Abstract
The cyanobacterium Synechococcus elongatus expresses robust circadian (daily) rhythms under the control of the KaiABC-based core clockwork. Unlike eukaryotic circadian systems characterized thus far, the cyanobacterial clockwork modulates gene expression patterns globally and specific clock gene promoters are not necessary in mediating the circadian feedback loop. The oscilloid model postulates that global rhythms of transcription are based on rhythmic changes in the status of the cyanobacterial chromosome that are ultimately controlled by the KaiABC oscillator. By using a nonessential, cryptic plasmid (pANS) as a reporter of the superhelical state of DNA in cyanobacteria, we show that the supercoiling status of this plasmid changes in a circadian manner in vivo. The rhythm of topological change in the plasmid is conditional; this change is rhythmic in constant light and in light/dark cycles, but not in constant darkness. In further support of the oscilloid model, cyanobacterial promoters that are removed from their native chromosomal locations and placed on a plasmid preserve their circadian expression patterns.
Keywords: DNA topology, gene expression, supercoiling, biological clock
Eukaryotes and prokaryotic cyanobacteria display daily rhythms in gene expression, biochemistry, physiology, and/or behavior that are controlled by circadian biological clocks (1). The circadian regulation of these processes is thought to aid in the adaptation of organisms to daily changes in light, temperature, and other factors in their environment. Circadian regulation of gene expression in different organisms is now known to occur at various cellular levels including control of promoter activity, mRNA stability, translation, and protein degradation.
A promoter trap analysis revealed that a circadian system regulates global transcriptional activity in the prokaryotic cyanobacterium Synechococcus elongatus PCC7942 (2). In S. elongatus, the activities of essentially all promoters are rhythmically orchestrated, whereas in eukaryotes, microarray analyses suggest that only ≈5–15% of genes display circadian rhythms of mRNA abundance (3–11). Although microarrays are informative, this technique is not sensitive to small changes in mRNA abundance and is not a quantitative measure of rhythmic transcriptional activity for mRNAs that are either very unstable or very stable. Because of these limitations of microarrays, the degree to which circadian clocks control promoter activity is likely to have been underestimated in eukaryotes (12, 13). Because the promoter trap technique measures promoter activity, its application in S. elongatus may explain why pervasive clock control over gene expression has been found in cyanobacteria, but not in eukaryotic circadian systems.
In S. elongatus, transcriptional activity is regulated by a molecular clockwork encoded by three genes, kaiA, kaiB, and kaiC; inactivation of any of the kai genes abolishes clock function (14). The proteins encoded by these genes interact with one another (15–17) to form large protein complexes in vivo with KaiC as the core (18, 19). KaiC is found in both phosphorylated and nonphosphorylated forms in vivo, and its phosphorylation status is correlated with clock speed (20–23). The rhythm of KaiC phosphorylation remains intact for several cycles even when cyanobacterial cells are maintained in constant darkness (DD), which is a condition in which transcription and translation are absent or greatly reduced (24). Furthermore, a rhythm of KaiC phosphorylation can be reconstituted in vitro by using only purified KaiA, KaiB, and KaiC proteins and ATP (25). Taken together, these observations indicate that the core circadian oscillator of cyanobacteria does not require a transcription–translation feedback loop.
Several observations suggest a global mechanism for clock control of promoter activities in cyanobacteria. Continuous overexpression of KaiC represses rhythmic transcription of all cyanobacterial promoters (26), including of its own kaiBC promoter (14). Furthermore, the replacement of the kaiA and/or kaiBC promoters with a heterologous promoter, the inducible Escherichia coli trcp promoter, permits rhythmic transcription of the kaiABC cluster, global gene expression, and repression by KaiC (23, 26). Therefore, none of those three key properties depend on specific cyanobacterial promoters mediating transcription of the kai genes. The “oscilloid model” has been proposed to explain the all-encompassing circadian regulation of gene expression observed in cyanobacteria (27). This model posits that KaiC mediates both its own negative-feedback regulation and global regulation of transcription throughout the genome by orchestrating circadian oscillations in the structure of cyanobacterial chromosomes (21, 27). Bacterial chromosomes are compacted and coiled into highly organized structures called “nucleoids” (28); changes in the local supercoiling status of DNA within nucleoids can affect the rate of transcription (29–32). Inhibitors of DNA gyrase cause a dramatic alteration of chromosomal supercoiling status that triggers genome-wide changes in transcription in E. coli (33). The oscilloid model proposes that rhythmic changes of chromosome topology promote the cyclic modulation of the transcriptional rates of all genes in cyanobacteria, accounting at least in part for global regulation of gene expression. Therefore, cis-elements that mediate rhythmic gene expression are globally modulated by chromosomal status in addition to being targeted on a promoter-by-promoter basis by specific trans-acting factors. Moreover, heterologous promoters from other species of bacteria that are integrated into the cyanobacterial chromosome are rhythmically active because they are also subject to the rhythmic chromosomal status (23, 26).
As a test of the oscilloid model, we assayed for changes in the topology of a nonessential plasmid in S. elongatus and found that plasmid topology changes in a circadian manner. We show that there is a conditional coupling between the circadian clock as measured by the rhythm of KaiC phosphorylation and the rhythm of plasmid topology change. Furthermore, cyanobacterial promoters removed from their native chromosomal surroundings and introduced into plasmids show similar circadian promoter activities that are KaiC dependent and are repressed by KaiC overexpression. We suggest that KaiC-containing protein complexes either directly (or indirectly by regulating the activities of nucleoid-associated proteins) modulate chromosome structure leading to the global regulation of promoter activities in cyanobacteria.
Results
A Cyanobacterial Plasmid Displays Circadian Rhythms of Topological Change.
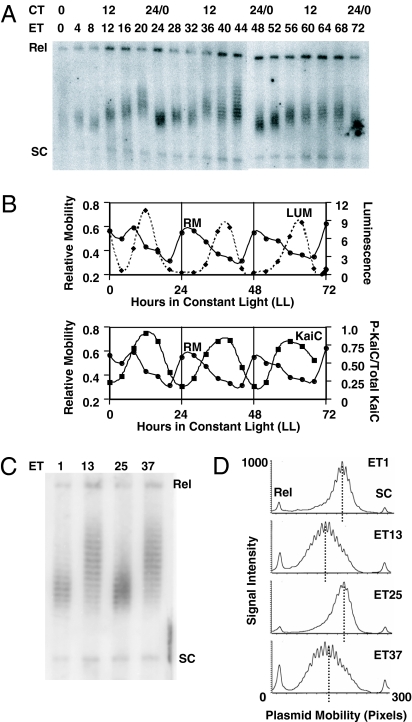
To test the oscilloid model in S. elongatus, we used chloroquine agarose gel electrophoresis (CAGE) to assess topological changes in the nonessential native plasmid, pANS (34). CAGE separates plasmid topoisomers based on their linking number (35, 36). During electrophoresis in the presence of the intercalating agent, chloroquine, relaxed open circular plasmid DNA molecules migrate more slowly than do supercoiled DNA molecules allowing plasmid topoisomers that differ in their linking number to be separated. Relaxed, open circular (Rel) and supercoiled (SC) forms of the plasmid were identified by varying the concentration of chloroquine during electrophoresis and observing the changes in migration patterns of these species (data not shown). Fig. 1A shows the time-dependent change in the plasmid topoisomer population from cyanobacterial cells maintained in constant light (LL). The population of plasmid molecules isolated from these cells displayed a change in the linking number of the plasmid, with the topoisomer population having a higher overall linking number (higher mobility) during the subjective day [circadian time 0 (CT0) to CT12] and a lower overall linking number (lesser mobility) during the subjective night (CT12–CT24). When plotted as a function of CT, the relative mobility of the topoisomer population revealed a robust rhythm of topological change with a period of ≈24 h that was in antiphase to the activity rhythm of the psbAIp promoter (as assessed by luminescence recorded from the chromosomal psbAIp:luxAB reporter) assayed from the same cell population (Fig. 1B Upper). During the subjective day, the relative mobility of the plasmid was generally high (when the activity of psbAIp is increasing), whereas during the subjective night, plasmid mobility decreased. This rhythm of topological change in the plasmid was approximately in antiphase to the rhythm of KaiC phosphorylation (Fig. 1B Lower). To quantify this rhythm of topological change, densitometry of CAGE patterns of plasmid DNA from subjective day [experimental time 1 (ET1) and ET25] and night (ET13 and ET37) phases showed that the population of topoisomers isolated during the day phase displayed not only a higher linking number (greater mobility), but also a smaller range in the linking number of topoisomers than does the population isolated from the night phase (Fig. 1 C and D). This observation suggests that DNA topology in cyanobacteria is controlled by the circadian clock, complementing a recent report that compaction of the cyanobacterial chromosome is under circadian control (37). A clock-disrupted strain in which the kaiC gene was deleted (ΔkaiC) displayed no rhythm of topological change in the plasmid population (data not shown). Moreover, cells treated with novobiocin, an inhibitor of DNA gyrase activity, showed an increase in the linking number of the population of plasmid topoisomers [supporting information (SI) Fig. 5A] and the amplitude of the chromosomal psbAI promoter activity rhythm was reduced by treatment with novobiocin (SI Fig. 5B).
Fig. 1.
The topological state of an endogenous plasmid exhibits a circadian rhythm. (A) Separation of plasmid topoisomers by CAGE. Plasmid DNA was isolated every 4 h from cyanobacterial cells maintained in LL, separated by CAGE, and detected as described. (B) Comparison of plasmid mobility with chromosomal psbAI promoter activity and KaiC phosphorylation status. Densitometric analyses (as in D) of data in A allowed the determination of the positions of the relaxed (Rel) and supercoiled (SC) forms as well as the median position in the plasmid topoisomer population. Relative mobility (RM) was calculated as follows: RM = (Median − Rel)/(SC − Rel). In the upper graph, RM (solid curve) is plotted versus time in LL; for comparison, the luminescence from the chromosomal psbAIp:luxAB reporter (dashed curve) from the same culture of cells is also plotted. In the lower graph, RM (solid curve) is replotted along with the phosphorylation status of KaiC protein (dashed curve) isolated from the same culture of cells. (C) Separation of plasmid topoisomers from peak and trough phases of the circadian cycle. Plasmid DNA was isolated at peak (ET1, ET25, corresponding to CT1) and trough (ET13, ET37, corresponding to CT13) phases from cells maintained in LL, and the plasmid topoisomers were separated by CAGE. (D) Densitometric analysis of CAGE data in C. The median position of the plasmid topoisomer population is indicated with a dashed line. In all images, “Rel” indicates relaxed, open circular plasmid form, and “SC” indicates fully supercoiled plasmid DNA. Subjective day phases are ET0–ET12, ET24–ET36, and ET48–ET60, and subjective night phases are ET12–ET24, ET36–ET48, and ET60–ET72.
Plasmid Topology also Changes Rhythmically in Light/Dark Cycles.
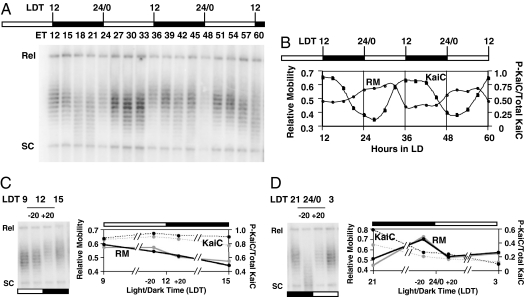
When cyanobacterial cells were grown in a 12-h light/12-h dark cycle (LD 12:12), the topology of the plasmid cycled rhythmically with approximately the same period and phase as in LL. The amplitude of the rhythm of topological change in the plasmid population was not as robust in LD as in LL (Fig. 2 A and B), possibly because of a dark-related process that attenuates the rhythm of topological change in LD. During the transition from light to darkness at LDT12, we observed a small, but measurable, decrease in the overall linking number (lower mobility) of the topoisomer population (Fig. 2C and SI Fig. 6). In contrast, during the transition from darkness to light (LDT24/0) there was a much larger and more rapid decrease in the relative mobility (from ≈0.7 to 0.5) of the plasmid topoisomers, indicating a rapid change in the linking number of the plasmid (Fig. 2D and SI Fig. 6). As in LL, the rhythm of plasmid topology is roughly in antiphase to the rhythm of KaiC phosphorylation, with the increase of phosphorylated KaiC correlated to a decrease in plasmid mobility late in the day (Fig. 2B and SI Fig. 6). The prompt, transient change in plasmid topology at the dark-to-light transition (LDT24/0) suggests either (i) that the circadian clock may control a mechanism that is capable of quickly changing the topological state of DNA in cyanobacteria or (ii) that environmental changes such as light intensity may directly modulate topological status in a way that could potentially provide entraining signals to the central clock.
Fig. 2.
Plasmid topology changes rhythmically in LD cycles. (A) Separation of plasmid topoisomers by CAGE. Plasmid DNA was isolated every 3 h starting at the onset of darkness from cells maintained in a LD 12:12 cycle; day and night are indicated by the bars above (white and black bars, respectively). (B) Comparison of relative plasmid mobility with KaiC phosphorylation status. RM (solid curve, determined as in Fig. 1) and the phosphorylation status of KaiC (dashed line) are plotted versus time in LD 12:12. (C and D) Changes in plasmid mobility and KaiC phosphorylation status during the light-to-dark and dark-to-light transitions. Plasmid DNA and total protein were isolated 20 min before and after the transition from light to dark (C) and from dark to light (D). Plasmid topoisomers were separated by CAGE; a comparison of RM (solid lines) to the KaiC phosphorylation status (dashed lines) is shown on an expanded timescale. RM and P-KaiC/Total KaiC are plotted in black for the first transition periods (ET12 in C and ET24 in D) and in gray for the second transition periods (ET36 in C and ET48 in D). LDT indicates light–dark time; intervals of light (white) and darkness (black) are indicated by the bars above the images; other labels are as in Fig. 1. See SI Fig. 6 for the densitometric analysis of data shown in C and D.
The Plasmid Topology Rhythm Is Conditionally Coupled to the Central Kai Clockwork.
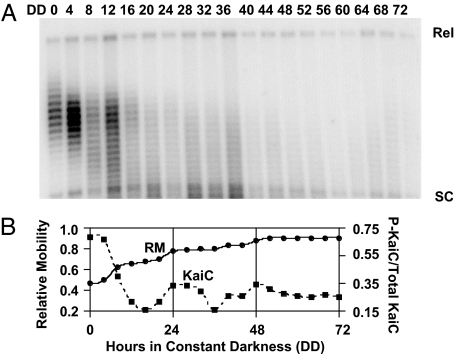
Although S. elongatus is an obligate photoautotroph, when cells are maintained in DD, they remain viable and the KaiC phosphorylation rhythm continues for several cycles; however, transcription and translation is absent in S. elongatus in DD (24). If the rhythm of KaiC phosphorylation directly drives rhythmic changes in plasmid topology, one would expect that the topology rhythm would be maintained in DD. Contrary to this prediction, we found that the relative mobility of the topoisomer population does not oscillate in DD (Fig. 3A). In fact, the relative mobility of the plasmid population progressively increased for ≈36 h after cells were transferred to DD even though there was still a detectable rhythm in the phosphorylation status of KaiC (Fig. 3B; ref. 24). This observation suggests that there is a “conditional linkage” between the central KaiABC clockwork and the output mechanism that mediates DNA topology. In DD, this linkage is uncoupled such that plasmid DNA molecules, and possibly the chromosome itself, become progressively more supercoiled in DD.
Fig. 3.
The plasmid topology rhythm is uncoupled from the central clock in DD. (A) Plasmid DNA and total protein were isolated every 4 h for 72 h from cyanobacterial cells maintained in DD, and topoisomers were separated by CAGE. Cells were grown in LD 12:12 before the transfer to DD. (B) Comparison of the relative mobility of the plasmid topoisomers to KaiC phosphorylation status. RM (solid curve), determined as in Fig. 1, and the KaiC phosphorylation status (dashed curve) are plotted versus time in DD.
Cyanobacterial Promoters Display Circadian Characteristics When Incorporated into Plasmids.
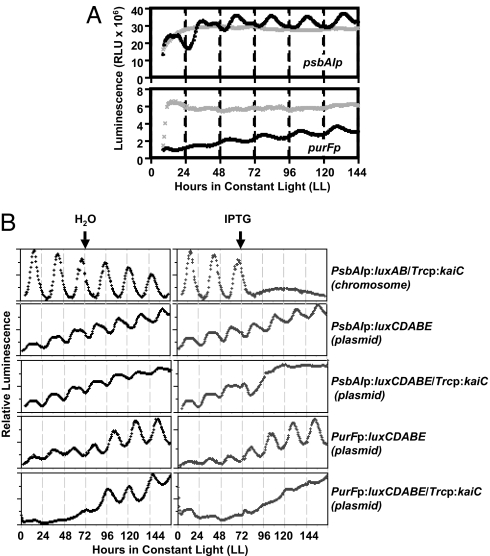
Because both plasmid topology and chromosome compaction (37) appear to be modulated by the circadian clock in cyanobacteria, we assessed whether promoters driving luxCDABE expression are regulated similarly on a plasmid as when they are located in the chromosome. Two promoters that have different circadian expression profiles were chosen. The psbAI promoter is a class I promoter whose peak activity is observed near subjective dusk, whereas the purF promoter is a class II promoter whose activity peaks near subjective dawn. The luminescence produced by the plasmid reporter constructs was recorded in both wild-type cyanobacterial cells and in ΔkaiC cells (Fig. 4A). Both the psbAI (Fig. 4A Upper) and purF (Fig. 4A Lower) promoters drove rhythmic expression of luciferase in wild-type cells with the same phasing and period as when these promoters are located in the chromosome (38), indicating that these promoters remain under clock control even in absence of a chromosomal context. In contrast to wild-type cells, there was no detectable rhythm of expression in ΔkaiC cells from these promoters (Fig. 4A). The activity of the psbAI and purF promoters was not abolished in the absence of a functional circadian clock; only the rhythmic component of the promoter activity was affected. Together with the results shown in Fig. 1, this observation implies that clock-modulated changes in plasmid topology could drive rhythmic transcriptional activity.
Fig. 4.
Cyanobacterial promoters display circadian properties on plasmids. (A) (Upper) Luminescence versus time in LL is plotted for the psbAI promoter (class I) driving expression of luxCDABE cloned into the plasmid pUHEX in wild-type cells (black) and in a ΔkaiC strain (gray). (Lower) Luminescence versus time in LL is plotted for the purF promoter (Class II) driving luxCDABE in pUHEX in wild-type cells (black) and in a ΔkaiC strain (gray). (B) Overexpression of KaiC represses rhythmic promoter activity on plasmids. In a KaiC overexpression strain, luminescence from a chromosomal psbAI promoter driving luxAB expression is compared with the psbAI promoter or purF promoter driving luxCDABE cloned into pUHEX. For each graph, luminescence is plotted versus time in LL in the absence of KaiC overexpression (left graphs; treatment with water) or with KaiC overexpression [right graphs; treatment with the inducer isopropyl β-d-thiogalactoside (IPTG) was initiated at the time shown by the arrow].
As previously reported (26), the rhythmic component of chromosomal psbAIp activity, but not its total activity, is repressed when KaiC is overexpressed (Fig. 4B, row 1). We observed that rhythmic expression of psbAIp when located on the plasmid has the same phase and period as when it is located on the chromosome, but it displayed higher basal activity with a lower peak-to-trough amplitude when on the plasmid (Fig. 4B, compare rows 1 and 2). The higher basal activity of promoters on the plasmid may reflect a plasmid copy number greater than one per cell; however, even the chromosome number is often two to eight per cell in S. elongatus (39). Furthermore, the rhythmic component, but not the basal activity of psbAIp on the plasmid was repressed by overexpression of KaiC (Fig. 4B, row 3). The oppositely phased purF promoter (38) also drove rhythmic expression of luxCDABE with the appropriate phase and period from a plasmid with a high basal level of promoter activity (Fig. 4B, row 4). As with psbAIp, the rhythmic component, but not the basal activity, of purFp was repressed by KaiC overexpression (Fig. 4B, row 5). This observation of higher basal activity of promoters on plasmids suggests that promoters on the chromosome are subjected to an additional layer(s) of repression. For example, the observed pattern of promoter expression from our plasmid reporters (i.e., higher basal activity and the activity increase by KaiC overexpression) is strikingly like that of chromosomal reporters in a labA− strain (40), implying that the labA pathway may be involved in this additional regulatory layer. Therefore, clock control of promoter activities in cyanobacteria show similar rhythmic characteristics regardless of whether the promoter is located in the chromosome or on a plasmid. Our plasmid reporter system, together with our assay for changes in plasmid DNA topology, seems to accurately reflect the mechanisms of clock control of promoter activities.
Discussion
By using a nonessential plasmid as a reporter of the superhelical state of DNA in cyanobacteria, we have shown that the circadian oscillator controls an in vivo rhythm of plasmid topological change. This topological rhythm is detectable in both LL and in LD, and rhythmic changes in plasmid topology cycle in antiphase to the rhythm of phosphorylation of the KaiC clock protein. For example, in both LL and LD, the linking number of the plasmid topoisomers is greatest early in the subjective day, which correlates with hypophosphorylated KaiC. Thus, as levels of phosphorylated KaiC decrease in cells, there is a concomitant increase in the amount of unconstrained supercoils in the plasmid (seen as an increase in the linking number).
What is the basis of this rhythm of supercoiling? It is possible that the topology rhythm we observe is merely a consequence of rhythmic transcriptional activity, because RNA polymerase introduces supercoils as it transcribes DNA templates (41). We favor the interpretation suggested by the oscilloid model, namely that the central oscillator orchestrates global gene expression by regulation of chromosomal topology (27). In bacteria, chromosomal and plasmid DNA is maintained under negative superhelical tension and a number of DNA transactions including gene expression are facilitated by negative supercoiling of the bacterial genome (42, 43). Rhythmic regulation of DNA supercoiling could explain the global circadian control of promoter activity in cyanobacteria because the topological state of DNA is known to influence promoter activity in bacteria (29–32). How might this regulation be accomplished? One possible explanation is that KaiC-containing protein complexes directly interact with DNA to alter the supercoiling status because KaiC binds DNA with low affinity (44). Alternatively, KaiC-containing protein complexes may regulate DNA supercoiling indirectly, for example, by rhythmically altering (i) the balance between topoisomerase I and DNA gyrase activities, (ii) the activities of DNA binding proteins that constrain supercoils in DNA, or (iii) the activity of proteins that mediate attachment of the nucleoid to the bacterial membrane (SI Fig. 7).
In addition, we have shown that plasmids isolated from cyanobacteria maintained in DD do not display a rhythm of topological change even though the phosphorylation rhythm of KaiC continues. Therefore, the plasmid topology rhythm is conditionally coupled to the core KaiABC circadian oscillator in a light-dependent manner (12). In DD, the core oscillator can continue to “tick” but is uncoupled from its ability to modulate changes in plasmid topology and other clock outputs as well, most significantly, the global rhythm of gene expression (24). The progressive increase in plasmid supercoiling in DD is not due to continued transcription because transcription and translation are inhibited in darkness (24). This observation, together with the results obtained with the DNA gyrase inhibitor (SI Fig. 5), argue strongly that the rhythmic changes in DNA topology in LL and LD are not a direct consequence of rhythmic transcription as mentioned above, but rather are due to some clock-controlled mechanism that modulates DNA topology as suggested by the oscilloid model.
The oscilloid model elegantly accounts for the observation that heterologous promoters (23, 26), including one that is well-known to be responsive to supercoiling (ref. 45 and data not shown), that are integrated into the cyanobacterial chromosome are rhythmically active. We also found that cyanobacterial promoters can be placed on plasmids, completely removed from their native chromosomal context, and still retain their circadian characteristics presumably because of the oscillation of plasmid topology. Our results, together with the report of a clock-controlled rhythm of chromosome compaction (37), provide further support for the oscilloid model for global regulation of promoter activity by the circadian clock in cyanobacteria, However, the simplistic interpretation that supercoiling is directly equivalent to chromosome compaction is inaccurate; in bacteria, chromosome compaction is partially due to the action of nucleoid-associated proteins such as HU or HNS in addition to supercoiling of the DNA (46, 47).
Recently a two-component signal transduction pathway composed of the sensory histidine kinase, SasA, and its cognate response regulator, RpaA, has been implicated as the pathway by which the KaiABC oscillator globally regulates promoter activity in cyanobacteria (48). Inactivation of either sasA or rpaA greatly reduces the amplitude of oscillations of clock-controlled genes, producing an arhythmic phenotype. RpaA was proposed to be a “master” circadian transcriptional regulator, but RpaA fails to bind to the kaiBC promoter in gel shift experiments (mentioned in ref. 48). An alternative hypothesis, however, would be that RpaA is not a master circadian transcription factor, but rather regulates the activity/expression of chromosomal proteins such as HU, HNS, or topoisomerases that are responsible for altering the status of the cyanobacterial chromosome. Thus, the activities of SasA/RpaA can be incorporated into an updated version of the oscilloid model (SI Fig. 7). In this model, proteins of the input pathway provide temporal information to the KaiABC oscillator; output from the central oscillator regulates promoter activity globally by rhythmically altering chromosome topology either directly or via the two-component SasA/RpaA pathway. This updated version of the model can account for our observation of rapid changes in plasmid topology associated with the transition of cells between darkness and light (Fig. 2D and SI Fig. 6), because clock-mediated changes in the activities of chromosome remodeling proteins could quickly change the topological state of the plasmid. On the other hand, rapid changes in plasmid topology are more difficult to explain by postulating that a master transcription factor alone mediates global regulation of promoter activities.
Our results and those of Smith and Williams (37) provide evidence of chromosome topological alterations that link the cyanobacterial circadian clock to rhythmic promoter activity. In contrast to the cyanobacterial clock, eukaryotic clock control of gene expression is usually described in terms of regulation by the rhythmic activity of specific transcription factors. However, recent evidence (49–51) may bring these two different mechanisms closer together. In eukaryotes, activation and repression of transcription are often associated with histone modifications. In mammals, activation of several CLOCK-BMAL1 regulated genes requires histone acetylation, which activates transcription by decondensing chromatin. Interestingly, CLOCK has now been shown to have a histone acetyltransferase activity that is necessary to rescue circadian rhythmicity in Clock mutant cells (51). The histone acetyltransferase activity of CLOCK and its apparent requirement for rhythmicity suggests that regulation of chromatin structure is an important mechanism for clock control of transcription in eukaryotes. Thus, regulation of chromosome structure may be a conserved feature of circadian clocks in general and not merely limited to the cyanobacterial circadian system.
Materials and Methods
Bacterial Strains, Growth Conditions, and in Vivo Luminescence Rhythm Assays.
All strains were grown as described elsewhere (23, 52) (also see SI Materials and Methods). For time course studies, the strain AMC149 was grown in standard batch cultures maintained at OD750 between 0.15 and 0.2 by continuous dilution throughout the experiment. Cultures were synchronized by growth in three cycles of LD 12:12 before release into LL or DD. Duplicate 30-ml culture samples were harvested at each time point; one aliquot of cells was used to isolated plasmid DNA, and the other was used to isolate protein. Luminescence assays were conducted as described previously (23, 52); for observation of the effect of trcp-driven KaiC on rhythmicity, water or 1 mM isopropyl β-d-thiogalactoside was applied under the agar on the third day of the assay, and then measurements were continued for 4–5 d.
Isolation of Plasmid DNA from Cyanobacterial Cells, CAGE, and Southern Blotting.
Plasmid DNA was harvested from cyanobacteria by using QIAprep Spin Miniprep kits (Qiagen, Valencia, CA) as recommended by the manufacturer. One-half of each DNA sample (25 μl) was loaded into the well of a 0.8% agarose/0.5× TBE (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.3) gel containing 10 μg/ml chloroquine; plasmid topoisomers were separated by electrophoresis at 40 V with buffer circulation for 24 h in the dark. After electrophoresis, agarose gels were soaked in distilled water for 1 h to remove the chloroquine, and plasmid DNA was transferred to BioBond Plus nylon membrane (Sigma-Aldrich, St. Louis, MO) by Southern blotting (53). Plasmid DNA was detected by hybridization using a DNA probe derived from pANS (see SI Materials and Methods). Probe hybridization was performed at 52°C in 10% (wt/vol) dextran sulfate (≈500,000 molecular weight)/1% (wt/vol) SDS/1 M NaCl with 50 μg/ml denatured herring sperm DNA for 16–24 h. Membranes were washed twice in 2× standard saline phosphate/EDTA (SSPE) (0.18 M NaCl, 10 mM phosphate, pH 7.4, 1 mM EDTA) at 23°C, twice in 2× SSPE/1% SDS at 50°C, and twice in 0.1× SSPE at 23°C. Membranes were dried and radioactivity was detected by phosphoimaging. Phosphoimages were analyzed by densitometry using IP Lab Gel H software (BD Biosciences, Rockville, MD). From densitometric traces, positions of the relaxed open circular (Rel), supercoiled (SC), and the median signal from the separated topoisomer population were determined. Relative mobility for the plasmid topoisomer population at each time point was then calculated as follows: RM = (median − Rel)/(SC − Rel).
Immunoblot Analyses of KaiC Protein.
Total protein was extracted from cells, separated by electrophoresis, and blotted as described previously (23). Antisera to KaiC (23) was used to detect both phosphorylated and nonphosphorylated KaiC. Quantification of KaiC was performed by using ImageJ software (National Center for Biotechnology Information).
Construction of Plasmid Reporters.
To create luminescent reporter plasmids, a 5.85-kb BamHI/SacI fragment containing the luxCDABE operon of Photorhabdus luminescens from pXen-13 (Xenogen Corporation, Alameda, CA) was inserted into the EcoRV site of a cyanobacterial neutral site II (NSII) vector, pAM1579 (gift from S. Golden, Texas A&M University, College Station, TX) creating pLux-NSII. A 416-bp SalI fragment with the psbAI promoter from pAM1583 (gift from Susan Golden) was inserted into the StuI site of pLux-NSII to make pPsbAI-LuxNSII. A 670-bp DNA fragment containing the purF promoter was amplified by PCR using the following primer pair: 5′-AGGCCT (StuI) CAACATCGATCGCGTCTGTGGT-3′ and 5′-GAATTC (EcoRI) CTTGCAGAGCTCCTGATTGGCA-3′. This DNA fragment was cloned into the StuI/EcoRI sites of pLux-NS II to yield pPurF-LuxNS II. A 2.4-kb XhoI/XbaI DNA fragment containing the cpc promoter region from the cyanobacterial expression plasmid pUHEX (GenBank accession no. AB084895; a gift from T. Onizuka, Toray Research Center, Kanagawa, Japan) was replaced with the 6.2-kb fragment containing psbAIp:CDABE from pPsbAI-LuxNS II or the 6.5-kb NotI/NheI fragment containing purFp:CDABE from pPurF-Lux-NS II to produce pCyanoPsbAIp·luxCDABE and pCyanoPurFp·luxCDABE, respectively.
Construction of Cyanobacterial Plasmid Reporter Strains.
To make bioluminescent plasmid reporter strains, pCyanoPsbAIp·luxCDABE or pCyanoPurFp·luxCDABE were transformed into a wild-type S. elongatus R2 strain to produce {PsbAIp·luxCDABE}p and {PurFp·luxCDABE}p, respectively. To evaluate the role of KaiC, an in-frame deletion of the endogenous kaiC containing the inducible trcp:kaiC was constructed as described in refs. 14 and 52 and transformed with the reporter plasmids to create plasmid reporter strains with KaiC overexpression PsbAIp:luxCDABE/Trcp:kaiC and PurFp:luxCDABEp/Trcp:kaiC. For comparison, trcp:kaiC was introduced into a neutral site II of wild-type (AMC 149) or kaiC-null strain (23) whose NS I harbored a psbAIp·luxAB to create the strain PsbAIp:luxAB/Trcp:kaiC.
Supplementary Material
Acknowledgments
We thank Drs. Vladimir Podust and Dong-Eun Chang for assistance, Dr. Neil Osheroff helpful suggestions, Dr. Susan Golden for pAM1579 and pAM1583, and Dr. Takuo Onizuka for pUH-EX. This research was supported by National Institute of General Medical Sciences Grant R01 GM067152.
Abbreviations
- DD
constant darkness
- LL
constant light
- LD
light/dark cycle
- CAGE
chloroquine agarose gel electrophoresis
- CT
circadian time
- ET
experimental time.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706069104/DC1.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 2.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowrousian M, Duffield GE, Loros JJ, Dunlap JC. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D. Proc Natl Acad Sci USA. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald MJ, Rosbash M. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 8.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith GA, Gant TW, Hastings MH, Kyriacou CP. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 9.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 10.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Nature. 2002;417:8–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 11.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 12.Woelfle MA, Johnson CH. J Biol Rhythms. 2006;21:419–431. doi: 10.1177/0748730406294418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michael TP, McClung CR. Plant Physiol. 2003;132:629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki H, Taniguchi Y, Kondo T, Ishiura M. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Piston D, Johnson CH. Proc Natl Acad Sci USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kageyama H, Kondo T, Iwasaki H. J Biol Chem. 2003;278:2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 19.Mori T, Williams DR, Byrne MO, Qin X, Mchaourab HS, Egli M, Stewart PL, Johnson CH. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH. Genome Biol. 2004;5:217.1–217.4. doi: 10.1186/gb-2004-5-4-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Mori T, Johnson CH. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomita J, Nakajima M, Kondo T, Iwasaki H. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 26.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Proc Natl Acad Sci USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori T, Johnson CH. Semin Cell Dev Biol. 2001;12:271–278. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]
- 28.Trun NJ, Marko JF. ASM News. 1998;64:276–283. [Google Scholar]
- 29.Pruss GJ, Drlica K. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 30.Straney R, Krah R, Menzel R. J Bacteriol. 1994;176:5999–6006. doi: 10.1128/jb.176.19.5999-6006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider R, Travers A, Muskhelishvilli G. Mol Microbiol. 2000;38:167–175. doi: 10.1046/j.1365-2958.2000.02129.x. [DOI] [PubMed] [Google Scholar]
- 32.Unniraman S, Nagaraja V. J Genet. 2001;80:119–124. doi: 10.1007/BF02717907. [DOI] [PubMed] [Google Scholar]
- 33.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson ML, Gaasenbeek M, Laudenbach DE. Mol Gen Genet. 1995;247:623–632. doi: 10.1007/BF00290354. [DOI] [PubMed] [Google Scholar]
- 35.Ogata Y, Mizushima T, Kataoka K, Miki T, Sekimizu K. Mol Gen Genet. 1994;244:451–455. doi: 10.1007/BF00583895. [DOI] [PubMed] [Google Scholar]
- 36.Ogata Y, Mizushima T, Kataoka K, Kita K, Miki T, Sekimizu K. J Biol Chem. 1996;271:29407–29414. doi: 10.1074/jbc.271.46.29407. [DOI] [PubMed] [Google Scholar]
- 37.Smith RA, Williams SB. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Tsinoremas NF, Golden SS, Kondo T, Johnson CH. Mol Microbiol. 1996;20:1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 39.Mori T, Binder B, Johnson CH. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu LF, Wang JC. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang HY, Fang M. Prog Nucleic Acid Res Mol Biol. 2003;73:43–68. doi: 10.1016/s0079-6603(03)01002-x. [DOI] [PubMed] [Google Scholar]
- 43.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Proc Natl Acad Sci USA. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Min HY, Liu Y, Johnson CH, Golden SS. J Biol Rhythms. 2004;19:103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- 46.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azam TA, Ishihama A. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 48.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Etchegaray J-P, Lee C, Wade PA, Reppert SM. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 50.Ripperger JA, Schibler U. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 51.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Mori T, Johnson CH. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning—A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.