Abstract
Cryptochrome blue-light receptors mediate many aspects of plant photomorphogenesis, such as suppression of hypocotyl elongation and promotion of cotyledon expansion and root growth. The cryptochrome 1 (cry1) protein of Arabidopsis is present in the nucleus and cytoplasm of cells, but how the functions of one pool differ from the other is not known. Nuclear localization and nuclear export signals were genetically engineered into GFP-tagged cry1 molecules to manipulate cry1 subcellular localization in a cry1-null mutant background. The effectiveness of the engineering was confirmed by confocal microscopy. The ability of nuclear or cytoplasmic cry1 to rescue a variety of cry1 phenotypes was determined. Hypocotyl growth suppression by blue light was assessed by standard end-point analyses and over time with high resolution by a custom computer-vision technique. Both assays indicated that nuclear, rather than cytoplasmic, cry1 was the effective molecule in these growth inhibitions, as was the case for the mechanistically linked membrane depolarization, which occurs within several seconds of cry1 activation. Petiole elongation also was inhibited by nuclear, but not cytoplasmic, cry1. Conversely, primary root growth and cotyledon expansion in blue light were promoted by cytoplasmic cry1 and inhibited by nuclear cry1. Anthocyanin production in response to blue light was strongly stimulated by nuclear cry1 and, to a lesser extent, by cytoplasmic cry1. An important step toward elucidation of cry1 signaling pathways is the recognition that different subcellular pools of the photoreceptor have different functions.
Keywords: blue light, hypocotyl, nuclear localization, membrane depolarization, anion channel
Plants employ at least three families of photoreceptor proteins to monitor light. Through signaling pathways that are being intensively studied, these photoreceptors convert the quality, quantity, and direction of incident light into developmental and physiological responses that best adapt the plant to the prevailing light environment. The cryptochrome photoreceptors of plants absorb UV-A/blue wavelengths (1) and mediate the suppression of seedling stem growth (2), promotion of leaf and cotyledon expansion (3, 4), flowering time (5, 6), resetting of the circadian oscillator (7), chlorophyll and anthocyanin synthesis (8), programmed cell death (9), and other processes. Widespread in biology, cryptochromes are known to regulate circadian rhythms in many organisms, including mammals (10).
The subject of this article is the cryptochrome 1 (cry1) of Arabidopsis thaliana. It was the founding member of the cryptochrome family of photoreceptors, originally identified by a genetic lesion that impaired blue-light suppression of hypocotyl elongation. Although some mechanistic details of cry1 action have been determined, an integrated view of how its absorption of blue light results in the various previous responses is lacking. The structural similarity between cry1 and DNA photolyases, light-activated enzymes that repair DNA dimers, indicated a nuclear localized function (11). Indeed, cry1 is present in the nucleus (12), but it is not known to bind or repair DNA (1). At least some of the cry1 action has been attributed to its physical interaction with the COP1 E3 ligase, a regulator of seedling development (13, 14) that is present in the nucleus until light stimulates its export to the cytoplasm (15). As a COP1 interactor, cry1 is believed to exert at least some of its effects by influencing the levels of the HY5 transcription factor, which interacts with the promoters of some light-regulated genes (16). Comparative transcript-profiling studies performed on cry1 mutant and wild-type seedlings exposed to blue light identified a large number of genes exhibiting cry1-dependent expression at the point in time when cry1 begins to influence the rate of hypocotyl elongation, which is ≈45 min after the onset of irradiation (17). Another microarray expression study identified suites of genes that changed expression levels after 6 days of blue light in a manner that depended on cry1 and/or cry2 (18). Proteomic analysis identified 61 specific proteins that were present at different levels in a cry1 mutant, compared with the wild type, after a blue-light treatment (19). Thus, there is ample evidence consistent with cry1 action being nuclear-localized and manifested by changes in gene expression. Phosphorylation of cry1, either by an autocatalytic mechanism or a separate kinase, seems to be a required element of the response mechanism (20–22).
Other findings about cry1 action do not as easily fit a scenario in which a nuclear-localized photoreceptor fairly directly affects gene expression to affect photomorphogenesis. The presence of cry1 in the cytoplasm is one such observation (23). Another is that blue light activates anion channels at the plasma membrane, causing a depolarization after a lag time of only a few seconds in a cry1/cry2-dependent fashion (24, 25). This channel activation has been causally linked to the onset of cry1-dependent growth inhibition, which occurs 30–40 min after the onset of irradiation and involves changes in auxin and gibberellin levels and/or signaling (17). Determining the contribution of cytoplasmic or nuclear cry1 to these processes is the main theme of the present work, which is analogous to previous studies of the phytochrome B photoreceptor (26). The general experimental approach is to (i) engineer cry1 to contain a short sequence of amino acids that has a nuclear-localizing effect or a different short sequence that results in export from the nucleus, and (ii) determine the extent to which the differently localized photoreceptors restore function to a cry1 mutant in a number of different photomorphogenesis assays.
Results
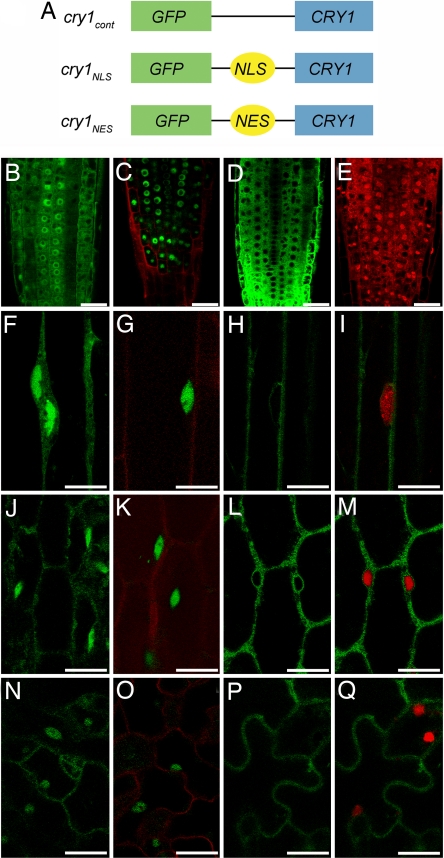
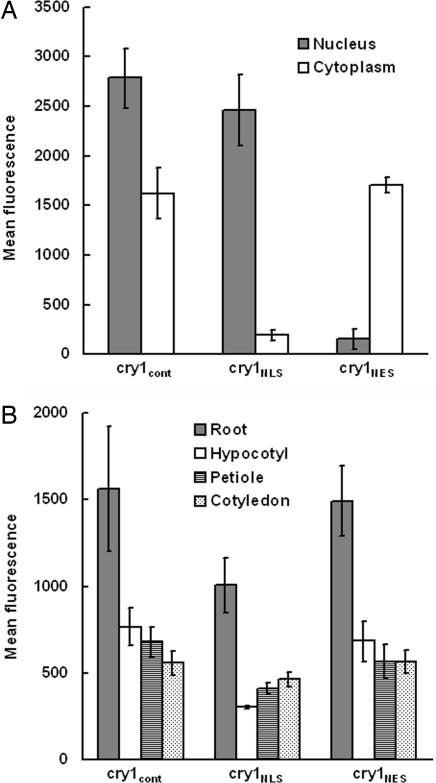
To visualize the nuclear versus cytoplasmic distribution of the cry1 photoreceptor, the DNA-coding sequence of GFP was attached to the N terminus of the CRY1 coding sequence (Fig. 1A). The N terminus was selected because previous work indicated that modifying the C terminus was likely to affect function (23). The decision to place the chimeric gene under the control of the 35S promoter was based on the need for a strong GFP signal to visualize the low-abundance cry1 protein (27) and the previous finding that cry1 expressed by this strong promoter functions well without severe pleiotropic effects in the seedlings (27). To manipulate the cytoplasmic distribution of the cry1 photoreceptor, a nuclear localization signal (NLS) or nuclear export signal (NES) used by Matsushita et al. (26) for a similar purpose was inserted between the GFP and the cry1-coding sequences (Fig. 1A). Not shown in Fig. 1A is the negative control construct in which Arg-611 of cry1 was changed to lysine. Previous studies showed that this mutation (hy4-24) blocks function without affecting protein levels (8). The cry1-304 allele, previously demonstrated to lack cry1 protein (20), was transformed with these various constructs. Fig. 1B shows that, in the root apex, GFP-cry1 (hereafter cry1cont for control) was present in both the cytoplasm and nucleus, as reported before for the native protein (28). Fig. 1C shows that the GFP-NLS-cry1 (hereafter cry1NLS) was concentrated in the nucleus, and levels in the cytoplasm were below the detection limit. Fig. 1D shows that GFP-NES-cry1 (hereafter cry1NES) was expressed well in the cytoplasm, but could not be detected in nuclei, which were visualized by staining the roots with propidium iodide (Fig. 1E). Essentially the same results were observed in cells of etiolated hypocotyls (Fig. 1 F–I), light-grown hypocotyls (Fig. 1 J–M), and light-grown cotyledons (Fig. 1 N–Q). These localization patterns were independent of blue-light treatment. They represent the steady-state levels of cry1 in the different compartments, but do not provide information about the rates of exchange between compartments. An analysis of GFP fluorescence intensity was performed to quantify the cytoplasmic and nuclear cry1 levels in the root tips of plants expressing the different constructs. The results, shown in Fig. 2A, are consistent with the confocal images, in that the NLS sequence tag had a profound nuclear-accumulation effect. The NES tag effectively excluded cry1 from the nucleus. Importantly, the level of cry1 in the nucleus of cry1NLS plants was similar to the levels of cry1 in the cytoplasm of cry1NES plants, and the cry1cont situation was similar to the sum of the cry1NES and cry1NLS levels. At the tissue level, rather than the subcellular level, cry1 expression levels in the three lines were similar in each of the organs studied (Fig. 2B). Homozygous lines that faithfully maintained these desired subcellular cry1 distributions in the T4 generation were used to compare the extent to which cry1NES and cry1NLS rescued the cry1–304 defects in blue-light responses.
Fig. 1.
Schematics of CRY1 transgene constructs and subcellular localization of the proteins. (A) Schematics of the GFP-tagged chimeric genes with which cry1 was transformed to test the function of nuclear and cytoplasmically localized cry1. (B–D) Root tips of 4-day-old light-grown seedlings expressing cry1cont (B), cry1NLS (C), or cry1NES (D). (E) Propidium iodide staining highlights the nuclei in red. (F–H) Dark-grown hypocotyls expressing cry1cont (F), cry1NLS (G), or cry1NES (H). (I) Cell in H stained with Vybrant DyeCycle Orange to show the nucleus in red. (J–L) Light-grown hypocotyls expressing cry1cont (J), cry1NLS (K), or cry1NES (L). (M) Cell in L stained with Vybrant DyeCycle Orange to show the nucleus in red. (N–P) Light-grown cotyledons expressing cry1cont (N), cry1NLS (O), or cry1NES (P). (Q) Cell in P stained with Vybrant DyeCycle Orange to show the nucleus in red. Propidium iodide staining in C, G, K, and O shows the cell structure in red. (Scale bars: B–Q, 20 μm.)
Fig. 2.
GFP-cry1 quantification in the transgenic seedlings. (A) Nuclear and cytoplasmic GFP-cry1 levels in root apices measured as fluorescence intensity by confocal microscopy. (B) Overall GFP-cry1 levels in the indicated organs in the three transgenic lines. The roots were imaged with a C-Apochromat ×40 objective lens so their fluorescence levels in B are not comparable to the other organs, which were imaged with a Plan-Aprochromat ×20 objective lens. Each measurement in A or B is the mean of measurements obtained from four to seven seedlings. Error bars represent standard errors.
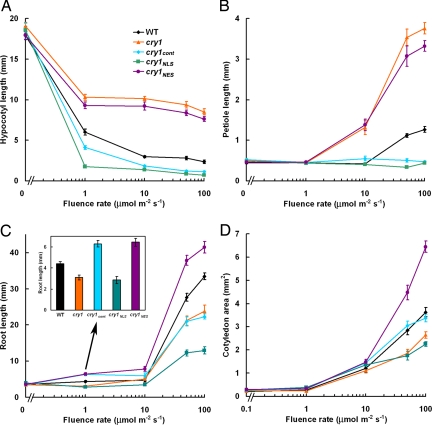
Among the obvious changes in growth and shape that an etiolated seedling undergoes in response to light are suppression of hypocotyl elongation, expansion of the cotyledons, elongation of the cotyledon petioles, and promotion of root elongation (1). The cry1 photoreceptor participates in all of these responses, so each was quantified in the different mutant and transgenic lines after 7 days of growth in a range of blue photon fluence rates to determine whether cry1NLS or cry1NES was as able to rescue the cry1–304 defects more or less well as the cry1cont control. Fig. 3A shows that blue light suppressed hypocotyl elongation in wild-type seedlings and that, at the higher fluence rates of 10, 50, and 100 μmol·m−2·s−1, cry1 seedlings were ≈4-fold taller than wild type. This iconic cry1 phenotype was completely suppressed by cry1cont, showing that the GFP-cry1 molecule was functional. The results obtained with the cry1NLS and cry1NES lines were unequivocal. Complete suppression of the phenotype was observed in cry1NLS seedlings; however, cry1NES was ineffective. Therefore, nuclear, but not cytoplasmic, cry1 can explain the long-term result of cry1 action on hypocotyl growth in blue light. Opposite to its effect on hypocotyl elongation, blue light promoted cotyledon petiole elongation in wild-type seedlings. The promotive effect of blue light was much greater in cry1 than wild type (Fig. 3B), indicating that cry1 negatively regulates this photomorphogenic response, which is presumably initiated by a different photoreceptor. Suppression of the cry1 long petioles beyond the wild-type levels was achieved by cry1cont and cry1NLS, but cry1NES produced no suppression (Fig. 2B).
Fig. 3.
Phenotypic analysis of CRY1 transgenic plants. Plants were grown in darkness or under continuous blue light with various intensities for 7 days. Then hypocotyl lengths (A), cotyledon petiole lengths (B), root lengths (C), and cotyledon areas (D) were measured. (C Inset) Important differences in root lengths at 1 μmol·m−2·s−1 blue light. The mean values from >15 plants are shown. Error bars represent standard errors.
Some of the root growth promotion by blue light is because of cry1 (29). Fig. 3C shows the root of cry1 is shorter than wild type. Shifting the distribution of cry1 to the cytoplasm stimulated root growth (Fig. 3C) at all fluence rates. Conversely, concentrating cry1 in the nucleus inhibited root growth (Fig. 3C). The presence of cry1 in both compartments (cry1cont) produced promotion at low fluence rates and inhibition at the higher fluence rates, relative to the wild type. The main conclusion to draw from Fig. 3C is that cry1NES promotes root growth in blue light (not in darkness) and that cry1NLS has the opposite effect.
Blue light promotes cotyledon expansion by cry1 signaling (3, 4). The effect is evidenced here by the reduced cotyledon area of cry1 seedlings grown in blue light, especially at higher fluence rates (Fig. 3D). Unlike the other processes presented in Fig. 3, cry1NES was most effective at promoting cotyledon expansion and correcting the cry1 defect. Concentrating cry1 in the nucleus and excluding it from the cytoplasm (cry1NLS) prevented the photoreceptor from rescuing the mutation. As in roots, cry1NES promoted cotyledon expansion relative to wild type, cry1NLS, or the control line (cry1cont), which takes into account the effect of the 35S promoter. The stimulatory effect of cytoplasmic cry1 on cotyledon expansion is probably because of a true function of the photoreceptor; disabling cry1 function through the R611K mutation (cry1*) abolished its ability to modify any of the cry1 phenotypes tested in any of the subcellular contexts [supporting information (SI) Fig. 8]. Thus, all of the effects in Fig. 3 are more likely related to cry1 signaling, rather than indirect effects such as titrative reduction of interacting proteins or nonspecific interferences caused by extraordinary levels or locales of expression.
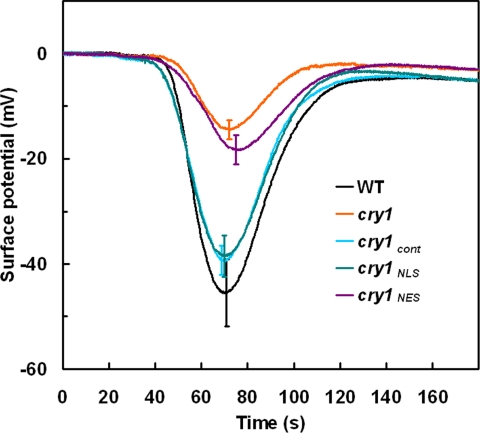
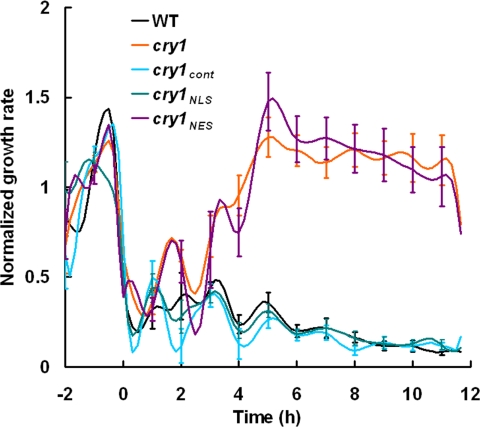
Previous studies using high-resolution tools designed for studying Arabidopsis seedling growth demonstrated that growth inhibition induced by blue light is initiated not by cry1, but by phototropin 1 (phot1), an unrelated photoreceptor (25). After 30–40 min of blue-light irradiation, a cry1-mediated phase of growth inhibition replaces the phot1 phase (25). The onset of the cry1 phase, but not the initial phot1 phase, has been mechanistically linked to an anion channel-mediated depolarization of the plasma membrane that is complete ≈2 min after the onset of blue-light irradiation (25). It seemed possible that some portions of these earliest effects of cry1 on cell physiology and hypocotyl growth may be more dependent on cytoplasmic cry1 than nuclear cry1. Therefore, electrophysiology and the latest computer-vision technique for measuring hypocotyl growth from time series of electronic images (30) were used to investigate the functions of cry1NLS and cry1NES. Fig. 4 shows that the membrane depolarization defect of cry1 was not rescued by cry1NES, but was completely rescued by cry1NLS and cry1cont. If cry1NLS mediates the depolarization and the depolarization is causal to the onset of cry1-dependent growth inhibition (25), then both should be rescued by the same form of cry1. If the ionic events are rescued by one form and the growth response by the other, the two will have been uncoupled. Hypocotyl growth rates were determined by using the morphometric algorithms developed by Miller et al. (30). The abrupt escape from inhibition after ≈30–40 min of irradiation typical of cry1 was not corrected by cry1NES (Fig. 5). However, cry1cont and cry1NLS completely restored wild-type growth inhibition to cry1. Thus, the electrophysiological results and high-resolution growth measurements (Figs. 4 and 5) are consistent with the model that rapid ionic responses to blue light and the subsequent cry1-mediated phase of growth inhibition are causally connected and both mediated by nuclear cry1. Variations of this conclusion are considered in Discussion.
Fig. 4.
Blue light-induced membrane depolarizations in wild type, cry1, and transgenic plants. Membrane depolarization induced by a 20-s pulse of 200 μmol·m−2·s−1 blue light in hypocotyls started at 20 s and ended at 40 s. The mean values from more than eight responses were plotted. Error bars at the peak of each line were shown.
Fig. 5.
Morphometric analysis of seedling hypocotyl growth in blue light. The growth kinetics of 2-day-old etiolated wild type, cry1, cry1cont, cry1NLS, and cry1NES respond to 50 μmol·m−2·s−1 blue-light treatment. All growth rates were normalized to the average dark growth rate during the 2 h preceding blue-light treatment. Data represent the average of more than eight plants per genotype. Standard error was given every hour.
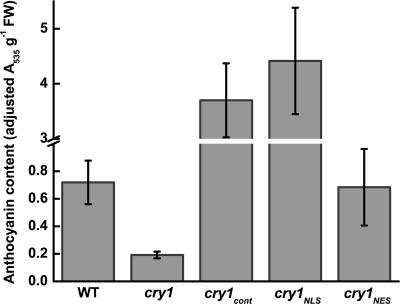
Anthocyanin pigments accumulate in Arabidopsis seedlings in response to light primarily through a cry1-dependent blue-light pathway. Genes in the biosynthetic pathway are up-regulated in a cry1-dependent manner. However, Noh and Spalding (31) found that a posttranslational step involving cry1-dependent anion channel activation was required for proper anthocyanin accumulation. To test the possibility that this complexity was the result of combinations of nuclear and cytoplasmic cry1 activities, anthocyanin accumulation in the transgenic lines grown in blue light was measured. As expected, cry1 seedlings were unable to accumulate anthocyanin (Fig. 6). Wild-type anthocyanin levels were restored by cry1NES, but cry1NLS and cry1cont were much more potent, promoting anthocyanin accumulation at least 5-fold higher than the wild-type level.
Fig. 6.
Anthocyanin levels in 7-day-old seedlings grown under 100 μmol·m−2·s−1 blue light.
Discussion
The majority of available evidence from animal and plant studies before this study pointed to the nucleus as the primary site of cryptochrome action. However, evidence of a nuclear function for these blue-light receptors typically does not exclude a cytoplasmic function. For example, in animals, cryptochromes heterodimerize in the cytoplasm with PERIOD (PER) proteins (encoded by the period genes) before the complex translocates to the nucleus, where it represses key clock genes (32, 33). Although the nuclear gene-regulatory role is clearly important, the cryptochromes also may perform a yet-to-be discovered function while in the cytoplasm. Likewise, the evidence that cry1 of Arabidopsis interacts with the COP1 E3 ligase to control levels of the HY5 transcription factor points to primary cry1 action being nuclear localized, but COP1 moves to the cytoplasm in response to light where, in combination with cytoplasmic cry1, it could conceivably perform a photomorphogenic function (13–15). Indeed, a C-terminal portion of cry1 fused to β-glucuronidase (GUS) was cytoplasmic in the light and nuclear in the dark (23). The presence of an effective NLS in the amino acid sequence of cry2 in Arabidopsis (28, 34) and the absence of one in cry1 may indicate that cry1 in the cytoplasm is there for a functional reason. The two cry1-like cryptochromes in rice when fused to GFP and overexpressed were present in both the nucleus and cytoplasm (35). The five different cryptochromes in the fern Adiantum capillus-veneris when fused to GUS and expressed in gametophytic cells displayed a range of nuclear:cytoplasm distributions, some of which of were light-dependent (36). Thus, cytoplasmic cryptochrome is not an anomaly, and studies aimed at determining its relevance to seedling photomorphogenesis could produce a better understanding of how this photoreceptor operates across the biological spectrum. The results presented here demonstrate that cytoplasmic cry1 functions during seedling photomorphogenesis in ways that can be separated from the functions of nuclear cry1.
The long hypocotyl displayed by cry1 mutants grown in blue light for prolonged periods was the phenotype that led to the discovery of cryptochromes. The cry1-dependent phase of growth inhibition depends on the activation of anion channels at the plasma membrane, which depolarizes the membrane (37). Blocking the anion channels pharmacologically in wild-type seedlings phenocopies the cry1 mutant for a few hours, but not for the long term (37). This finding raised the possibility of their being at least two cry1-dependent phases–the first one dependent on anion channels and the second not. Although cytoplasmic cry1 would seem to be better positioned to activate anion channels at the plasma membrane than nuclear cry1, the data clearly indicated otherwise. Cytoplasmic cry1 suppressed neither the membrane depolarization defect nor the defect in hypocotyl growth control. Complementary to this result is the finding that cry1NLS restored both the membrane depolarization and the initial cry1 phase of growth inhibition. It seems somewhat counterintuitive, but nuclear localized cry1 is responsible for the activation of plasma membrane anion channels and the attendant phase of cry1-dependent growth inhibition.
One direct model that could accommodate these results is that nuclear cry1 generates a signal that reaches the anion channels at the membrane within the several seconds lag time. The timing is too fast to be mediated by gene expression, but light-driven redox changes (38) or ion fluxes could conceivably connect nuclear and plasma membrane activities. A much less direct model also can be envisioned. Perhaps cry1 mutants grown in the dark underexpress the anion channels or some component of the signaling chain so that blue light results in a weaker depolarization. This indirect model depends on cry1 influencing the expression of genes in dark-grown seedlings [i.e., in the absence of the signal (blue light) believed to be necessary for cry1 activation].
The present study found no evidence of a role for cytoplasmic cry1 in the control of hypocotyl or cotyledon petiole elongation, but cotyledon expansion, root growth, and anthocyanin accumulation all displayed a dependence on cry1NES. In some cases, the cry1 phenotypes could be completely explained by a particular form of cry1, but other cases were more complicated. In the case of cotyledon expansion, cytoplasmic cry1 appeared to oppose the action of nuclear cry1. Cotyledon expansion in cry1 was faithfully rescued by cry1cont, not by cry1NLS, and was stimulated beyond the wild-type level by cry1NES. The anthocyanin assays also provided evidence of different forms contributing in different ways. Anthocyanin accumulation was rescued by cry1NES, yet hyperstimulated by cry1NLS, a complexity that is perhaps related to the previous finding that transcriptional control and posttranslational steps regulated the biosynthetic pathway in response to blue light (31).
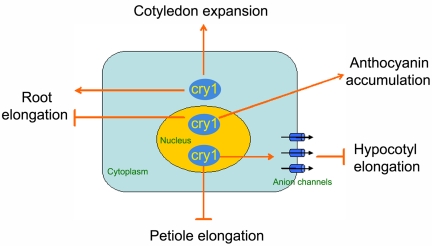
The present work (Fig. 3C) indicates that the promotive effect of cry1 on root growth noted by Canamero et al. (29) was due to cytoplasmic cry1. At high fluence rates, blue light inhibits root elongation, which is caused by nuclear cry1 (Fig. 3C). Redistribution of cry1 between the nucleus and cytoplasm was not observed in response to changes in light conditions (data not shown). A better explanation is that cytoplasmic cry1 dominates the signaling pathway to promote root growth in low fluence-rate blue light, whereas the inhibitory effect of nuclear cry1-mediated signal transduction predominates at high fluence rates. These and the other results presented here can be summarized in a diagrammatic model (Fig. 7) that integrates cry1 subcellular localization and the effect the photoreceptor has on seedling photomorphogenesis.
Fig. 7.
Model integrating cry1 localization with cry1-mediated signal transduction induced by blue light.
Methods
Engineering of the CRY1 Gene and Generation of Transgenic Plants.
To generate the chimeric fusion construct GFP-CRY1, the complete CRY1 coding sequence was amplified by PCR from an Arabidopsis cDNA clone (U12079) by using a forward primer 5′-GGCTCGAGTCTGGTTCTGTATCTGGTTGTGGTTC-3′ and a reverse primer 5′-GGCTCGAGTTACCCGGTTTGTGAAAGCCGTCTCC-3′. The italicized regions denote XhoI sites that were introduced to both primers to facilitate the cloning of amplified DNA. To introduce NLS or NES signal peptide DNA into the construct, the forward primers 5′-GGCTCGAGcctaagaagaagagaaaggttTCTGGTTCTGTATCTGGTTGTGGTTC-3′ or 5′-GGCTCGAGcttgctcttaagttggctggacttgatattTCTGGTTCTGTATCTGGTTGTGGTTC-3′, respectively, were used. The signal sequences are shown in lowercase. After digestion with XhoI, the fragment was fused in frame to the C-terminal end of eGFP in pEGAD vector.
A point mutation in the CRY1-coding region that substitutes Arg-611 with a lysine residue (R611K) in the protein sequence was previously shown to create a protein that accumulates normally, but has no detectable function (8). This point mutation was introduced into the cDNA clone (U12079) by a QuikChange II site-directed mutagenesis kit (Stratagene) by using the primers 5′-CCAGAATTTAATATCAAAATTGTTGCAGAGAGC-3′ and 5′-GCTCTCTGCAACAATTTTGATATTAAATTCTGG-3′. The resulting plasmid was used as the template to clone the mutated CRY1 cDNA with NLS or NES tag into pEGAD as described before. The mutated, nonfunctional cry1 produced is designated cry1*.
The resulting constructs were introduced into Agrobacterium tumefaciens GV3101 and used to transform cry1–304 mutant plants by using the floral dip method (39). Plants carrying the transgene were isolated through basta selection. The homozygous lines for the transgene were obtained in the T4 generation.
Plant Materials and Growth Analysis.
A. thaliana ecotype Columbia was used as a wild type in all experiments. Seeds were surface-sterilized with 75% ethanol and plated on half-strength Murashige and Skoog agar plates [2.15 g/liter MS salts, 0.8% bacto-agar (pH 5.7)] (Sigma–Aldrich). The plates were maintained in darkness at 4°C for 2–3 days and then placed under various intensities of constant blue light at 22°C for 7 days. To quantify growth rates, seedlings were rearranged on agar plates, and images were obtained with a flatbed scanner. Hypocotyls, petioles primary roots, and the cotyledon area were determined by using National Institutes of Health IMAGE software.
Subcellular Localization Studies.
Confocal microscopy was performed with a Zeiss LSM 510 laser scanning confocal microscope equipped with a meta-detector. Propidium iodide was used to stain the nucleus and the cell wall to delineate the root structure. Vybrant DyeCycle Orange stain (Invitrogen) was used to stain the nucleus in the hypocotyl and cotyledon. Either a C-Apochromat ×40 water immersion lens or a Plan-Apochromat ×63 oil immersion lens was used. The sample was excited with the 488-nm laser line from a 30-mW argon gas laser. The fluorescence was captured in 10-nm bandwidths, and then linear unmixing was performed to isolate the GFP signal from the stained or endogenous fluorescence background.
Mean fluorescence intensities were calculated within manually selected regions of interest with Zeiss LSM 510 software. In the primary root nucleus, a circular area of ≈50 μm2 covering the whole nucleus was selected. One representative nucleus per root was recorded, and the values reported are the means of at least four separate roots. For the cytoplasmic values, a field of similar size adjacent to the nucleus was chosen. To measure overall GFP intensity in the different organs, regions of ≈0.01–0.02 mm2 for the root, 0.1–0.15 mm2 for the hypocotyl, 0.05–0.1 mm2 for the petiole, and 0.15–0.2 mm2 for the cotyledon were used. Seedlings were 4 days old.
Morphometric Analysis of Hypocotyl Growth.
Two-day-old seedlings grown in the dark on 1% agar containing 1 mM KCl and 1 mM CaCl2 were used to measure hypocotyl growth with a recently developed computer-vision method (30). Briefly, electronic images acquired at 10-min intervals by CCD cameras using infrared light were analyzed with custom morphometric algorithms designed to extract information such as growth rate (30). Hypocotyl inhibition was induced by 50 μmol·m−2·s−1 blue light.
Surface Potential Measurements.
Four-day-old etiolated seedlings grown on 1% agar containing 1 mM KCl and 1 mM CaCl2 were used to measure surface potential induced by a pulse of 200 μmol·m−2·s−1 blue light as described previously (24), except that the xenon arc light source and delivery mechanism were those used by Folta et al. (40).
Anthocyanin Extraction and Quantification.
Anthocyanin measurement was conducted according to a previously published method (31). Seven-day-old seedlings grown under 100 μmol·m−2·s−1 blue light were harvested from the agar plates, quickly weighed, and placed into microcentrifuge tubes containing 350 μl of 18% 1-propanol, 1% HCl, and 81% water. The tubes were placed in boiling water for 3 min and then incubated in darkness overnight at room temperature. After a brief centrifugation to pellet the tissue, 300 μl of the solution was removed and brought to a final volume of 600 μl by adding solvent. The amount of anthocyanins in the resulting extract was quantified spectrophotometrically. The values are reported as (A535–A650) per gram of fresh weight.
Supplementary Material
Acknowledgments
We thank Tessa L. Durham for assistance with membrane potential measurements, Daniel R. Lewis for assistance with confocal microscopy, and Nathan D. Miller and Candace R. Moore for assistance with the computerized growth measurements. This work was supported by National Science Foundation Grants IOB-0517350 and DBI-0421266 (to E.P.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705082104/DC1.
References
- 1.Lin C, Shalitin D. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad M, Cashmore AR. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 3.Jackson JA, Jenkins GI. Planta. 1995;197:233–239. doi: 10.1007/BF00202642. [DOI] [PubMed] [Google Scholar]
- 4.Neff MM, Chory J. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagnall DJ, King RW, Hangarter RP. Planta. 1996;200:278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Yang H, Mockler TC, Lin C. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 7.Somers DE, Devlin PF, Kay SA. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad M, Lin C, Cashmore AR. Plant J. 1995;8:653–658. doi: 10.1046/j.1365-313x.1995.08050653.x. [DOI] [PubMed] [Google Scholar]
- 9.Danon A, Sanchez Coll N, Appel K. Proc Natl Acad Sci USA. 2006;103:17036–17041. doi: 10.1073/pnas.0608139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancar A. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- 11.Lin C. Plant Cell. 2002;14:S207–S225. doi: 10.1105/tpc.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cashmore AR, Jarillo JA, Wu YJ, Liu D. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 13.Yang HQ, Tang RH, Cashmore AR. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 15.von Arnim AG, Deng XW. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 16.Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 17.Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP. Plant J. 2003;36:203–214. doi: 10.1046/j.1365-313x.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Plant Cell. 2001;13:2589–2607. doi: 10.1105/tpc.010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phee BK, Park S, Cho JH, Jeon JS, Bhoo SH, Hahn TR. Mol Cell. 2007;23:154–160. [PubMed] [Google Scholar]
- 20.Shalitin D, Yu X, Maymon M, Mockler T, Lin C. Plant Cell. 2003;15:2421–2429. doi: 10.1105/tpc.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozgur S, Sancar A. Biochemistry. 2006;45:13369–13374. doi: 10.1021/bi061556n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouly JP, Giovani B, Djamei A, Mueller M, Zeugner A, Dudkin EA, Batschauer A, Ahmad M. Eur J Biochem. 2003;270:2921–2928. doi: 10.1046/j.1432-1033.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- 23.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 24.Cho MH, Spalding EP. Proc Natl Acad Sci USA. 1996;93:8134–8138. doi: 10.1073/pnas.93.15.8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folta KM, Spalding EP. Plant J. 2001;26:471–478. doi: 10.1046/j.1365-313x.2001.01038.x. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita T, Mochizuki N, Nagatani A. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 27.Lin C, Ahmad M, Cashmore AR. Plant J. 1996;10:893–902. doi: 10.1046/j.1365-313x.1996.10050893.x. [DOI] [PubMed] [Google Scholar]
- 28.Guo H, Duong H, Ma N, Lin C. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- 29.Canamero R, Bakrim N, Bouly JP, Garay A, Dudkin E, Habricot Y, Ahmad M. Planta. 2006;224:995–1003. doi: 10.1007/s00425-006-0280-6. [DOI] [PubMed] [Google Scholar]
- 30.Miller ND, Parks BM, Spalding EP. Plant J. 2007;52:374–381. doi: 10.1111/j.1365-313X.2007.03237.x. [DOI] [PubMed] [Google Scholar]
- 31.Noh B, Spalding EP. Plant Physiol. 1998;116:503–509. doi: 10.1104/pp.116.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowrey PL, Takahashi JS. Annu Rev Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- 33.Panda S, Hogenesch JB, Kay SA. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 34.Kleiner O, Kircher S, Harter K, Batschauer A. Plant J. 1999;19:289–296. doi: 10.1046/j.1365-313x.1999.00535.x. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto N, Hirano T, Iwasaki T, Yamamoto N. Plant Physiol. 2003;133:1494–1503. doi: 10.1104/pp.103.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imaizumi T, Kanegae T, Wada M. Plant Cell. 2000;12:81–96. doi: 10.1105/tpc.12.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks BM, Cho MH, Spalding EP. Plant Physiol. 1998;118:609–615. doi: 10.1104/pp.118.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 39.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 40.Folta KM, Lieg EJ, Durham T, Spalding EP. Plant Physiol. 2003;133:1464–1470. doi: 10.1104/pp.103.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.