Abstract
Plants are unique in their ability to store proteins in specialized protein storage vacuoles (PSVs) within seeds and vegetative tissues. Although plants use PSV proteins during germination, before photosynthesis is fully functional, the roles of PSVs in adult vegetative tissues are not understood. Trafficking pathways to PSVs and lytic vacuoles appear to be distinct. Lytic vacuoles are analogous evolutionarily to yeast and mammalian lysosomes. However, it is unclear whether trafficking to PSVs has any analogy to pathways in yeast or mammals, nor is PSV ultrastructure known in Arabidopsis vegetative tissue. Therefore, alternative approaches are required to identify components of this pathway. Here, we show that an Arabidopsis thaliana mutant that disrupts PSV trafficking identified TERMINAL FLOWER 1 (TFL1), a shoot meristem identity gene. The tfl1-19/mtv5 (for “modified traffic to the vacuole”) mutant is specifically defective in trafficking of proteins to the PSV. TFL1 localizes to endomembrane compartments and colocalizes with the putative δ-subunit of the AP-3 adapter complex. Our results suggest a developmental role for the PSV in vegetative tissues.
Keywords: protein trafficking, vegetative tissue, adaptin
There are at least two kinds of plant vacuoles: lytic vacuoles (LVs) of acidic pH that are functionally equivalent to yeast vacuoles and mammalian lysosomes, and protein storage vacuoles (PSVs) of neutral pH (1–5) that store reserve proteins, minerals, and defense proteins such as lectins. Besides pH, which is not a reliable indicator of vacuole type (6), they are categorized by their luminal contents, processing enzymes, and tonoplast proteins (1–3, 7–10). In the classic model of sorting to LVs, cargo proteins contain an N-terminal NPIR motif that is recognized by a sorting receptor like BP-80. Upon receptor binding, cargo proteins are packaged into clathrin-coated vesicles (CCVs) at the trans-Golgi network (TGN). CCVs travel to the prevacuolar compartment where receptors dissociate from cargoes in the low-pH environment then recycle to the TGN. Proteins destined for PSVs may also have NPIR-like motifs; however, they often feature a C-terminal vacuolar sorting signal (ctVSS) or internal targeting signal motifs. Neither signal is found in LV-targeted proteins, and they are less defined than NPIR motifs.
It is well established that LVs are important for homeostasis, turgor pressure maintenance, and other functions. In contrast, PSVs are present in tissues and organs other than seeds, but their functions in vegetative tissues are poorly understood. One known role for PSVs is storage of defense proteins, such as lectins (11), chitinase, and glucanase (4). Some Arabidopsis vegetative storage proteins are anti-insect acid phosphatases (12). PSVs are found in roots, tubers, bark parenchyma, and paraveinal parenchyma (13–15) and are probably in the perennial root systems of plants whose shoots die in winter (e.g., perennial grasses) (16–20). Accumulation of proteins into PSVs and later degradation for renewed growth is important physiologically for many plant cells and organs. In the past, PSVs of cotyledons and cereal endosperm have received attention (7, 21, 22), but vegetative storage compartments may be important for tree growth and in annual shoot regrowth of perennial grasses like Miscanthus. It is unknown whether vegetative PSVs store pigments and proteins together or in distinct cargo-specific PSVs. The mechanistic details of trafficking to seed and vegetative PSVs are poorly understood.
Although we are addressing PSV trafficking in the model plant Arabidopsis, which has a short cycle and probably does not exchange fixed nitrogen among tissues to the extent of trees or other perennials, ctVSS trafficking does occur in vegetative tissues and is clearly distinct from LV trafficking pathways (23, 24). Arabidopsis genes for vegetative storage proteins are expressed in nonseed tissues (25), and PSVs are present in protoplasts from Arabidopsis leaves (26).
We are addressing these questions by developing genetic and biochemical approaches to dissect the PSV pathway. First is the creation of an Arabidopsis line, Vac2, and the subsequent generation of mutants from this line that are impaired in trafficking to PSVs, and not to LVs (ref. 24 and our own unpublished data). In yeast, genes required for vacuole trafficking are known primarily via screens for mutants that secrete vacuolar proteins (27). However, evidence linking genes to trafficking to PSVs in plants is very limited. Recently, ours and other laboratories have started to address this question genetically (refs. 24 and 28–31 and our own unpublished data). Assays for yeast vacuolar trafficking mutants detected vacuolar enzyme activities in the growth media. Disruption of PSV trafficking in plants also results in secretion of storage cargo (30, 32–34), but the presence of proteins in the apoplasm cannot be measured readily. However, vacuolar sorting signals (VSSs) from PSV proteins such as barley lectin (BL) target chimeric proteins to the vacuole (35). Thus, we used this VSS to develop a storage vacuole marker whose secretion was easily detectable. Combined with an endogenous apoplastic regulatory protein, we were able to isolate mutants. The CLV3 protein negatively regulates proliferation of the shoot apical meristem (SAM) (36–39). As an extracellular ligand, it activates the CLV1/2 LRR kinase receptor. The BL VSS fused to CLV3 reroutes the resulting VAC2 fusion protein to the vacuole, preventing activation of the receptor (37) (Fig. 1). These results suggested we could isolate mutants defective in trafficking to PSVs on the basis of the degree of meristem proliferation. Previously, we demonstrated that the Arabidopsis SNARE VTI12 mediates trafficking of storage proteins in seeds and vegetative tissues, but not trafficking to the LV (24), thus validating the Vac2 assay to isolate mutants in this important pathway.
Fig. 1.
Floral meristems from wild-type Landsberg erecta, clavata3-2, Vac2, mtv5, and mtv5 complemented with TFL1 driven by its native promoter. (Scale bars, 2 mm.)
Because the assay is not restricted to seeds, it should identify trafficking components potentially specific to vegetative tissues. Therefore, we can delineate the functions of PSVs in these tissues, which is completely unexplored. Mutants from the screen may also highlight the consequences of PSV trafficking defects at the whole-plant level. Using an EMS-mutagenized Vac2 mutant, we demonstrate a role for the shoot meristem identity factor TFL1 in endomembrane trafficking.
Results and Discussion
TFL1 Is Identified in a Screen for Proteins That Mediate Trafficking to the PSV.
We developed an Arabidopsis thaliana line, Vac2 [supporting information (SI) Fig. 5A], which allows the identification of proteins that mediate trafficking to the PSV. In previous work, we demonstrated that the CLAVATA3 (CLV3) protein is exported to the apoplast (37). Extracellular localization of CLV3 protein is necessary for activation of the CLV1/2 LRR kinase receptor, and subsequently, restriction of the stem cells at the SAM. Wild-type CLV3 fused to a C-terminal propeptide (CTPP) signal redirects CLV3 to the PSV instead of the apoplasm (37). Consequently, there is no complementation in a clv3-2 mutant background. Here, we describe the mutagenesis of Vac2 and characterize a component of the PSV trafficking machinery (SI Fig. 5B).
The Vac2 line was treated with EMS, and M2 plants were screened for complementation of the clv3-2 phenotype. We isolated putative mutants, outcrossed them to the Columbia ecotype, and mapped them (40). One line, designated mtv5 (for “modified traffic to the vacuole”), was tightly linked to the CTR1 marker on chromosome 5 (SI Fig. 5 D and E). TFL1 is tightly linked to CTR1, so we sequenced the TFL1 gene from the mutant and found a mutation (R143K) in a highly conserved domain (SI Fig. 5F). This allele of tfl1 was designated tfl1-19. We refer to the tfl1-19 VAC2 clv3-2 genotype as mtv5 and to tfl1-19 CLV3/CLV3 without VAC2 as tfl1-19. Transformation of the mtv5 mutant with a native promoter-driven TFL1 construct restored the parental phenotype (Fig. 1 and SI Fig. 6).
tfl1-19 Plants Are Defective in Trafficking to the PSV.
TFL1 is a member of the CETS (CEN, TFL1, and SP) gene family (SI Fig. 5F) and was reported to be a mobile signal that mediates the transition to flowering (41, 42). CLV proteins are responsible for controlling the size of the SAM; they restrict the population of stem cells by limiting the expression of the transcription factor WUSCHEL (WUS) in the peripheral zone of the meristem (36, 43, 44). Thus, both TFL1 and CLV3 act in different circuits of the gene regulatory network that controls shoot development (45).
Nevertheless, upon identification of mtv5 as TFL1, we were concerned that it was acting as a bypass suppressor mutant of clv3-2. Analysis of populations in which various tfl1 alleles and clv3-2 were segregating showed that none of the tfl1 alleles were epistatic to clv3-2 (SI Fig. 7, SI Table 1, and SI Text), and thus the floral phenotype indicated a trafficking defect in the tfl1-19 mutants.
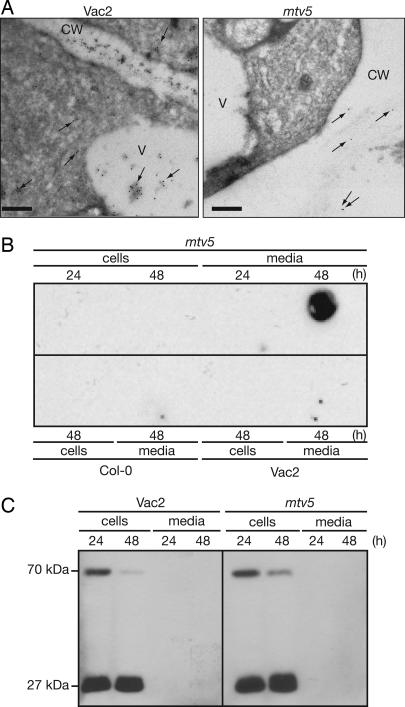
To delineate whether the tfl1-19 mutation was responsible for the trafficking defect, immunoelectron microscopy localization of the CLV3:T7:CTPPBL fusion protein in Vac2 and mtv5 was performed. The results demonstrated that the T7 label was localized to the apoplasm in the mutant and strictly to the vacuole and cytoplasm in the Vac2 parent (Fig. 2A). Furthermore, the localization of aleurain, a LV cargo, was unchanged in mtv5 (SI Fig. 8). These results demonstrated that complementation of the clv3-2 phenotype was due to a trafficking defect, not to an epistatic interaction between the clv3-2 and tfl1-19 mutations, and the defect was specific to the PSV pathway.
Fig. 2.
Trafficking defects in mtv5 mutants. (A) CLV3:T7:CTPPBL relocalizes in mtv5. Electron micrographs show that the chimeric protein is trafficked to the vacuole in the Vac2 parental line but trafficked to the apoplasm in mtv5. Arrows designate the locations of gold particles. V, vacuole; CW, cell wall. (Scale bars, 200 nm.) (B) Col-0, Vac2, and mtv5 protoplasts. Protoplasts were incubated for 24 or 48 hours as indicated. Cells and media were separated, and proteins were analyzed in a dot-blot by using an α-T7 antibody. (C) Trafficking of a transiently expressed Arabidopsis aleurain-like protein (AtALP:GFP) is not affected by the mtv5 mutation. The 70-kDa protein is the AALP:GFP fusion, and the 27-kDa protein is GFP that was released from the 70-kDa chimeric protein upon correct delivery to the vacuole. The experiment was carried out as in B, except that protein extracts were resolved using SDS/PAGE and visualized using an α-GFP antibody.
The CLV3:T7:CTPPBL chimera was too small to detect by SDS/PAGE (46, 47), so we examined the impact of the tfl1-19 mutation on trafficking by dot-blot analysis of protoplasts from Vac2 and mtv5 lines. After 24 and 48 h of incubation, the cells and media were probed with T7 antibody (Fig. 2B). The T7 signal was detected only in the mtv5 media fraction after 48 h of incubation. Therefore, the T7-tagged construct entered the default secretion pathway only in the mtv5 mutant. In a parallel experiment, we examined whether mtv5 was deficient in trafficking to the LV. Vac2 and mtv5 protoplasts transfected with an Arabidopsis aleurain-like protein fused to GFP (AtALP:GFP), a marker for LV traffic, showed normal trafficking (Fig. 2C) (48). Therefore, mtv5 plants were deficient specifically in traffic to the PSV.
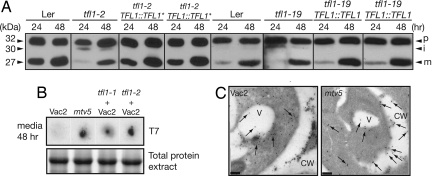
Further evidence that loss of TFL1 negatively affects traffic to the PSV was provided by examination of a GFP:CTPPBL construct transfected into Landsberg erecta (Ler), tfl1-2 (49, 50), and tfl1-19 protoplasts. The tfl1-19 protoplasts were isolated from a line in which VAC2 and clv3-2 had been outcrossed (SI Fig. 6). The CTPP motif was derived from BL, which is modified in vivo by a high-mannose oligosaccharide at a site within the CTPP (51). In wild-type protoplasts, GFP:CTPPBL was detected as a 32-kDa precursor protein (p) and a 27-kDa mature protein (m) (Fig. 3A). In the tfl1-2 protoplasts, we observed precursor (p) and 30-kDa size intermediate forms (i) of the protein. It was not clear whether the 30-kDa intermediate protein is a nonglycosylated form or a partially processed form of the protein; nevertheless, this was also observed in tfl1-1 and tfl1-11, but not in Col-0 protoplasts (data not shown). In tfl1-19 protoplasts, there were small amounts of the intermediate form and little observable processed protein at 24 hours, indicating that tfl1-19 also had a defect in trafficking, although it is a weaker allele with respect to trafficking than tfl1-2. Therefore, the trafficking defect in mtv5 was neither allele-specific nor a result of the Vac2 genetic background. Protoplasts isolated from two independent tfl1-2 and tfl1-19 lines transformed with TFL1::TFL1 constructs had no observable intermediate form, and the amounts of mature protein were comparable to those from wild-type Ler protoplasts at 24 hours, indicating that the trafficking defect in tfl1-19 plants was a direct effect of the mutation.
Fig. 3.
Multiple alleles of TFL1 have trafficking defects. (A) Protoplasts were isolated from Ler, tfl1-2, tfl1-2 complemented with pASM4 (TFL1::TFL1*), tfl1-19, or tfl1-19 complemented with pNVR3068 (TFL1::TFL1). Cells were transformed with GFP:CTPPBL and collected after 24 and 48 hours; the proteins were analyzed by SDS/PAGE and Western blotting by using α-GFP antibodies. (B) (Upper) Dot-blots of media collected after a 48-hour incubation of protoplasts from Vac2, mtv5, and tfl1-1 and tfl1-2 crossed into the Vac2 line. (Lower) Shown is a Coomassie-stained SDS/PAGE gel of the corresponding cell extracts as a loading control. (C) Vacuolar peroxidases are partially relocalized in mtv5. Immunogold-labeling experiments show that Arabidopsis vacuolar peroxidases containing CTPPs are trafficked to the vacuole and cytoplasm in the Vac2 parental line but trafficked to the cell wall and apoplasm in mtv5. Arrows designate the locations of gold particles. V, vacuole; CW, cell wall. (Scale bars, 200 nm.)
Further evidence that mutations in TFL1 cause trafficking defects to the PSV was provided by tfl1-1 and tfl1-2 (49, 50) protoplasts in the Vac2 background (Fig. 3B). The CLV3:T7:CTPPBL chimeric protein was detected in media after a 48-hour incubation of protoplasts for all three tfl1 alleles. The plants from which the protoplasts were isolated also had a complemented floral phenotype (data not shown).
Finally, we recently described a six-member subfamily of Arabidopsis peroxidases that contained ctVSS and were found in enriched vacuolar fractions (52). Using immunoelectron microscopy, we localized these peroxidase family members to the vacuoles of wild-type plants (24). Furthermore, we observed the presence of these vacuolar peroxidases in the apoplastic fluid of CTPP trafficking mutants (24). Quantitative analysis of immunogold labeling using antibodies raised against a conserved domain of this family showed that, in Vac2, >99% of the label was localized to the vacuole and cytoplasm; however, in mtv5 mutants, >50% of the label relocalized to the cell wall and apoplasm (Fig. 3C and SI Fig. 9). Thus, an endogenous Arabidopsis PSV protein is mistargeted in mtv5, supporting our hypothesis that TFL1 is necessary for proper trafficking of PSV-targeted proteins.
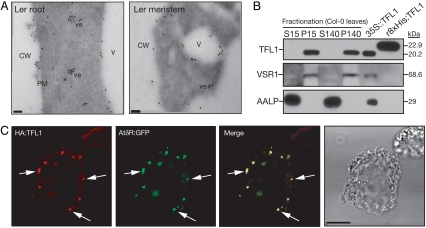
In plants, TFL1 and FLOWERING LOCUS T (FT) have antagonistic roles in control of flower development (53–55) and have been shown in yeast two-hybrid experiments to interact with FD, a bZIP transcription factor that promotes the floral transition in the shoot apex (56, 57). Aside from these heterologous data, the molecular function of TFL1 is poorly understood. The expression of both the TFL1 gene and its protein product are not restricted to the SAM, as demonstrated by gene expression and proteomic analysis (SI Fig. 10 and SI Text). We determined the subcellular localization of TFL1 by using immunoelectron microscopy and confocal microscopy (Fig. 4 A and C and SI Figs. 11 and 12). Immunogold labeling in the shoot apex and root tissue of wild-type plants showed that TFL1 was localized to the plasma membrane, tonoplast, and 100-nm-sized dense vesicles/compartments (Fig. 4A). Fractionation of wild-type protoplasts into soluble and membrane fractions demonstrated that TFL1 was associated with membranes (Fig. 4B). This result is in contrast with previous results (42) in which TFL1 was localized to the soluble fraction of ap1 cal double-mutant extracts. We believe that this result was due to overexpression of TFL1, because we have detected partial soluble localization of the TFL1 protein in extracts isolated from both 35S::TFL1 plants and protoplasts transfected with overexpression constructs (data not shown).
Fig. 4.
Subcellular localization of TFL1. (A) Immunoelectron microscopy of root tips and shoot meristems demonstrated that TFL1 was localized to the plasma membrane, vacuole, and dense vesicles ≈100 nm in diameter. Localization was identical in mtv5 (data not shown). V, vacuole; CW, cell wall; ve, vesicle; PM, plasma membrane. (Scale bars, 100 nm.) (B) Subcellular distribution of TFL1 protein. Proteins were extracted from protoplasts and centrifuged; the soluble and pellet fractions were analyzed by Western blotting using α-TFL1 antibodies. Anti-VSR1 and anti-aleurain antibodies were used as controls for the membrane and soluble fractions, respectively. Total protein extracts from 2-week-old 35S::TFL1 seedlings and purified recombinant 8XHis:TFL1 from Escherichia coli were used as positive controls for the α-TFL1 antibodies. (C) Protoplasts were transformed with either HA:TFL1 or AtδR:GFP constructs. The protoplasts were fixed and visualized with a TRITC-conjugated secondary antibody for HA:TFL1. GFP signals were captured directly from fixed protoplasts. (Scale bar, 20 μm.)
To assign TFL1 to a specific trafficking complex, we considered recent evidence that AtVTI12 mediates trafficking to the PSV and that VTI12 and the AP-3 δ-adaptin partially colocalize to the same compartment (24, 58). Col-0 protoplasts transformed with marker constructs showed that HA:TFL1 and HA:tfl1-19 colocalize with AtδR:GFP, a marker for the putative δ-subunit of the AP-3 adapter complex (Fig. 4C and SI Fig. 11) (59). We did not observe colocalization of HA:TFL1 with the DIP:myc (34) or AtSYP21:myc (59) markers, which are found at the prevacuolar compartments of the PSV and LV, respectively (SI Fig. 11).
The finding that TFL1 colocalizes with δ-adaptin is intriguing. The Drosophila δ-adaptin homolog is garnet, a gene product thought to function as a coat protein in a vesicular trafficking pathway responsible for the formation of eye pigment storage granules (60). In mammals, the AP-3 pathway mediates the biogenesis of lysosome-related organelles (LROs) (such as melanosomes, platelet-dense granules, cytotoxic T lymphocyte granules, and lamellar bodies of lung type-2 cells) (61). Although the PSV has been regarded as a structure unique to plants, it appears that the AP-3 adapter complex may function in various pathways responsible for trafficking to storage compartments in eukaryotes.
The presence of LROs in lysosome-containing cells leads to the hypothesis that the PSV may serve a similar role in plant cells. Both compartments have storage functions; furthermore, PSVs are not limited to seed tissues, and therefore must have additional functions in the plant. We propose a model where PSVs in meristematic cells store factors necessary for control of flowering and meristem maintenance, until the proper combination of developmental and environmental cues trigger their secretion and activate the flowering pathway. The subcellular localization of developmental regulators adds a new layer of complexity and a different view of the control of plant organ initiation and differentiation.
Materials and Methods
Plant Materials and Growth Conditions.
Wild-type Arabidopsis thaliana plants were either in the Columbia (Col-0) or Landsberg erecta (Ler) ecotypes as designated. The Vac2 line that we used as the parental for EMS mutagenesis was a class 2 line with a single homozygous insert at the bottom of chromosome 3. This line, L1, expresses amounts of VAC2 that are close to the empirically determined saturation levels and exhibits a clv3 phenotype (24, 37). Vac2 seeds were treated with EMS according to standard protocols (62). The clv3-2, tfl1-1, tfl1-2, tfl1-11, and tfl1-14 alleles were obtained from the Arabidopsis Biological Resource Center (ABRC). Plants were grown under standard long-day conditions (16 h of light, 8 h of dark) at 22 ± 2°C.
Mapping of the mtv5 Mutation.
F2 seeds resulting from a cross between mtv5 and Col-0 wild-type plants were broadcast in flats and grown under standard long-day conditions. Plants that had a wild-type Col-0 phenotype were removed, and tissue samples were taken from 58 plants with the mtv5 phenotype. Genomic DNA was isolated using published protocols (62), and rough mapping was carried out using a pooled recombinant DNA strategy (SI Fig. 5D) (40). The mtv5 mutation was very tightly linked to the CTR1 marker at the top of chromosome 5 (SI Fig 5E). We had noted when screening for F2 recombinants from this line that a subpopulation of plants segregating into the Columbia background had a phenotype similar to that described for terminal flower 1 (tfl1) (SI Fig. 5C) (49, 63). TFL1 is tightly linked to CTR1, so the TFL1 gene from the mutant line was sequenced and found to contain a R143K mutation in a highly conserved domain of the TFL1 protein (SI Fig. 5F). This allele of tfl1 was designated tfl1-19.
Construction of Plasmids.
The tfl1-19/mtv5 mutation was complemented with a native construct amplified from Ler genomic DNA by using the following primers: forward, 5′-TTCTCTAGACATCAGAATCACTTTCTTCTACCC-3′ (XbaI) and reverse, 5′-TTCGGTACCAACTATCCTTTTCCGTATCTCCAC-3′ (KpnI). The resulting 3.6-kb product was digested with XbaI and KpnI and cloned into the pCAMBIA1300MCS binary vector (64) to produce pNVR3068. This plasmid was used for transformation of the mtv5 or tfl1-19 mutants as described below.
The genomic construct used to transform tfl1-2 mutants was pASM4 and included 1.035 kb of TFL1 coding sequence, 2.195 kb upstream of the ATG codon, and 4.613 kb of the genomic region downstream of the stop codon. This construct was made by subcloning a 7.843-kb XhoI/EcoRI fragment from a genomic clone in the λFixII vector (Stratagene) into the SalI/EcoRI sites of the binary vector pBIN19.
The TFL1 cDNA was obtained from the ABRC, and the tfl1-19/mtv5 mutant cDNA was created using a commercially available site-directed mutagenesis kit (Stratagene). The sequence was confirmed at the University of California, Riverside, Core Instrumentation Facility.
To generate the HA:TFL1 construct, the TFL1 cDNA was amplified by PCR using the following primers: 5′-GGAATTCCATATGATGGAGAATATGGGAACT-3′ (NdeI) and 5′-TTCGGTACCCTAGCGTTTGCGTGCAGCGGTTTC-3′ (KpnI). The resulting product was digested with NdeI and KpnI and cloned into the HA:Rha1 vector (48) that was also digested with NdeI and KpnI.
To make the GFP:CTPPBL fusionconstruct, a GFP sequence, including the signal peptides from SP-GFP-2SC (65) was amplified using the following primers: 5′-CGCGGATCCATGGCCAGACTCACAA-3′ (BamHI) and 5′-CCCCCGGGATCTCCCTTGTACAGCTCG-3′ (XmaI). The PCR product was ligated into 326 GFP3G (a gift from Inhwan Hwang, Center for Plant Intracellular Trafficking, Pohang University of Science and Technology, Pohang, Korea) that was digested with BamHI and XmaI. The CTPP sequence of BL was amplified from pGA643 VAC2 construct (37) by using the primers 5′-CCCCCGGGACCGGTCTTCGCCGGGGCCATC-3′(XmaI) and 5′-CCGCTCGAGCGGAGAATTATTAAGGACTAG-3′ (XhoI) and was ligated into the 326SPGFP construct by using the XmaI and XhoI sites.
Complementation of the mtv5, tfl1-19, and tfl1-2 Mutations.
To obtain tfl1-19 mutants, mtv5 was crossed with Ler wild type. F2 plants with tfl1 phenotype were selected and genotyped for the VAC2 construct and the clv3-2 mutation. The VAC2 construct was detected by PCR using VAC2 forward (5′-GAGGGGGAAATGTTTCGAGT-3′), VAC2 reverse (5′-ATACATGGGGGAACGCTTTT-3′) and TR3 (5′-ACGTGACTCCCTTAATTCTCC-3′) primers. The genotype of the CLV3 locus was determined by Southern blot analysis as described in ref. 66. Genomic DNA was digested with HindIII, and a PCR product that contains CLV3 was amplified from genomic DNA by using CLV3 forward (5′-GTTGTGAACCTCCACAGCAT-3′) and CLV3 reverse 2 (5′-GCGTTATTTGAGGTGGGAAA-3′) to use as probe. The complementation construct pNVR3068 was transformed into the GV3101 strain of Agrobacterium tumefaciens and used for transformation of mtv5 and tfl1-19 by using published protocols (67). Resistant plants were selected on solid media containing 25 μg/ml of hygromycin B.
The tfl1-2 plants were transformed with pASM4 in a similar manner, and resistant plants were selected in media containing 50 μg/ml kanamycin.
Microscopy.
For transmission electron microscopy, Arabidopsis root tips and shoot apices were cryosectioned as described in ref. 68. All immunogold labeling was carried out as described in refs. 68 and 69.
For analysis of single and double mutants, flowers were imaged under a Leica MZ FLIII stereomicroscope fitted with a SPOT RT digital camera (Diagnostic Instruments). For SEM, fresh flowers were mounted on SEM stubs and imaged in a XL30 FEG scanning electron microscope (Phillips) at an accelerating voltage of 5 kV (62).
Transient Expression and in Vivo Trafficking Assays.
Protoplasts were isolated from two week-old seedlings grown on B5 agar media and transformed by a polyethylene glycol-mediated procedure as described in ref. 70. The SYP21:myc, AtδR:GFP, AtALP:GFP, and DIP:myc constructs were the kind gifts of Inhwan Hwang.
Protein extracts from protoplasts were prepared at either the 24- or 48-h time points after transformation, as described in ref. 48. Protoplast incubation media was collected after centrifugation at 6,000 × g for 5 min. One mg of BSA was added to the media, which was then precipitated with 10% trichloroacetic acid and followed by centrifugation at 10,000 × g for 5 min. Precipitated protein aggregates were dissolved in 0.1 M NaOH. Immunoblots were developed using an ECL detection kit (Pierce Biotechnology).
Immunocytochemistry.
Immunocytochemistry was carried out as described in refs. 33, 48. Briefly, protoplasts were resuspended in W6 buffer [154 mM NaCl, 125 mM CaCl2, 2.5 mM maltose, 5 mM KCl, and 10 mM Hepes (pH 7.2)]. The protoplasts were distributed on charged slides and incubated for 1 hour, followed by a 1-hour fixation in 3% PFA. The protoplasts were then washed three times with TBW buffer [10 mM Tris (pH 7.4), 0.9% NaCl, 0.25% gelatin, 0.02% SDS, and 0.1% Triton X-100]. The fixed protoplasts were incubated with the appropriate antibodies overnight at 4°C. The α-TFL1 and α-myc antibodies were purchased from Santa Cruz Biotechnology, and the α-HA antibody was purchased from Roche Diagnostics. The signals were visualized using the described fluorescent conjugated secondary antibodies. The FITC-conjugated α-goat IgG and Cy3-conjugated α-rabbit IgG were purchased from Kirkegaard & Perry Laboratories, and the TRITC-conjugated α-rat IgG was purchased from Invitrogen.
Fractionation of Protoplasts.
Total proteins from the protoplasts were extracted using a lysis buffer that consisted of 10 mM Hepes (pH 7.7) and 1X protease inhibitor mixture (Roche Diagnostics) followed by repeated freeze–thaw cycles. The total protein was fractionated into soluble and membrane fractions by centrifugation at either 15,000 × g for 15 min or 140,000 × g for 1 hour. Aliquots from the membrane and soluble fractions were resolved using SDS/PAGE and were transferred to a nitrocellulose membrane for Western blot analysis using the α-TFL1 antibodies (Santa Cruz Biotechnology). The soluble fraction was analyzed using an anti-aleurain antibody (71).
Supplementary Material
Acknowledgments
We thank Drs. X. Chen, G. Hicks, S. Robert, H. Smith, I. Lee, and J. Zouhar for critical reading of the manuscript; Drs. D. Carter and V. Kovaleva for microscopy; Dr. I. Hwang for constructs; and M. Brown for technical assistance. This work was funded by Department of Energy, Division of Energy Biosciences, Grant DE-FG03-02ER15295/A000 (to N.V.R.) and a National Institutes of Health National Research Service postdoctoral fellowship (to C.C.). The work at Instituto de Biología Molecular y Celular de Plantas was funded by Spanish Ministry of Education and Science Grant BIO2006-10994.
Abbreviations
- BL
barley lectin
- CTPP
C-terminal propetide
- ctVSS
C-terminal vacuolar sorting signal
- LRO
lysosome-related organelle
- LV
lytic vacuole
- PSV
protein storage vacuole
- SAM
shoot apical neristem.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708236104/DC1.
References
- 1.Paris N, Stanley CM, Jones RL, Rogers JC. Cell. 1996;85:563–572. doi: 10.1016/s0092-8674(00)81256-8. [DOI] [PubMed] [Google Scholar]
- 2.Jauh GY, Fischer AM, Grimes HD, Ryan CA, Rogers JC. Proc Natl Acad Sci USA. 1998;95:12995–12999. doi: 10.1073/pnas.95.22.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jauh GY, Phillips TE, Rogers JC. Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuhaus JM, Rogers JC. Plant Mol Biol. 1998;38:127–144. [PubMed] [Google Scholar]
- 5.Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA. Plant Cell. 2006;18:2567–2581. doi: 10.1105/tpc.106.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara-Nishimura I, Nishimura M. Plant Physiol. 1987;85:440–445. doi: 10.1104/pp.85.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoh B, Hinz G, Jeong BK, Robinson DG. J Cell Sci. 1995;108:299–310. doi: 10.1242/jcs.108.1.299. [DOI] [PubMed] [Google Scholar]
- 8.Swanson SJ, Bethke PC, Jones RL. Plant Cell. 1998;10:685–698. doi: 10.1105/tpc.10.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinz G, Hillmer S, Baumer M, Hohl I. Plant Cell. 1999;11:1509–1524. doi: 10.1105/tpc.11.8.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang LW, Phillips TE, Rogers SW, Rogers JC. J Cell Biol. 2000;150:755–769. doi: 10.1083/jcb.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peumans WJ, Van Damme EJM. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Ahn J-E, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K. Plant Physiol. 2005;139:1545–1556. doi: 10.1104/pp.105.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman EM, Hankins CN, Shannon LM. Plant Physiol. 1988;86:1027–1031. doi: 10.1104/pp.86.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wetzel S, Greenwood JS. Plant Physiol. 1991;97:771–777. doi: 10.1104/pp.97.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence SD, Greenwood JS, Korhnak TE, Davis JM. Planta. 1997;203:237–244. doi: 10.1007/s004250050187. [DOI] [PubMed] [Google Scholar]
- 16.Cyr DR, Bewley JD. Planta. 1990;182:370–374. doi: 10.1007/BF02411387. [DOI] [PubMed] [Google Scholar]
- 17.Gomez L, Faurobert M. J Exp Bot. 2002;53:2431–2439. doi: 10.1093/jxb/erf098. [DOI] [PubMed] [Google Scholar]
- 18.Justes E, Thiebeau P, Avice JC, Lemaire G, Volenec JJ, Ourry A. J Exp Bot. 2002;53:111–121. [PubMed] [Google Scholar]
- 19.Wisniewski M, Bassett C, Arora R. Tree Physiol. 2004;24:339–345. doi: 10.1093/treephys/24.3.339. [DOI] [PubMed] [Google Scholar]
- 20.Dhont C, Castonguay Y, Nadeau P, Belanger G, Drapeau R, Laberge S, Avice JC, Chalifour FP. Ann Bot (London) 2006;97:109–120. doi: 10.1093/aob/mcj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohl I, Robinson DG, Chrispeels MJ, Hinz G. J Cell Sci. 1996;109:2539–2550. doi: 10.1242/jcs.109.10.2539. [DOI] [PubMed] [Google Scholar]
- 22.Hillmer S, Movafeghi A, Robinson DG, Hinz G. J Cell Biol. 2001;152:41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitale A, Raikhel NV. Trends Plant Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- 24.Sanmartin M, Ordonez A, Sohn EJ, Robert S, Sanchez-Serrano JJ, Surpin MA, Raikhel NV, Rojo E. Proc Natl Acad Sci USA. 2007;104:3645–3650. doi: 10.1073/pnas.0611147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utsugi S, Sakamoto W, Murata M, Motoyoshi F. Plant Mol Biol. 1998;38:565–576. doi: 10.1023/a:1006072014605. [DOI] [PubMed] [Google Scholar]
- 26.Park M, Kim SJ, Vitale A, Hwang I. Plant Physiol. 2004;134:625–639. doi: 10.1104/pp.103.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowers K, Stevens TH. Biochim Biophys Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Fuji K, Shimada T, Takahashi H, Tamura K, Koumoto Y, Utsumi S, Nishizawa K, Maruyama N, Hara-Nishimura I. Plant Cell. 2007;19:597–609. doi: 10.1105/tpc.106.045997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Shimada T, Takahashi H, Ueda H, Fukao Y, Kondo M, Nishimura M, Hara-Nishimura I. Plant Cell. 2006;18:3535–3547. doi: 10.1105/tpc.106.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. Proc Natl Acad Sci USA. 2003;100:16095–16100. doi: 10.1073/pnas.2530568100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I. Plant Cell Physiol. 2006;47:1187–1194. doi: 10.1093/pcp/pcj103. [DOI] [PubMed] [Google Scholar]
- 32.Craig S, Goodchild DJ. Protoplasma. 1984;122:91–97. [Google Scholar]
- 33.Frigerio L, de Virgilio M, Prada A, Faoro F, Vitale A. Plant Cell. 1998;10:1031–1042. doi: 10.1105/tpc.10.6.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park M, Lee D, Lee GJ, Hwang I. J Cell Biol. 2005;170:757–767. doi: 10.1083/jcb.200504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bednarek SY, Raikhel NV. Plant Cell. 1991;3:1195–1206. doi: 10.1105/tpc.3.11.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 37.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. Plant Cell. 2002;14:969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenhard M, Laux T. Development (Cambridge, UK) 2003;130:3163–3173. doi: 10.1242/dev.00525. [DOI] [PubMed] [Google Scholar]
- 39.Fiers M, Ku KL, Liu CM. Current Opinion in Plant Biology. 2007;10:39–43. doi: 10.1016/j.pbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Lukowitz W, Gillmor CS, Scheible WR. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti L, Bradley D. Plant Cell. 2007;19:767–778. doi: 10.1105/tpc.106.049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark SE, Williams RW, Meyerowitz EM. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 44.Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 45.Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 47.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 48.Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, Hwang I. Plant Cell. 2003;15:1057–1070. doi: 10.1105/tpc.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- 50.Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F. Mol Gen Genet. 1997;254:186–194. doi: 10.1007/s004380050407. [DOI] [PubMed] [Google Scholar]
- 51.Wilkins TA, Bednarek SY, Raikhel NV. Plant Cell. 1990;2:301–313. doi: 10.1105/tpc.2.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 54.Hanzawa Y, Money T, Bradley D. Proc Natl Acad Sci USA. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. EMBO J. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 57.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 58.Lee G-J, Kim H, Kang H, Jang M, Wook Lee D, Lee S, Hwang I. Plant Physiol. 2007;143:1561–1575. doi: 10.1104/pp.106.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song J, Lee MH, Lee GJ, Yoo CM, Hwang I. Plant Cell. 2006;18:2258–2274. doi: 10.1105/tpc.105.039123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ooi CE, Moreira JE, DellAngelica EC, Poy G, Wassarman DA, Bonifacino JS. EMBO J. 1997;16:4508–4518. doi: 10.1093/emboj/16.15.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Odorizzi G, Cowles CR, Emr SD. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- 62.Weigel D, Glazebrook J. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2002. [Google Scholar]
- 63.Shannon S, Meeks-Wagner DR. Plant Cell. 1991;3:877–892. doi: 10.1105/tpc.3.9.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McElroy D, Chamberlain DA, Moon E, Wilson KJ. Mol Breed. 1995;1:27–37. [Google Scholar]
- 65.Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I. Plant J. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- 66.Rojas-Pierce M, Springer PS. Methods Mol Biol. 2003;236:221–240. doi: 10.1385/1-59259-413-1:221. [DOI] [PubMed] [Google Scholar]
- 67.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 68.Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. Proc Natl Acad Sci USA. 1998;95:9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng H, von Mollard GF, Kovaleva V, Stevens TH, Raikhel NV. Mol Biol Cell. 1999;10:2251–2264. doi: 10.1091/mbc.10.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. Plant Cell. 2001;13:1511–1525. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV. J Cell Biol. 2000;149:1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.