Abstract
The MYB-related gene ASYMMETRIC LEAVES 1 (AS1) and its orthologs have an evolutionarily conserved role in specification of leaf cell identity. AS1 is expressed in leaf founder cells, where it functions as a heterodimer with the structurally unrelated AS2 proteins to repress activity of KNOTTED 1-like homeobox (KNOX) genes. AS1 therefore confines KNOX activity to the shoot apical meristem, where it promotes stem cell function through the regulation of phytohormone activities. Here, we show that loss-of-function mutations in AS1 unexpectedly convey heightened protection against necrotrophic fungi. AS1 operates as a negative regulator of inducible resistance against these pathogens by selectively binding to the promoters of genes controlled by the immune activator, jasmonic acid (JA), damping the defense response. In contrast, AS1 is a positive regulator of salicylic acid (SA)-independent extracellular defenses against bacterial pathogens. Neither the absence of AS2 nor ERECTA function, which enhances the morphological phenotype of as1, nor the conditional or constitutive expression of KNOX genes impacted disease resistance. Thus, the function of AS1 in responses to phytopathogens is independent of its AS2-associated role in development. Loss of function in the AS1 orthologs PHAN in Antirrhinum majus and NSPHAN in Nicotiana sylvestris produced pathogen-response phenotypes similar to as1 plants, and therefore the defense function of AS1 is evolutionarily conserved in plant species with a divergence time of ≈125 million years.
Keywords: Arabidopsis thaliana, asymmetric leaves 1, disease resistance, plant defense response
An evolutionarily conserved genetic pathway controls specification of leaf cell fate at the shoot apical meristem and involves the Arabidopsis ASYMMETRIC LEAVES1 (AS1), Antirrhinum PHANTASTICA (PHAN), Nicotiana NSPHAN, and maize ROUGH SHEATH2 (RS2) genes. These genes encode orthologous MYB transcription factors that are expressed only in leaf founder cells and leaf primordia, where they are needed to repress KNOTTED1-like homeobox (KNOX) genes (1–4). KNOX genes are expressed within the shoot apical meristem, where they regulate the activity of gibberellin and cytokinin hormones to maintain indeterminacy (5–9). AS1 forms heterodimers with the structurally unrelated protein AS2 (10), and both proteins are required to prevent KNOX expression in developing leaves (9, 10). Ectopic KNOX expression produces lobed leaves, similar to those of as1 and as2 loss-of-function mutants (11), suggesting that it is responsible for this aspect of the as1 or as2 mutant phenotype.
A small number of genes that are needed for more general aspects of plant development have also been found to regulate disease resistance. For example, a potential transcriptional cofactor encoded by AtTIP49a promotes the plant defense response but is also needed for viability of female gametophytes and seedlings (12). Similarly, a potential component of a ubiquitin protein ligase complex encoded by SGT1b (SUPPRESSOR OF THE G2 ALLELE OF skp1-4 1b) is required for both plant disease resistance and auxin signaling (13).
The emerging evidence suggests that plant immune function discriminates between microbes that exhibit distinct pathogenic lifestyles (14). Thus, salicylic acid (SA)-dependent defenses largely underpin resistance against biotrophic pathogens, whereas jasmonic acid (JA)- and ethylene (ET)-based responses are required for protection against necrotrophic pathogens (15–18). Furthermore, these signaling networks are thought to be regulated in a mutually antagonistic fashion, suggesting that plants can prioritize specific defense mechanisms (19–20). Recently, a series of loss-of-function mutations have been uncovered that convey enhanced susceptibility to necrotrophic fungi (21–23). However, only ocp3 (overexpressor of cationic peroxidase 3) and jin1 (jasmonate-insensitive 1) have been reported to increase resistance against this class of pathogen (24, 25).
Here, we show that loss of AS1 function conveys increased protection against necrotrophic fungal pathogens. Our findings suggest that AS1 operates as a negative regulator of inducible defense responses against these pathogens by occupying the promoters of a subset of defense genes, damping their expression. In contrast, AS1 operates as a positive regulator of disease resistance against bacterial pathogens. These AS1 functions are independent of its AS2- and KNOX-associated roles in development and are conserved in plant species with a divergence time of ≈125 million years.
Results
Forward Screen to Uncover Mutations that Convey Resistance Against Botrytis cinerea.
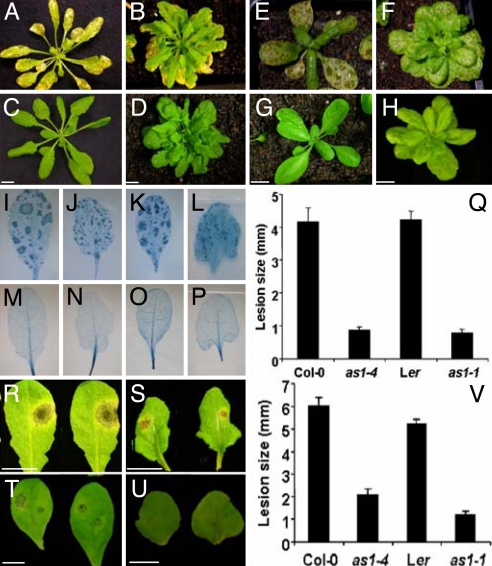
To identify Arabidopsis mutants expressing increased resistance against the necrotrophic fungal pathogen B. cinerea, we screened the progeny of ≈10,000 plants carrying T-DNA insertions (26) by spraying with a suspension of B. cinerea spores. One mutant was found with increased resistance to B. cinerea and lobed leaves (Fig. 1 A–D). Increased pathogen resistance and altered leaf morphology behaved as a single recessive trait and cosegregated with insensitivity to glufosinate herbicide encoded by the T-DNA. Sequences flanking this insert were recovered by thermal asymmetric interlaced (TAIL)-PCR, revealing that the insertion was within the only intron of the AS1 gene, and that the T-DNA prevented accumulation of AS1 transcripts detectable by RT-PCR, consistent with a null mutation (data not shown). The mutation was confirmed as an allele of AS1 in crosses to the classical mutation as1-1 (27) and was therefore named as1-4. Furthermore, as1-1 also conferred heightened resistance against B. cinerea, confirming the pleiotropic roles of AS1 in pathogen resistance and plant development (Fig. 1 E–H).
Fig. 1.
Loss-of-function mutations in AS1 increase disease resistance against necrotrophic fungi. (A–H) The Arabidopsis lines Col-0 (A and C), Col-0 as1-4 (B and D), Ler (E and G), and Ler as1-1 (F and H) were challenged with a spore suspension of B. cinerea (A, B, E, and F). For mock controls, the inoculation solution lacked spores (C, D, G, and H). All plants were scored for disease development at 96 hpi. (I–P) Col-0 (I and M), as1-4 (J and N), Ler (K and O), and as1-1 plants (L and P) were either challenged with a spore suspension of B. cinerea (I–L) or mock inoculated (M–P) as described above. Subsequently, these lines were stained with trypan blue, which marks dead plant cells, revealing lesion development. (Q) Quantification of the extent of B. cinerea-generated lesions in the indicated plant genotypes. (R–U) The Arabidopsis lines Col-0 (R), Col-0 as1-4 (S), Ler (T), and Ler as1-1 (U) were drop-inoculated with an A. brassicicola spore suspension and scored at 96 hpi. (V) Size of A. brassicicola-generated lesions in the indicated plant genotypes. Error bars represent 95% confidence limits. All experiments were repeated at least twice with similar results. (Scale bars, 1 cm.)
We monitored expression of the B. cinerea ActA gene, which correlates directly with the extent of B. cinerea growth (28). At 96 h postinoculation (hpi), ActA transcript accumulation in as1-1 plants was reduced by 71% compared with wild type (data not shown). Furthermore, trypan blue staining of dead cells after B. cinerea inoculation revealed that as1 mutants developed strikingly smaller lesions (reduced in size by ≈80% compared with wild type; Fig. 1 I–Q). Scoring overall symptom development with a disease index also showed as1 mutations conveyed robust resistance against B. cinerea (data not shown). The level of protection against B. cinerea infection in as1 plants was similar to that established by exogenous application of JA to wild-type plant lines [supporting information (SI) Fig. 5]. To investigate whether as1 can restrict the growth of other necrotrophic fungi, as1 and wild-type plants were challenged with Alternaria brassicicola. Both as1-1 and as1-4 mutants showed increased resistance, marked by a conspicuous decrease in lesion size (Fig. 1 R–V).
as1-Mediated Resistance Requires COI1 and EIN2.
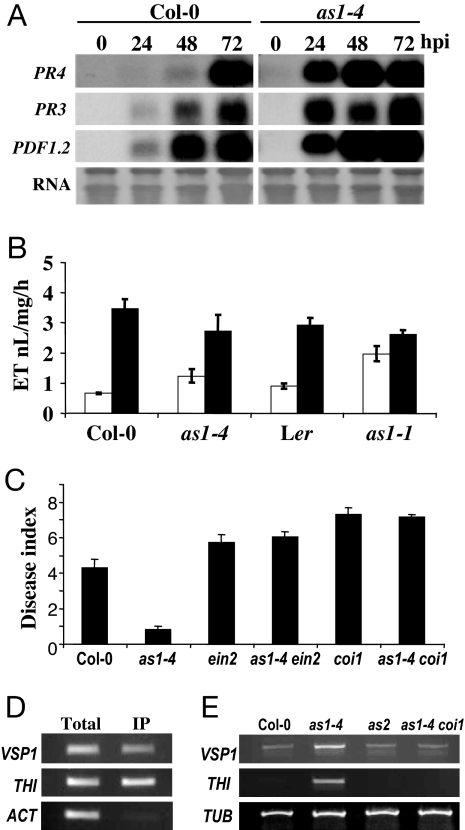
Resistance to necrotrophic pathogens is typically dependent on the plant hormones JA and ET (15–18). To examine whether increased resistance in as1 mutants involved altered JA or ET responses, we monitored expression of defense marker genes that are up-regulated by these hormones. In as1-4 mutants, induction of the JA/ET-dependent PLANT DEFENSIN1.2 (PDF1.2), PATHOGENESIS-RELATED3 (PR3), and PR4 genes (15) was accelerated in response to B. cinerea infection (Fig. 2A). We therefore determined the concentrations of JA and ET during infection with B. cinerea. In noninoculated leaves, levels of ET were twice as high in as1-1 mutants compared with wild type and 1.3 times higher in as1-4 mutants but induced to the same levels in as1 and wild type upon infection with B. cinerea (Fig. 2B). JA levels in uninfected and infected as1 mutants were similar to wild type (data not shown). To examine if ET or JA signaling is required for as1-mediated disease resistance, we generated as1 double mutants with either ein2 (29) or coi1 (30), which cause insensitivity toward ET or JA, respectively, and result in enhanced susceptibility to some necrotrophic pathogens (15, 16). In response to B. cinerea, as1 ein2 and, particularly, as1 coi1 plants exhibited increased lesion development relative to the as1 line (Fig. 2C) and were compromised in JA/ET-dependent gene expression (data not shown). Therefore, as1-mediated resistance against B. cinerea requires ET and, more significantly, JA signaling.
Fig. 2.
Misexpression of JA/ET-dependent defense response genes in as1 plants. (A) Northern blot analysis of PDF1.2, PR3, and PR4 transcript accumulation in the indicated plant genotypes challenged with B. cinerea at the stated hpi. (B) Determination of the rate of ET production in the indicated plant lines. Noninoculated plants (empty bars) and B. cinerea-inoculated plants (filled bars). (C) The stated plant genotypes were challenged with B. cinerea and scored for disease development at 96 hpi. (D) Detection of the indicated gene-regulatory sequences by PCR in total and immunoprecipitated (IP) chromatin extracts by using an anti-GFP antiserum to enrich for chromatin containing PHAN-GFP. The actin (ACT) gene is used as a nonbinding control. (E) Accumulation of the listed gene transcripts in Col-0 wild-type and as1 and as2 mutant plants determined by RT-PCR. Tubulin (TUB) is a constitutively expressed control. Error bars represent 95% confidence limits. These experiments were repeated at least twice with similar results.
Identification of Gene Promoters Occupied by AS1.
Because as1 mutants showed increased induction of pathogen-induced marker genes, we tested whether AS1 might have a direct role in regulation of defense genes by using a chromatin immuno-precipitation (ChIP)-gene chip approach (31). A transgene expressing a PHAN-green fluorescent protein (GFP) fusion complemented both the developmental and defense-related phenotypes of as1 mutants, revealing that PHAN is functionally equivalent to AS1 (SI Fig. 6). ChIP-chip utilizing antibodies against GFP suggested that AS1 occupied the promoters of at least 96 genes (SI Table 1). We analyzed the expression profiles of these genes, where possible, by interrogating array experiments documented in the TAIR database (www.arabidopsis.org/). The expression of 37% of interrogated genes was changed by at least 2-fold in response to B. cinerea/JA/ET (SI Table 2), consistent with AS1 having a direct role in regulation of genes that are involved in defense.
The well characterized JA-regulated genes THIONIN (THI) and VEGETATIVE STORAGE PROTEIN (VSP1) (17) were among the most significantly enriched target genes in ChIP-chip. Maximum VSP1 and THI enrichment occurred with DNA sequences −350 to 400 and −150 to 250 bp, respectively, upstream of their respective start codons. In the case of VSP1, this sequence is 89 bp upstream of a known JA-responsive element (32). Primers designed to amplify these sequences were used to verify PHAN binding in three independent ChIP experiments, in which THI and VSP1 were enriched 5.39- and 5.42-fold, respectively (Fig. 2D), and similar enrichment was obtained when a different antibody against GFP was used in immunoprecipitation (SI Fig. 7). THI and VSP1 transcript accumulation was greater in as1 compared with wild type, suggesting that AS1 binding is needed to repress these two target genes (Fig. 2E). THI and VSP1 were not misexpressed in an as1 coi1 double mutant, indicating that JA signaling is still required to induce THI and VSP expression in the absence of AS1 activity. In addition, as2 mutants did not show increased THI and VSP1 transcript levels, suggesting that the role of AS1 in disease resistance might be independent of AS2.
Response of as1 Plants to Diverse Pathogens.
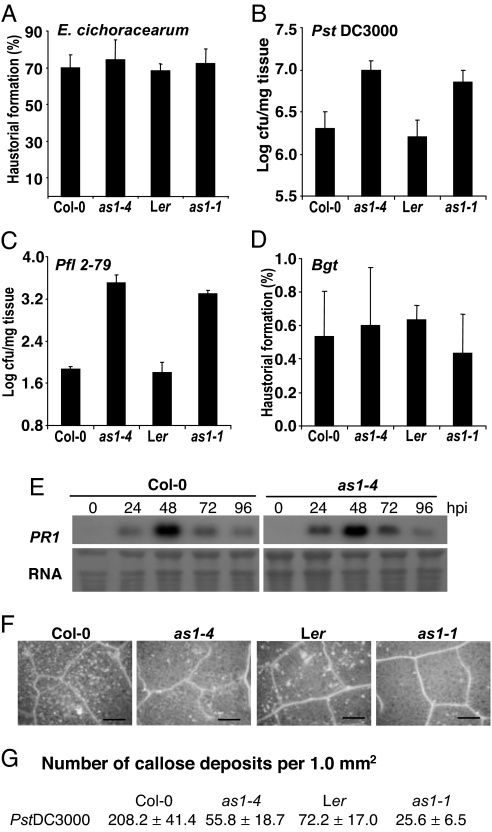
We tested whether AS1 function affected responses to other pathogens. Resistance to a virulent biotrophic powdery mildew fungus, Erysiphe cichoracearum (33), was unaffected by the as1 mutation (Fig. 3A). In contrast, as1 mutants supported growth of the virulent hemibiotrophic bacterium, Pseudomonas syringae pv tomato (Pst) DC3000 (34), to levels ≈7 times greater than wild type (Fig. 3B). Similarly, as1 mutants were more susceptible to Pseudomonas fluorescens (Pfl) 2-79, for which wild-type Arabidopsis is a nonhost (35) (Fig. 3C). Arabidopsis is also a nonhost for the wheat powdery mildew fungus Blumeria graminis f.sp. tritici (Bgt) (36). In contrast to Pfl2-79, however, nonhost resistance against Bgt was not compromised in as1 mutants (Fig. 3D). Basal and resistance (R) gene-mediated protection against two isolates of Hyaloperonospora parasitica, Noco2 (37) and Cala2 (38), also appeared not to be impacted by mutations in AS1 (SI Fig. 8). Neither PR1 expression (Fig. 3E) nor SA accumulation (data not shown) was affected in as1 plants inoculated with PstDC3000. However, callose deposition, a marker for SA-independent extracellular defense (39), was compromised in response to either PstDC3000 (Fig. 3 F and G) or Pfl2-79 (data not shown).
Fig. 3.
Both nonhost and basal resistance is compromised in as1 plants against attempted bacterial infection. (A) Extent of E. cichoracearum infection measured by determining the percentage of conidia that form a haustorium on the indicated plant lines. (B) Quantification of PstDC3000 growth in the stated plant genotypes at 5 days postinoculation. (C) Growth of Pfl2-79 in the stated plant genotypes at 5 days postinoculation. (D) Extent of Bgt infection measured by determining the percentage of conidia that form a haustorium on the indicated plant lines. (E) Accumulation of PR1 transcripts in wild-type Col-0 and Col-0 as1-4 plants at the given times after B. cinerea inoculation. (F) Portions of leaves from the indicated plant genotypes stained with Aniline blue to detect callose deposition (white dots) after inoculation with PstDC3000. (Scale bars, 250 μm.) (G) Quantification of callose deposition. Error bars represent 95% confidence limits. These experiments were repeated at least three times with similar results.
AS1 Function in Responses to Phytopathogens Is Independent of Its AS2-Associated Role in Development.
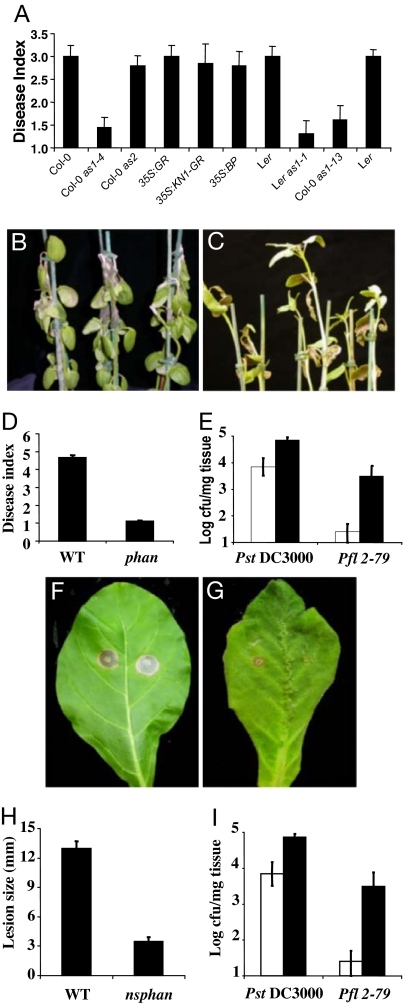
AS2, a member of the LATERAL ORGAN BOUNDARIES (LOB) family of plant-specific proteins, forms heterodimers with AS1 and is likely to function in the same developmental process (9, 10). The as2 and as1 lines have similar morphological phenotypes (10), but whereas as1 plants were more resistant to B. cinerea, as2 mutants were as susceptible as wild type (Fig. 4A). Furthermore, analysis of the weak as1-13 allele, which shows only mild morphological defects, revealed that this line still exhibited significant protection against B. cinerea (Fig. 4A). These findings suggest, firstly, that the resistance effects of as1 mutations are not due to altered morphology and, secondly, that the role of AS1 in repressing JA-induced defense genes is separate from its AS2-dependent role in development.
Fig. 4.
AS1 is an evolutionarily conserved regulator of plant immunity that functions independently of AS2 and KN1. (A) The given Arabidopsis genotypes were spray-inoculated with a B. cinerea spore suspension. The 35S:GR and 35S:KN1-GR lines were exogenously treated with 1 μM dexamethasone 48 h before B. cinerea inoculation and disease symptoms scored at 96 hpi by using a disease index. (B and C) Wild-type A. majus (B) and phan (C) plants were challenged with B. cinerea and scored at 96 hpi. (D) Quantification of B. cinerea disease phenotypes in the stated plant genotypes by using a disease index. (E) Wild-type (empty bars) and phan (filled bars) A. majus plants were inoculated with either PstDC3000 or Pfl2-79, and bacterial growth was determined at 5 days after inoculation. (F and G) Wild-type N. sylvestris plants (F) and an N. sylvestris antisense line (G) depleted in NSPHAN RNA were inoculated with B. cinerea and scored at 96 hpi. (H) Quantification of B. cinerea disease phenotypes in the stated plant genotypes. (I) Wild-type N. sylvestris (empty bars) and an N. sylvestris antisense line depleted in NSPHAN RNA (nsphan) (filled bars) were challenged with either PstDC3000 or Pfl2-79 and bacterial growth was determined 5 days after inoculation. Error bars represent 95% confidence limits. These experiments were repeated three times with similar results.
The only recognized developmental function of AS1 and AS2 is to prevent expression of KNOX genes (1, 10), and misexpression of the KNOX gene KN1 is sufficient to cause lobed leaves similar to those of as1 or as2 (11). Because the roles of AS1 in development and defense could be separated by their dependence on AS2, we predicted that KN1 misexpression would not cause increased resistance to B. cinerea. To test this, we induced ectopic KNOX activity by using a fusion of KN1 to the rat glucocorticoid receptor under control of the constitutive 35S promoter. This activity causes rapid dexamethasone-dependent changes in gibberellin and cytokinin levels and development of as1-like lobed leaves (11). Inducing KN1 activity before B. cinerea inoculation reduced the expression of a KN1 target gene, which encoded a gibberellin 20-oxidase (11) (data not shown) but did not confer increased resistance to B. cinerea (Fig. 4A). Constitutive expression of another KNOX gene, BREVIPEDICELLUS (BP) (40), that is an endogenous target of AS1 and AS2, gave a similar result. We also tested potential involvement of the ERECTA receptor-like kinase, which has multiple developmental roles mainly associated with cell growth. Although er mutations enhance the as1 and as2 mutant phenotype (10) and cause increased susceptibility to some bacterial pathogens (41), er mutants showed the same response to B. cinerea as wild type (Fig. 4A).
The Defense Function of AS1 Is Evolutionarily Conserved.
AS1 is orthologous to PHAN in Antirrhinum (2) and NSPHAN in tobacco (4), which have conserved roles in repressing KNOX genes in leaves. We therefore determined whether the functions of AS1 in plant–pathogen interactions were also conserved. Wild-type A. majus showed strong disease development 10 days after spraying with a suspension of B. cinerea spores, whereas phan mutants had significantly reduced levels of disease (Fig. 4 B–D). Conversely, phan mutants supported an increase in PstDC3000 and Pfl2-79 growth relative to wild type (Fig. 4E). Similarly, an antisense N. sylvestris line depleted in NSPHAN RNA (4) showed reduced lesion development in response to B. cinerea relative to wild type (Fig. 4 F–H) and increased PstDC3000 and Pfl2-79 growth (Fig. 4I). Collectively, these data suggest that AS1 has an evolutionarily conserved role in plant–pathogen interactions.
Discussion
We have uncovered an unexpected role for the developmental regulator, AS1, in plant–pathogen interactions. Loss-of-function mutations in this gene convey strikingly increased resistance against both B. cinerea and A. brassicicola. Therefore, AS1 may repress the inducible defense response against a relatively broad range of necrotrophic fungi. Our findings suggest that AS1 acts to suppress resistance to nectrotrophs by repressing the expression of JA-induced resistance proteins, including THI, which when overexpressed is sufficient to confer enhanced resistance to the necrotropic pathogen Fusarium oxysporum in Arabidopsis (42). Because AS1 associates physically with the promoters of JA-induced genes, this repression is likely to be direct. Target gene expression and increased resistance, however, remain dependent on functional JA signaling in the absence of AS1 activity. For at least one target gene, the promoter region occupied by AS1 is distinct from that required for response to JA, and AS1 expression is not affected when JA signaling is disrupted. This observation suggests that induction by JA signaling and repression by AS1 act in parallel, converging on target gene promoters. One rationale for the repressive role of AS1 is that it damps the JA response, preventing premature deployment of the immunity response and its associated costs.
Epistasis analysis revealed that EIN2 and, particularly, COI1 were required for as1-mediated resistance against B. cinerea. This finding contrasts with resistances established by opr3, the only other reported loss-of-function mutation that conveys robust resistance against B. cinerea (24) or by overexpression of ETHYLENE RESPONSE FACTOR1 (ERF1), which were both independent of EIN2 (24, 43). AS1 is a known repressor of developmental target genes (1) and is implicated in epigenetic control because it interacts with a HIRA-like protein (44), homologues of which are needed to maintain silent chromatin in yeast and animals. JA/ET-dependent gene expression in Arabidopsis involves epigenetic control through histone modification (45), and therefore AS1 might act in this process. AS1, however, appears unlikely to function as a corepressor of many genes given the relatively low number of potential targets detected in ChIP-chip experiments and the limited pleiotropic effects of as1 mutations.
Our findings suggest that AS1 is a positive regulator of disease resistance against bacterial pathogens. Callose deposition in response to attempted bacterial infection was compromised in as1 plants. However, neither PR1 expression nor SA accumulation was altered. Thus, AS1 appears to regulate bacterially triggered SA-independent extracellular defenses (which are known to be targets for suppression by effector proteins delivered via the type III secretory system) during bacterial pathogenesis (39). AS1 may therefore regulate defense responses against two distinct classes of phytopathogens.
AS2 functions as a heterodimer with AS1 in leaf development (9, 10). Significantly, as2 mutants did not show increased resistance against B. cinerea, implying that AS1 acts independently of AS2 in pathogen responses but not in development. The only known role of AS1 and AS2 in leaf morphogenesis is in repression of KNOX genes (1, 10). Ectopic KNOX activity encoded by BP, a target of AS1 repression (40), caused an as1-like morphology but did not increase resistance against B. cinerea, further supporting a function for AS1 in disease resistance that is independent of its AS2-associated developmental role.
AS1 orthologs in Antirrhinum (2) and tobacco (4) have a conserved role in repressing KNOX genes in leaves. We have shown that their role in repressing resistance against B. cinerea is also conserved, and that they also have similar effects on resistance to hemibiotrophic bacterial pathogens. The role of AS1 in immunity is therefore conserved in eudicot species with a divergence time of ≈125 million years. The ability to repress KNOX gene expression is likely to be more ancient because the morphological effects of as1 mutations can be complemented by an AS1-like gene from Selaginella (46), which last shared a common ancestor with eudicots ≈400 million years ago. Although we have not tested when the role of AS1-like genes in disease resistance evolved, it is attractive to speculate that the ancestral role was developmental, and that AS1-like genes that were already expressed in lateral organs were later coopted to regulate defense responses.
A dual role in development and immunity is paralleled in animals in which the Spätzle-Toll-Cactus signaling pathway that controls dorsoventral patterning in Drosophila also regulates the innate immune response (47). However, it differs in involving a common transcriptional regulator in plants rather than redeployment of a signaling cassette as in Drosophila.
Methods
Plant Genotypes.
The original as1-1 mutant was identified in a Col-0 genetic background (1); this mutation was also introgressed into Ler. The generation of activation T-DNA-tagged lines and as1 double mutants was as described in ref. 26. The A. majus (2) and N. sylvestris (4) lines were reported previously.
Pathogen Maintenance and Inoculations.
B. cinerea (PJH2) (26) and A. brassicola (MUCL20297) (15) were grown on oat meal and ½ potato dextrose agar media, respectively. Fungal spore density was adjusted to 5 × 105 spores per ml in ½ potato dextrose broth for B. cinerea and water for A. brassicicola. The resulting B. cinerea spore suspensions were sprayed onto 4-week-old Arabidopsis plants that were maintained at 100% humidity for 4 days. A. brassicicola was inoculated by applying single drops of spore suspension onto detached plant leaves. The disease index used to score B. cinerea infection was based on previously reported methodology (26). PstDC3000 and Pfl2-79 were inoculated in 10 mM MgCl2 at 5 × 105 cfu/ml (35). The fungal pathogens E. cichoracearum and Bgt were maintained and disease assays performed as described in refs. 26 and 36. H. parasitica inoculations were undertaken as reported in refs. 37 and 38.
RNA Analysis.
Northern blot analysis was undertaken as reported in ref. 26. The PR1, PR3, PR4, and PDF1.2 probes were amplified as described in refs. 15 and 26. The PCR primers for AS1 were 5′-GCCTATTGACGAGAGTAAGTAC-3′ and 5′-CCACAAGCTCTGACAAGAACAC-3′. The PCR primers for AtGA20ox1 were as reported in ref. 11.
Histochemical Analysis and Chemical Treatments.
Lesion development was scored by staining leaves with lactophenol blue and clearing with saturated chloral hydrate as described in ref. 36. For callose deposition, Arabidopsis leaves were infiltrated with a bacterial suspension of 2.5 × 105 cfu/ml and detached after 20 h. Epifluorescence microscopy was used to detect callose after staining with aqueous aniline blue as described in ref. 39. To transiently express KN1, a conditional expression system was used in a Col-0 background. This system consisted of a C-terminal fusion of the KN1 cDNA with the steroid-binding domain of the rat glucocorticoid receptor from pBI-ΔGR (11). A line expressing only ΔGR was used as a control.
Determination of SA, JA, and ET Levels.
Free SA and SA-β-glucoside (SAG) levels were determined essentially as described in ref. 35. ET emission from 4-week-old plants was measured by gas chromatography with an HP5980 series II gas chromatograph with a flame ionization detector (Hewlett Packard) equipped with a Quadrex BTR-CW (50-m length, 0.32-mm diameter) and run at 80°C (48). JA concentrations were determined by gas chromatography-mass spectrometry as described previously (26).
ChIP and RT-PCR.
Procedures are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Miltos Tsiantis (Oxford University, Oxford, U.K.) for the 35S::KN1-GR line, Neil McHale (Connecticut Agricultural Experiment Station, New Haven, CT) for the antisense N. sylvestris line, Jian-Min Zhu (Institute of Biological Sciences, Beijing, China) for Pfl2-79, Bruno P. A. Cammue (Katholieke Universiteit Leuven, Heverlee-Leuven, Belgium) for A. brassicicola strain MUCL20297, Jane Glazebrook (University of Minnesota, St. Paul, MN) for providing the camalexin standard, Jan van Kan (University of Wageningen, Wageningen, The Netherlands) for the B. cinerea actin probe, Roger Innes (Indiana University, Bloomington, IN) for PstDC3000, John Turner (University of East Anglia, Norwich, U.K.) for the coi1 line, and NASC for the ein2 line. The PHAN-GFP as1 line was produced by Peter Newton (University of Edinburgh, Edinburgh, U.K.). P.L.N. was the recipient of a scholarship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (Brazil). This research was funded in part by Biotechnology and Biological Sciences Research Council Grants P20067 (to G.J.L.) and G19821 (to A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705586104/DC1.
References
- 1.Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 2.Waites R, Selvadurai HR, Oliver IR, Hudson A. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- 3.Timmermans MCP, Hudson A, Becraft PW, Nelson T. Science. 1999;284:151–153. doi: 10.1126/science.284.5411.151. [DOI] [PubMed] [Google Scholar]
- 4.McHale NA, Koning RE. Plant Cell. 2004;16:1251–1262. doi: 10.1105/tpc.019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volbrecht E, Veit B, Sinha N, Hake S. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- 6.Schneeburger RG, Becraft PW, Hake S, Freeling M. Genes Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- 7.Long JA, Moan EI, Medford JI, Barton KA. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 8.Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe Y, Soma T, Ikezaki M, Machida C, Machida Y. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 9.Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. Development (Cambridge, UK) 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 10.Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. Development (Cambridge, UK) 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- 11.Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. Curr Biol. 2002;12:1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- 12.Holt BF, Boyes DC, Ellerström M, Siefers N, Wiig A, Kauffman S, Grant MR, Dangl JL. Dev Cell. 2002;6:807–817. doi: 10.1016/s1534-5807(02)00174-0. [DOI] [PubMed] [Google Scholar]
- 13.Gray WM, Musket PR, Chuang H-W, Parker JE. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazebrook J. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 15.Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomma BPHJ, Eggermont K, Tierens KFMJ, Broekaert WF. Plant Physiol. 1999;121:1093–1101. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner JG, Ellis C, Devoto A. Plant Cell. 2002;14(Suppl):S153–S164. doi: 10.1105/tpc.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gfeller A, Farmer EE. Science. 2004;306:1515–1516. doi: 10.1126/science.1104352. [DOI] [PubMed] [Google Scholar]
- 19.Gupta V, Willits MG, Glazebrook J. Mol Plant–Microbe Interact. 2000;13:503–511. doi: 10.1094/MPMI.2000.13.5.503. [DOI] [PubMed] [Google Scholar]
- 20.Felton GW, Korth KL, Wesley SV, Huhman DV, Mathews MC, Murphy JB, Lamb C, Dixon RA. Curr Biol. 1999;9:317–320. doi: 10.1016/s0960-9822(99)80140-7. [DOI] [PubMed] [Google Scholar]
- 21.Mengiste T, Chen X, Salmeron J, Dietrich R. Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veronese P, Chen X, Bluhm B, Salmeron J, Dietrich R, Mengiste T. Plant J. 2004;40:558–574. doi: 10.1111/j.1365-313X.2004.02232.x. [DOI] [PubMed] [Google Scholar]
- 23.Veronese P, Nakagami H, Bluhm B, AbuQamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. Plant Cell. 2006;18:257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coego A, Ramirez V, José Gil M, Flors V, Mauch-Mani B, Vera P. Plant Cell. 2005;17:2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant JJ, Chini A, Basu D, Loake GJ. Mol Plant–Microbe Interact. 2003;16:669–680. doi: 10.1094/MPMI.2003.16.8.669. [DOI] [PubMed] [Google Scholar]
- 27.Rédei GP, Hirono Y. Arabid Inf Serv. 1964;1:9–10. [Google Scholar]
- 28.Benito EP, ten Have A, van't Klooster JW, van Kan JAL. Eur J Plant Path. 1998;104:207–220. [Google Scholar]
- 29.Guzman P, Ecker JR. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feys BJF, Benedetti CE, Penfold CN, Turner JG. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 32.Guerineau F, Benjdia M, Zhou DX. J Exp Bot. 2003;385:1153–1162. doi: 10.1093/jxb/erg123. [DOI] [PubMed] [Google Scholar]
- 33.Vogel JP, Raab TK, Schiff C, Somerville SC. Plant Cell. 2002;14:2095–2106. doi: 10.1105/tpc.003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feechan A, Kwon E, Yun B-W, Wang Y, Pallas JA, Loake GJ. Proc Natl Acad Sci USA. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun B-W, Atkinson HA, Gaborit C, Greenland A, Read ND, Pallas JA, Loake GJ. Plant J. 2003;34:768–777. doi: 10.1046/j.1365-313x.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 37.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holub EB, Beynon JL, Crute IR. Mol Plant–Microbe Interact. 1994;7:223–239. doi: 10.1094/mpmi-8-0916. [DOI] [PubMed] [Google Scholar]
- 39.Hauck P, Thilmony R, Yang SH. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y. Plant J. 2003;36:353–365. doi: 10.1046/j.1365-313x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- 42.Epple P, Apel K, Bohlmann H. Plant Cell. 1997;9:509–520. doi: 10.1105/tpc.9.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berrocal-Lobo M, Molina A, Solano R. Plant J. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- 44.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou C, Zhang L, Duan J, Miki B, Wu K. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison CJ, Corley B, Moylan EC, Alexander DL, Scotland RW, Langdale JA. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- 47.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 48.Locke JM, Bryce JH, Morris PC. J Exp Bot. 2000;51:1843–1849. doi: 10.1093/jexbot/51.352.1843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.