 2006;344:17–24. doi: 10.1016/j.virol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Roche S, Bressanelli S, Rey FA, Gaudin Y. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 6.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 7.Spear PG, Longnecker R. J Virol Methods. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian RP, Geraghty RJ. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galdiero S, Falanga A, Vitiello M, D'Isanto M, Collins C, Orrei V, Browne H, Pedone C, Galdiero M. ChemBioChem. 2007;8:885–895. doi: 10.1002/cbic.200700044. [DOI] [PubMed] [Google Scholar]
- 10.Cairns TM, Friedman LS, Lou H, Whitbeck JC, Shaner MS, Cohen GH, Eisenberg RJ. J Virol. 2007;81:5102–5111. doi: 10.1128/JVI.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne H, Bell S, Minson T, Wilson DW. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones NA, Geraghty RJ. Virology. 2004;324:213–228. doi: 10.1016/j.virol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 13.Earp LJ, Delos SE, Netter RC, Bates P, White JM. J Virol. 2003;77:3058–3066. doi: 10.1128/JVI.77.5.3058-3066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earp LJ, Hernandez LD, Delos SE, White JM. Methods Enzymol. 2003;372:428–440. doi: 10.1016/S0076-6879(03)72026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu CD, Chinenov Y, Kerppola TK. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 16.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 17.Shyu YJ, Liu H, Deng X, Hu CD. BioTechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 18.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 19.Pertel PE, Fridberg A, Parish ML, Spear PG. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 20.Miller CG, Krummenacher C, Eisenberg RJ, Cohen GH, Fraser NW. Mol Ther. 2001;3:160–168. doi: 10.1006/mthe.2000.0240. [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairns TM, Landsburg DJ, Whitbeck JC, Eisenberg RJ, Cohen GH. Virology. 2005;332:550–562. doi: 10.1016/j.virol.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Handler CG, Eisenberg RJ, Cohen GH. J Virol. 1996;70:6067–6070. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng T, Ponce-de-Leon M, Jiang H, Dubin G, Lubinski JM, Eisenberg RJ, Cohen GH. J Virol. 1998;72:65–72. doi: 10.1128/jvi.72.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roper RL, Payne LG, Moss B. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. Proc Natl Acad Sci USA. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodger G, Boname J, Bell S, Minson T. J Virol. 2001;75:710–716. doi: 10.1128/JVI.75.2.710-716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handler CG, Cohen GH, Eisenberg RJ. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Romero P, Perez A, Capul A, Montgomery R, Fuller AO. J Virol. 2005;79:4540–4544. doi: 10.1128/JVI.79.7.4540-4544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozalp C, Szczesna-Skorupa E, Kemper B. Drug Metab Dispos. 2005;33:1382–1390. doi: 10.1124/dmd.105.005538. [DOI] [PubMed] [Google Scholar]
- 31.Hynes TR, Tang L, Mervine SM, Sabo JL, Yost EA, Devreotes PN, Berlot CH. J Biol Chem. 2004;279:30279–30286. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 32.Zamyatnin AA, Jr, Solovyev AG, Bozhkov PV, Valkonen JP, Morozov SY, Savenkov EI. Plant J. 2006;46:145–154. doi: 10.1111/j.1365-313X.2006.02674.x. [DOI] [PubMed] [Google Scholar]
- 33.Shukla D, Spear PG. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roop C, Hutchinson L, Johnson DC. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner A, Bruun B, Minson T, Browne H. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isola VJ, Eisenberg RJ, Siebert GR, Heilman CJ, Wilcox WC, Cohen GH. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. J Virol. 2007;81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitbeck JC, Muggeridge MI, Rux A, Hou W, Krummenacher C, Lou H, van Geelen A, Eisenberg RJ, Cohen GH. J Virol. 1999;73:9879–9890. doi: 10.1128/jvi.73.12.9879-9890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 41.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Eisenberg RJ, Cohen GH. J Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avitabile E, Forghieri C, Campadelli-Fiume G. J Virol. 2007;20:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.