Abstract
Poxviruses are large enveloped viruses that replicate in the cytoplasm of vertebrate or invertebrate cells. At least six virus-encoded proteins are required for synthesis and processing of the double-stranded DNA genome of vaccinia virus, the prototype member of the family. One of these proteins, D5, is an NTPase that contains an N-terminal archaeoeukaryotic primase domain and a C-terminal superfamily III helicase domain. Here we report that individual conserved aspartic acid residues in the predicted primase active site were required for in vivo complementation of infectious virus formation as well as genome and plasmid replication. Furthermore, purified recombinant D5 protein synthesized oligoribonucleotides in vitro. Incorporation of label from [α-32P]CTP or [α-32P]UTP into a RNase-sensitive and DNase-resistant product was demonstrated by using single-stranded circular bacteriophage DNA templates and depended on ATP or GTP and a divalent cation. Mutagenesis studies showed that the primase and NTPase activities of the recombinant D5 protein could be independently inactivated. Highly conserved orthologs of D5 are present in all poxviruses that have been sequenced, and more diverged orthologs are found in members of all other families of nucleocytoplasmic large DNA viruses. These viral primases may have roles in initiation of DNA replication or lagging-strand synthesis and represent potential therapeutic targets.
Keywords: DNA replication, nucleocytoplasmic large DNA viruses, RNA primer, vaccinia virus
The poxviruses comprise a large family of DNA viruses that include the causal agent of smallpox (1). Unlike most other DNA viruses, poxviruses replicate entirely in the cytoplasm. To accommodate this unique lifestyle, they encode enzymes and factors needed for genome replication and transcription, which are potential targets for antivirals (2). Poxvirus genomes are 130,000–300,000 bp long and consist of two complementary strands of DNA that are covalently linked to form hairpins at each end. A transcription system is packaged in infectious virus particles allowing early mRNAs to be synthesized soon after cell entry. Early proteins are used for host defense, genome replication, and transcription of intermediate stage genes. Intermediate proteins include late-stage transcription factors, whereas late proteins are mostly involved in virus assembly and dissemination.
Most laboratory studies of poxviruses are carried out by using vaccinia virus (VACV). Studies with conditional lethal mutants indicate that five VACV early proteins are required for DNA replication, namely, E9 DNA polymerase, D4 uracil DNA glycosylase, A20 processivity factor, B1 protein kinase, and D5 NTPase (reviewed in ref. 3). The polymerase catalyzes primer- and template-dependent DNA synthesis and possesses 3′ to 5′ exonucleolytic activity (4, 5). The essential role of D4 in DNA replication (6) is independent of its uracil DNA glycosylase activity (7), which presumably has a facultative repair function. The A20 and D4 proteins physically interact (8, 9) and together provide processivity for the DNA polymerase (10). The B1 kinase phosphorylates a cellular DNA-binding protein called BAF and prevents the latter from blocking VACV DNA replication (11). The fast stop DNA replication phenotype of conditional lethal D5 mutants suggests a function at the replication fork (12). D5 also interacts with A20 (8, 9) and forms multimers (13). Extensive protein sequence analyses have indicated that the C-terminal region of the 90-kDa D5 protein belongs to the helicase superfamily III within the AAA+ class of NTPases, which includes the replicative helicases of numerous other DNA and RNA viruses (14, 15). Furthermore, the N-terminal domain of D5 has sequence and structural features that are common to the archaeoeukaryotic primase superfamily, the members of which have diverse roles in DNA replication and repair (16). Nevertheless, the only catalytic activity of D5 that has been demonstrated is nucleic acid-independent hydrolysis of rNTPs and dNTPs (17). However, transfection experiments have indicated that truncation of the N-terminal 300 amino acids of D5, as well as point mutations in the predicted helicase domain of D5, impair complementation of DNA replication (13).
DNA polymerases, unlike RNA polymerases, are incapable of initiating polynucleotide synthesis and therefore require a primer with a free 3′ hydroxyl end. Several different priming mechanisms are used in biological systems (18). The most common mechanism involves the synthesis of a short RNA primer that is then elongated by DNA polymerase. Other mechanisms include use of a tRNA primer, an amino acid side chain of a genome-attached protein, generation of a nick in one DNA strand, and DNA strand transfer. During parvovirus replication, a hairpin formed by a palindrome at the 3′ end of the single-stranded genome serves as a primer for unidirectional strand displacement synthesis called rolling hairpin replication (19, 20). Further rounds of replication require the generation of nicks near the ends of the parvovirus DNA with the resulting 3′ ends serving as primers. The similarity between the structure of the parvovirus replication intermediate and the mature poxvirus genome has led to a model in which replication is initiated at a putative nick proximal to the hairpin (21, 22). Strand elongation leads to the formation of concatemers that are resolved into unit length genomes with hairpin ends by a virus-encoded Holliday junction endonuclease (23).
Using a complementation assay, we now show that the conserved amino acids in the predicted primase active site of D5 are required for DNA replication in VACV-infected cells, and that purified recombinant D5 can synthesize oligoribonucleotides by using a single-stranded DNA template. The encoding of a primase by poxviruses has important implications for the mechanism of genome replication and provides a target for antivirals.
Results
The Predicted Primase Active Site of D5 Is Required for Virus Complementation.
The VACV D5 protein has a primase domain in the N-terminal region and an NTPase/helicase domain in the C-terminal region, an arrangement that is also found in some other archaeoeukaryotic primases (16). A multiple alignment of the primase domains of D5 orthologs from each poxvirus genus is shown in supporting information (SI) Fig. 5. To investigate the importance of the primase domain, we constructed plasmids encoding WT D5 and D5 with single alanine substitutions of D70, D72, and D170, which were predicted to be involved in the coordination of a Mg2+ cation for substrate binding and primer synthesis (16, 24) (SI Fig. 5). An additional mutant with an alanine substituted for an invariant glycine residue (G503) in the Walker A motif, predicted to be essential for NTPase activity (14, 15), was also constructed. Ten consecutive histidine codons were appended to the C terminus of the ORF encoding WT or mutated D5 for detection and isolation of the proteins, and a VACV intermediate stage promoter (25) was positioned upstream to allow transcription early during infection.
We determined the ability of the D5 constructs to complement VACV Cts24, a conditional lethal mutant defective in DNA replication at 40°C (12, 13, 26). Following a previously described protocol (13), cells were infected with Cts24 at the permissive temperature, transfected with D5 plasmids at 3 h after infection and shifted to the nonpermissive temperature at 5 h after infection or maintained at the permissive temperature. After 24 h, the formation of infectious virus was determined by a plaque assay. The virus yield was reduced ≈6-fold because of the shift to the nonpermissive temperature (Fig. 1A). The plasmid expressing WT D5 increased virus replication by ≈5-fold, whereas plasmids containing mutated D5 had minimal effects (Fig. 1A). The impairment of complementation caused by mutations in the predicted primase active site was similar to the effect of the mutation in the NTPase site. SDS/PAGE of the cell lysates, and Western blotting with antibody to the polyhistidine tag ruled out the possibility that the mutated proteins were less highly expressed or more unstable at 40°C compared with WT D5 (data not shown).
Fig. 1.
DNA replication depends on conserved amino acids in the primase domain of D5. (A) Virus complementation. BS-C-1 cells were infected in triplicate with Cts24 at a multiplicity of five plaque-forming units (PFU) per cell at 31°C. At 3 h after infection, cells were transfected with empty plasmid vector or plasmid that expressed WT D5, D5D70A, D5D72A, D5D170A, or D5G503A. Two hours later, the cells were shifted to 40°C or maintained at 31°C, as indicated. Cells were harvested at 24 h after infection and virus yields determined by plaque assay at 31°C. Error bars indicate range of one standard deviation. (B) Genome replication. Cells were infected in triplicate with Cts24, transfected and harvested as above. Total DNA was isolated and 2 μg transferred to a nylon membrane. The blot was hybridized to 32P-labeled VACV DNA and radioactivity was quantified with a PhosphorImager. (C) Viral origin-independent plasmid replication. Cells were uninfected or infected with Cts24 and transfected with plasmid p716 in addition to empty vector or D5 plasmids as above. At 5 h after infection, the temperature was raised to 40°C and at 24 h, the cells were harvested. Total DNA was isolated, digested with DpnI, and p716 replication was measured by real-time PCR.
Conserved Amino Acids Within the Predicted Primase Active Site of D5 Are Required for Genome and Plasmid Replication.
The above transfection protocol was also used to determine whether the defect in complementation was associated with impaired DNA replication. At 24 h after infection, the cells were harvested, and replication of the viral genome was determined by hybridization to a radioactively labeled viral DNA probe. Slot-blot analysis verified that Cts24 DNA replication was specifically impaired at 40°C, and that this could be rescued by transfection of a plasmid expressing WT D5 (Fig. 1B). In contrast, little or no increase in genome replication occurred when plasmids expressing mutated D5 were transfected (Fig. 1B). The mutated D5 proteins did not reduce genome replication at the permissive temperature, indicating they did not have a dominant negative effect under these conditions (data not shown).
Previous studies showed that a variety of circular DNAs lacking any poxvirus sequences replicate in cells infected with VACV, suggesting a mechanism that is independent of a specific origin sequence (27, 28). Furthermore, plasmid replication occurred in viral factories and depended on each of the known VACV replication proteins including D5 (29). It was of interest to determine whether the primase active site was also needed for plasmid replication. The protocol used above to measure genome replication in cells infected with Cts24 was modified by cotransfecting plasmid p716 (29, 30), which has no poxvirus sequences, along with a plasmid expressing WT or mutated D5. Total DNA was then isolated, and the transfected methylated plasmid was digested with DpnI. The amount of replicated DNA was then determined by real-time PCR by using primers 152 base pairs apart that flanked two DpnI sites in p716. This quantitative, low-background assay demonstrated that mutation of a single conserved amino acid in the putative primase active site reduced plasmid replication by ≈20-fold (Fig. 1C).
Recombinant D5 Catalyzes Oligoribonucleotide Synthesis.
The mutagenesis data indicated the importance of conserved amino acids in the predicted primase active site. However, to demonstrate that D5 has primase activity, we needed to purify the protein. For initial experiments, an inducible VACV system (31) was used to overexpress recombinant D5, containing a C-terminal 10-histidine tag, in HeLa cells. The protein was purified by using Ni-NTA beads, and samples taken during the isolation procedure were analyzed by SDS/PAGE and staining with Coomassie blue. The 90-kDa protein, eluting at 200 mM imidazole, was of the expected size for D5 (Fig. 2A) and reacted with antibody to the polyhistidine tag (not shown).
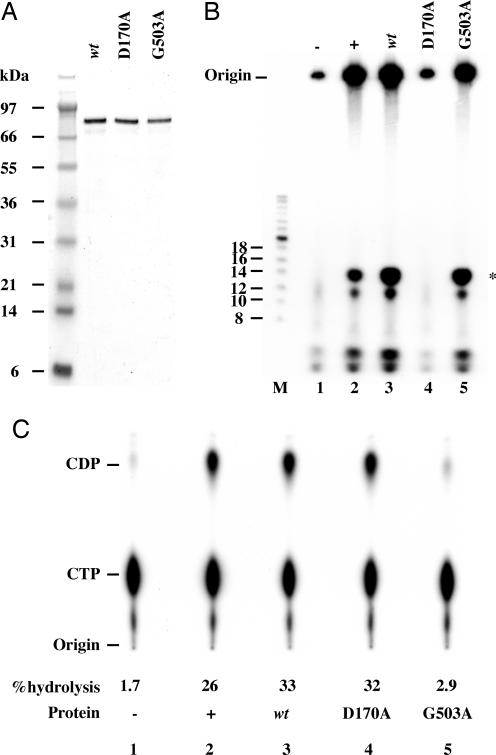
Fig. 2.
Primase activity of purified recombinant D5. (A) Affinity purification of D5. HeLa cells were infected with a recombinant VACV containing an inducible copy of D5 with a C-terminal 10-histidine tag. After 24 h, cell extracts were prepared, and the tagged D5 was purified on Ni-NTA beads. Samples were analyzed by SDS/PAGE and staining with Coomassie blue. Lanes: 1, cell extract; 2, flowthrough; 3, 20 mM imidazole wash; 4, 40 mM imidazole wash; and 5–9 fractions eluting with 200 mM imidazole. Molecular masses of protein markers (M) are shown on the left. (B) Comparison of herpes simplex virus (HSV) and VACV (D5) primase activities. Recombinant histidine-tagged D5 (1.1 pmols), HSV primase complex (3.2 pmols), or the equivalent volume of Ni-NTA eluate from cells infected with parental VACV (mock) was incubated in a reaction mixture containing rNTPs including [α-32P]CTP and 0.57 pmols φX174 DNA at 37°C for 30 min. Products were digested with calf intestinal alkaline phosphatase, denatured, and resolved by electrophoresis. Positions of 32P-end-labeled oligonucleotide markers (M) are indicated on the left. Asterisk indicates position of the reaction product migrating with 14-nt phosphorylated marker.
Affinity-purified D5 was tested for primase activity by using single-stranded φX174 phage DNA as a template in the presence of [α-32P]CTP and other unlabeled rNTPs. The herpes simplex type 1 primase complex (32) (kindly provided by Sandra Weller, University of Connecticut, Farmington, CT) was used as a positive control. Ni-NTA eluate from HeLa cells infected with the parental VACV lacking D5 with a polyhistidine tag served as a negative control. At the end of the reaction, the mixture was treated with phosphatase to degrade free nucleotides and the triphosphate end of synthesized oligoribonucleotides. A prominent radioactive band migrating near the 14-nt phosphorylated marker was detected in reactions that contained either the herpes simplex primase or D5 but not from the negative control (Fig. 2B). Additional material migrated more rapidly than the 14-nt marker and some remained at the top of the gel during electrophoresis. Similar results were obtained by using M13 single-strand DNA as the template and [α-32P]UTP as a substrate (data not shown).
Enzymatic Activity of Recombinant D5 from an Alphavirus Vector and Mutation of the Predicted Primase Active Site.
In the above experiments, attribution of primase activity to D5 was based on the affinity purification of the protein and the absence of such activity in protein “mock purified” by affinity chromatography in parallel from cells infected with VACV not expressing tagged D5. Nevertheless, the possibility remained that the primase activity was due to a minor VACV or cellular protein that associated with affinity purified D5. To address that possibility, we expressed 10-histidine-tagged versions of both the WT D5 and D5 with amino acid substitutions in either the predicted primase or NTPase active site. Aspartic acid 170 was changed to alanine in one construct and glycine 503 to alanine in another. In addition, an alphavirus expression system was used instead of the inducible VACV system, because of the potential of mutant D5 oligomerizing with endogenous WT D5. BHK cells were infected with the recombinant alphaviruses and the affinity purification procedure was monitored by SDS/PAGE followed by Coomassie blue staining. In each case, the predicted 90-kDa protein was the predominant band (Fig. 3A).
Fig. 3.
Primase and NTPase activities of WT and mutated D5 proteins. (A) Analysis of recombinant D5 proteins expressed by recombinant alphaviruses. Proteins were affinity purified on a Ni-NTA column, resolved by SDS/PAGE, and stained with Coomassie blue. Lanes containing histidine tagged WT D5, D5D170A, and D5G503A are indicated. Protein molecular mass standards are indicated at the left. (B) Oligoribonucleotide synthesis. Reaction conditions were the same as in Fig. 2B. Positions of end-labeled oligonucleotide markers (M) are indicated on the left. Lanes: 1, (−) Ni-NTA eluate from uninfected BHK cell extracts; 2, (+) 1.1 pmols WT D5 expressed by recombinant VACV; 3, 1.3 pmols WT D5 expressed by alphavirus; 4, 1.3 pmols D5D170A expressed by alphavirus; 5, 1.3 pmols D5G503A expressed by alphavirus. Asterisk indicates position of product migrating with 14-nt marker. (C) NTPase activity. Autoradiograph of thin-layer chromatography plate with positions of CDP and CTP markers. Percent hydrolysis of CTP to CDP is indicated. Lanes are the same as in B.
Primase activity was assayed as above by using D5 made by recombinant VACV as a positive control and Ni-NTA eluates of BHK cell extracts as a negative control. Labeled product migrating near the 14-nt marker was synthesized in reactions that contained either WT D5 or D5G503A but not D5D170A (Fig. 3B). Reactions with D5D170A also contained less radioactive material that remained near the top of the gel and that migrated faster than the 14-nt marker than did reactions containing WT D5 or D5G503A. These results demonstrated that the mutation in the putative primase domain abolished primase activity, whereas a mutation in the NTPase domain did not. Thus, the primase activity was due to D5 and not an enzyme contaminant.
NTPase assays, measuring the conversion of CTP to CDP, were performed to eliminate the possibility that the D170A mutation had a global effect on the structure of D5. As expected, the D5G503A had only trace NTPase activity (Fig. 3C). However, the NTPase activity of the D5D170A was similar to that of WT D5 (Fig. 3C). The latter result is consistent with a report that truncation of the N-terminal 300 amino acids does not impair NTPase activity (13). Thus the primase and NTPase activities of D5 are independent of each other.
Preliminary Characterization of the Primase Reaction and Product.
A time-course analysis indicated that the reaction products increased in amount from 1 through 45 min (Fig. 4A). However, only the products migrating near the 14-nt marker and the top of the gel were sensitive to RNaseA/T1 and nuclease P1 and insensitive to DNase I as expected for RNA (Fig. 4B). Similarly, only RNase-sensitive products depended on a DNA template (Fig. 4C). Synthesis of the product migrating with the 14-nt marker was diminished when GTP was omitted and not made when both ATP and GTP were absent, suggesting that synthesis is initiated with a purine as found for some other primases (18). The products were all greatly reduced in amount in the absence of Mg2+ (Fig. 4B). Further work is needed to determine the exact sizes of the RNA products and template preference.
Fig. 4.
Characterization of primase reaction and products. (A) Primase reaction time course. Primase reactions were carried out under standard conditions for the times indicated. (B) Effect of nuclease treatments on product. Primase reactions were carried out under standard conditions and the product was treated with calf intestinal phosphatase and (lane 1) nothing, (lane 2) nuclease P1, (lane 3) RNase A and T1, and (lane 4) DNase I. (C) Omission of reaction components. Primase reaction was carried out under standard conditions or with indicated component(s) missing using WT D5 expressed by alphavirus vector and φX174 DNA template. Asterisks indicate position of product migrating with the 14-nt marker. End-labeled oligonucleotide markers are on the left.
Discussion
We provided in vivo and in vitro data indicating that D5 is a DNA primase. First, a D5 ts mutant complementation assay (13) demonstrated the importance of conserved amino acids in the predicted primase active site for DNA replication. Furthermore, purified recombinant D5 exhibited primase activity that depended on conserved amino acids in the predicted active site.
Primases are DNA-dependent RNA polymerases that synthesize oligoribonucleotides 2–15 nt or longer, usually starting with ATP or GTP (18). Generally, any single-stranded DNA can serve as a template, although there may be preferential usage of some sequences. D5 primase activity was demonstrated by using single-stranded circular φX174 and M13 phage templates. A discrete RNase-sensitive band migrated near the 14-nt marker. We cannot be sure of the actual length of this oligoribonucleotide, because the markers were phosphorylated, and small oligonucleotides migrate anomalously in high percentage polyacrylamide gels (33). However, the product of the herpes simplex virus primase complex, which has been estimated to be 8 nt (32), comigrated with the major VACV product. We surmised that synthesis starts with ATP or GTP, because omission of both nucleotides abolished synthesis of the major product. Substitution of alanine for one of the conserved aspartic acid residues in the primase domain abolished primase activity. Mutation of the primase active site did not impair NTPase activity and vice versa, indicating these two domains of D5 function independently. Because D5 exists as an oligomer (13), it will be interesting to determine whether the primase and NTPase active sites can work in trans for in vivo complementation. An important future experiment will be to demonstrate that D5 can prime DNA replication in vitro.
The finding of primase activity has profound implications for understanding the mechanism of poxvirus DNA replication. D5 could function by synthesizing RNA primers for initiation of leading-strand DNA synthesis, although the current model involves nicking near the ends of the genome. That model is based on reports of a change in sedimentation of parental VACV DNA and labeling studies (34–37). However, the only VACV nicking enzyme that has been characterized is not essential for DNA replication, and the cleavage site produced contains a 3′ phosphate end, which would not serve as a primer for DNA polymerase (38, 39). Nevertheless, it is possible that nicking and RNA priming mechanisms are both used for initiation of replication. RNA primers could also allow lagging-strand DNA synthesis, which requires multiple initiations opposite the leading strand. In this regard, there have been reports of VACV DNA covalently linked to RNA (40) and of short nascent DNA resembling Okazaki fragments, which could be chased into larger molecules (41). However, these results have not been repeated or extended during the past 30 years, and a unidirectional strand displacement model analogous to parvovirus rolling hairpin replication has gained prominence despite the absence of direct evidence (21, 22, 42).
Lagging-strand synthesis would require a nuclease to remove the RNA primer and DNA ligase to covalently join the DNA fragments. A predicted Fen-1/Flap-like 5′→ 3′ endonuclease was detected in poxviruses and some other nucleocytopasmic DNA viruses (24, 43). Fen-1 degrades RNA primers in Okazaki fragments (44), and such a role for the VACV homolog, G5, would be anticipated. However, further studies are needed, because G5R ts mutants exhibited a block in the initial stage of morphogenesis rather than DNA replication (45). Although VACV encodes a DNA ligase that can substitute for the replicative yeast ligase, the gene is not essential and is not conserved in all poxviruses (46–48). This result is also perplexing, because there is no evidence for the recruitment of cellular DNA ligase I from the nucleus to cytoplasmic virus factories (48). The possibility of other cellular ligases substituting for the viral enzyme remains a possibility.
In conclusion, our demonstration that the VACV D5 protein can synthesize oligoribonucleotides functionally validates the computational study that placed the gene in the archaeoeukaryotic primase superfamily (16). The primase might be involved in either initiation of replication or lagging-strand synthesis. The result has broad significance, because uncharacterized D5 homologs are present in all poxviruses that have been sequenced and related, although highly diverged forms are present in members of the four other families of nucleocytoplasmic large DNA viruses (24).
Materials and Methods
Plasmid Constructions.
DNA encoding WT D5 and carrying a C-terminal 10-histidine tag was cloned from VACV genomic DNA by PCR using Accuprime Pfx (Invitrogen). Alanine substitutions were made by site directed mutagenesis as described in SI Text.
To construct replicon pERK3D5-WT, -D170A and -G503A, the coding sequence of D5 with a 10-histidine tag from the corresponding pG8D5 plasmids were amplified by PCR by using primers containing PacI and AscI restriction sites and cloned into the PacI and AscI sites of vector pERK3 (AlphaVax).
Construction of Virus Vectors.
An inducible recombinant VACV (vD5HISi) that overexpresses D5 with a C-terminal 10-histidine tag was formed by use of the VOTE system (49) as described in SI Text.
For the generation of recombinant alphaviruses based on the Venezuelan Equine Encephalitis virus replicon, the constructs pERK3D5-WT, -D170A, and – G503A were used as template DNA to generate the recombinant virus preparations VaxD5WT, VaxD5D170, and VaxD5D503, respectively. Transcription, electroporation and virus-like replicon particle production, using a two-helper system, were performed essentially as described (50) and as detailed in SI Text.
Purification of Recombinant Protein.
HeLa S3 cells (2 × 108) were infected with vD5HISi at a multiplicity of five plaque-forming units in the presence of 0.1 mM isopropyl β-d-thiogalactoside for 24 h at 31°C. For proteins expressed from recombinant alphavirus, BHK-21 (1 × 108) cells were infected at a multiplicity of two focus-forming units for 18 h at 31°C. Protein expressed from recombinant viruses was then prepared as described (51), with some modifications. Cells were harvested by low-speed centrifugation, washed twice with PBS, suspended in buffer C (20 mM Tris·HCl, pH 8.0, 10 mM NaCl, 1 mM 2-mercaptoethanol, 0.5 mM phenylmethylsulfonylfluoride, 2.5 mM MgCl2), placed on ice for 10 min, and disrupted with a Dounce homogenizer. An equal volume of buffer C plus 0.83 M NaCl, 30% glycerol, 0.02% Triton X-100, 2 mM imidazole was added to the lysate for 1 h, after which the mixture was centrifuged in a swinging bucket rotor at 21,000 × g for 30 min. The soluble material was mixed overnight with Ni-NTA agarose, which had been preequilibrated with the latter buffer. The column was sequentially washed with 10 volumes of buffer D (20 mM Tris·HCl, pH 8.0, 0.42 M NaCl, 15% glycerol, 1 mM 2-mercaptoethanol, 0.01% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride) plus 20 mM imidazole, 10 volumes of buffer D plus 40 mM imidazole and buffer D plus 0.2 M imidazole. Proteins that eluted with 0.2 M imidazole were pooled and dialyzed in 50 mM Tris·HCl, pH 7.4, 50 mM NaCl, 1 mM EDTA, 20% glycerol.
Transient Complementation Assay.
Complementation assays were performed as described (13) with some modifications. BS-C-40 cells were infected with Cts24 at a multiplicity of five plaque-forming units and incubated at 31°C. After 3 h, 100 ng of plasmid p716 (30) and 2 μg of pCR2.1-D5 and related plasmids or empty vector were transfected into cells using Lipofectamine 2000 reagent (Invitrogen). At 5 h after infection, cells were either maintained at 31°C or shifted to 40°C. At 24 h after infection, cells were harvested and DNA isolated by using the Qiamp DNA Blood Kit (Qiagen). Genome replication was evaluated by transferring 2 μg of DNA by using a slot-blot apparatus to an Immobilon-Ny+ membrane (Millipore). The blot was hybridized to a 32P-labeled VACV H6R DNA fragment and then exposed to a Phosphor screen. Data were acquired on a Storm 860 PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software (Molecular Dynamics).
Plasmid replication was monitored by real-time PCR as described (29). In addition, replicate sets of cells were harvested and virus yields determined by plaque assay on BS-C-40 cells as described (52).
Primase Reactions.
Reaction mixtures (5 μl) contained (unless stated otherwise) 25 mM Tris acetate (pH 7.5), 60 mM K acetate, 6 mM Mg acetate, 10 mM DTT, 100 μg/ml BSA, 1 mM ATP, 250 μM UTP, 250 μM GTP, 25 μM CTP, 0.66 μM [α-32P]CTP (3,000 Ci/mmol) (Perkin–Elmer Life Sciences), 1 μg (0.57 pmols) of φX174 phage DNA (New England Biolabs), 100–125 ng (1.1–1.3 pmols) recombinant protein. Reactions were incubated at 37°C for 30 min, then at 80°C for 10 min. Products were treated with 4 units of calf intestinal phosphatase (New England Biolabs) at 37°C for 1 h and denatured by adding an equal volume of formamide stop solution (95% deionized formamide, 20 mM EDTA, plus bromophenol blue and xylene cyanol). Reaction products were heated at 90°C for 2 min and analyzed on a denaturing 20% polyacrylamide, 7.5 M urea gel followed by autoradiography. Oligonucleotide size markers (GE Healthcare) were 32P-end-labeled using polynucleotide kinase (New England Biolabs) and [γ-32P]ATP [3,000 Ci/mmol] (Perkin–Elmer Life Sciences).
NTP Hydrolysis Assays.
NTPase assays were performed as described (17). Reaction mixtures contained 100–125 ng (1.1–1.3 pmols) of recombinant protein, 0.16 μM [α-32P]CTP (3,000 Ci/mmol) (Perkin–Elmer Life Sciences) and were incubated at 37°C for 1 h. Each reaction mixture (1 μl of a 1:50 dilution) was spotted onto plastic-backed polyethyleneimine–cellulose thin-layer chromatography sheets (J. T. Baker). Ascending chromatography in a solvent containing 0.8 M acetic acid and 0.8 M LiCl separated NTP hydrolysis products. The sheets were dried and analyzed by autoradiography. The blot was also exposed to a Phosphor screen and data acquired on a Storm 860 PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software (Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank L. Aravind for helpful discussions regarding the predicted primase domain of D5, R. Condit (University of Florida, Gainesville, FL) for VACV Cts24, and S. Weller for a sample of herpes simplex virus primase. N. Nossal (deceased) gave useful advice on the primase assay. D. Evans and S. Weller provided helpful reviews of the manuscript. This study was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709276104/DC1.
References
- 1.Moss B. In: Fields Virology. Knipe DM, editor. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- 2.Harrison SC, Alberts B, Ehrenfeld E, Enquist L, Fineberg H, McKnight SL, Moss B, O'Donnell M, Ploegh H, Schmid SL, et al. Proc Natl Acad Sci USA. 2004;101:11178–11192. doi: 10.1073/pnas.0403600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss B, De Silva F. In: DNA Replication and Human Disease. DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 707–727. [Google Scholar]
- 4.Challberg MD, Englund PT. J Biol Chem. 1979;254:7812–7819. [PubMed] [Google Scholar]
- 5.McDonald WF, Traktman P. J Biol Chem. 1994;269:31190–31197. [PubMed] [Google Scholar]
- 6.Millns AK, Carpenter MS, DeLange AM. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- 7.De Silva FS, Moss B. J Virol. 2003;77:159–166. doi: 10.1128/JVI.77.1.159-166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCraith S, Holtzman T, Moss B, Fields S. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii K, Moss B. Virology. 2002;303:232–239. doi: 10.1006/viro.2002.1721. [DOI] [PubMed] [Google Scholar]
- 10.Stanitsa ES, Arps L, Traktman P. J Biol Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- 11.Nichols RJ, Wiebe MS, Traktman P. Mol Biol Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans E, Traktman P. Chromosoma. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- 13.Boyle KA, Arps L, Traktman P. J Virol. 2007;81:844–859. doi: 10.1128/JVI.02217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya AE, Koonin EV, Wolf YI. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 15.Iyer LM, Leipe DD, Koonin EV, Aravind L. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Iyer LM, Koonin EV, Leipe DD, Aravind L. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans E, Klemperer N, Ghosh R, Traktman P. J Virol. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frick DN, Richardson CC. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 19.Cotmore SF, Tattersall P. In: DNA Replication and Human Disease. DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 593–608. [Google Scholar]
- 20.Berns K, Parrish CR. In: Field's Virology. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2437–2477. [Google Scholar]
- 21.Moyer RW, Graves RL. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- 22.Baroudy BM, Venkatesan S, Moss B. Cold Spring Harbor Symp Quant Biol. 1982;47:723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- 23.Garcia AD, Aravind L, Koonin EV, Moss B. Proc Natl Acad Sci USA. 2000;97:8926–8931. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer LA, Balaji S, Koonin EV, Aravind L. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Baldick CJ, Keck JG, Moss B. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seto J, Celenza LM, Condit RC, Niles EG. Virology. 1987;160:110–119. doi: 10.1016/0042-6822(87)90051-1. [DOI] [PubMed] [Google Scholar]
- 27.DeLange AM, McFadden G. Proc Natl Acad Sci USA. 1986;83:614–618. doi: 10.1073/pnas.83.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merchlinsky M, Moss B. In: Cancer Cells 6/Eukaryotic DNA Replication. Kelly T, Stillman B, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 87–93. [Google Scholar]
- 29.De Silva FS, Moss B. Virol J. 2005;2:23. doi: 10.1186/1743-422X-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winokur PL, McBride AA. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander WA, Moss B, Fuerst TR. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman G, Gottlieb J, Challberg MD. J Virol. 1992;66:4884–4892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holstege FC, Fiedler U, Timmers HT. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteban M, Holowczak JA. Virology. 1977;82:308–322. doi: 10.1016/0042-6822(77)90006-x. [DOI] [PubMed] [Google Scholar]
- 35.Pogo BT. Virology. 1980;101:520–524. doi: 10.1016/0042-6822(80)90466-3. [DOI] [PubMed] [Google Scholar]
- 36.Pogo BGT, O'Shea M, Freimuth P. Virology. 1981;108:241–248. doi: 10.1016/0042-6822(81)90543-2. [DOI] [PubMed] [Google Scholar]
- 37.Pogo BG, Berkowitz EM, Dales S. Virology. 1984;132:436–444. doi: 10.1016/0042-6822(84)90048-5. [DOI] [PubMed] [Google Scholar]
- 38.Merchlinsky M, Garon C, Moss B. J Mol Biol. 1988;199:399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- 39.Eckert D, Williams O, Meseda CA, Merchlinsky M. J Virol. 2005;79:15084–15090. doi: 10.1128/JVI.79.24.15084-15090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olgiati DD, Pogo BG, Dales S. J Cell Biol. 1976;68:557–566. doi: 10.1083/jcb.68.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esteban M, Holowczak JA. Virology. 1977;78:57–75. doi: 10.1016/0042-6822(77)90078-2. [DOI] [PubMed] [Google Scholar]
- 42.Baroudy BM, Moss B. Nucleic Acids Res. 1982;10:5673–5679. doi: 10.1093/nar/10.18.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva MD, Shen L, Tcherepanov V, Watson C, Upton C. Bioinformatics. 2006;22:2846–2850. doi: 10.1093/bioinformatics/btl506. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Kao HI, Bambara RA. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 45.Da Fonseca FG, Weisberg AS, Caeiro MF, Moss B. J Virol. 2004;78:10238–10248. doi: 10.1128/JVI.78.19.10238-10248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colinas RJ, Goebel SJ, Davis SW, Johnson GP, Norton EK, Paoletti E. Virology. 1990;179:267–275. doi: 10.1016/0042-6822(90)90295-3. [DOI] [PubMed] [Google Scholar]
- 47.Kerr SM, Smith GL. Virology. 1991;180:625–632. doi: 10.1016/0042-6822(91)90076-n. [DOI] [PubMed] [Google Scholar]
- 48.Kerr SM, Johnston LH, Odell M, Duncan SA, Law KM, Smith GL. EMBO J. 1991;10:4343–4350. doi: 10.1002/j.1460-2075.1991.tb05012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward GA, Stover CK, Moss B, Fuerst TR. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamrud KI, Custer M, Dudek JM, Owens G, Alterson KD, Lee JS, Groebner JL, Smith JF. Virology. 2007;360:376–387. doi: 10.1016/j.virol.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsafanas GC, Moss B. J Biol Chem. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- 52.Earl PL, Cooper N, Wyatt S, Moss B, Carroll MW. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. New York: Wiley; 1998. pp. 16.16.11–16.16.13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.