Abstract
Auxin is an essential regulator for plant development. To elucidate the mechanisms by which auxin regulates plant development, we isolated an Arabidopsis mutant naked pins in yuc mutants 1 (npy1) that develops pin-like inflorescences and fails to initiate any flowers in yuc1 yuc4, a background that is defective in auxin biosynthesis. The phenotypes of npy1 yuc1 yuc4 triple mutants closely resemble those of Arabidopsis mutants pin-formed1 (pin1), pinoid (pid), and monopteros (mp), which are defective in either auxin transport or auxin signaling. NPY1 belongs to a large family of proteins and is homologous to NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3), a BTB/POZ protein that regulates phototropic responses along with the protein kinase PHOT1 (Phototropin 1). We demonstrate that NPY1 works with the protein kinase PID, which is homologous to PHOT1, to regulate auxin-mediated plant development. The npy1 pid double mutants fail to form any cotyledons, a phenotype that is also observed in yuc1 yuc4 pid triple mutants. Interestingly, both auxin-regulated organogenesis and phototropic responses require an auxin response factor (ARF). Disruption of ARF7/NPH4 leads to nonphototropic hypocotyls and arf5/mp forms pin-like inflorescences. Whereas the PHOT1/NPH3 pathway is regulated by light, our data suggest that the PID/NPY1 pathway may be regulated by auxin synthesized by the YUC flavin monooxygenases. Our findings put YUCs, PID, and NPY1 into a genetic framework for further dissecting the mechanisms of auxin action in plant development.
Keywords: BTB domain, PINOID, YUCCA, flavin monooxygenase, AGC kinase
The plant hormone auxin was initially discovered because of its ability to regulate plant growth, which is also the basis of the bioassay for determining auxin activities and concentrations. In the past three decades, genetic screens for Arabidopsis mutants resistant to exogenous auxin were carried out on the basis of the observation that exogenous auxin inhibits Arabidopsis primary root growth (1, 2). Molecular analysis of the auxin resistant mutants has led to the discovery of an auxin signal transduction pathway starting from the auxin receptor TIR1 to the transcriptional regulation of auxin inducible genes (3–5).
Another key aspect of auxin function is to regulate various plant developmental processes, including the establishment of the apical-basal axis and initiation of embryonic and postembryonic organs (6–9). Unlike plant growth that is auxin concentration-dependent and that is proportional to auxin concentrations within the physiological range, plant development appears to require the establishment of a local auxin gradient (10, 11). At the cellular level, root elongation mainly involves cell division and cell elongation, whereas formation of a new organ requires cell differentiation and cell division. Genetic dissection of the mechanisms by which auxin regulates plant development has been difficult, because exogenous auxin treatments in Arabidopsis mainly inhibit growth and cannot rescue some auxin deficient mutants (12, 13).
Our current understanding of the mechanisms of auxin action in plant development mainly stems from the analysis of several developmental mutants, including pin-formed 1 (pin1) (6), pinoid (pid) (8, 14), and monopteros (mp) (9, 15). These mutants were initially isolated from genetic screens for developmental defects, not for defects in auxin pathways. The common feature for pin1, pid, and mp is that they all develop pin-like inflorescences and that they are defective either in auxin transport or signaling. PIN1 encodes an auxin efflux carrier for directional auxin transport (6, 16, 17), and PID is a Ser/Thr protein kinase that has been proposed to regulate auxin transport by modulating the localization of PIN proteins (14, 18). MP is the auxin response factor 5 (ARF5), a transcription factor that participates in auxin-regulated gene expression (15). It is proposed that the PIN-dependent auxin transport generates local auxin gradients and regulates plant development (10, 11).
We showed (12, 19) that auxin synthesized by the YUCCA (YUC) flavin monooxygenases is required for many developmental processes, including embryogenesis, seedling development, vascular differentiation, and flower development. Both YUC1 and YUC4 are only expressed in discrete groups of cells in the primordia of cotyledons, leaves, and flowers (12, 19). Moreover, different combinations of yuc mutants display different developmental defects, indicating that the temporal and spatial regulation of YUC expression plays a key role in shaping local auxin gradients (12, 19). We also showed that polar auxin transport and local auxin biosynthesis display synergistic genetic interactions (19).
The identification of YUC flavin monooxygenases as key auxin biosynthesis enzymes and the available yuc mutants provide a genetic foundation for us to further analyze the mechanisms of auxin in regulating developmental processes. Because the formation of pin-like inflorescences reflects the failure of organ formation and is a hallmark of defective auxin pathways, the phenotype serves as a useful trait for dissecting the molecular mechanisms by which auxin regulates organ initiation. Here, we report the isolation and molecular characterization of a pin1-like Arabidopsis mutant naked pins in yuc mutants 1 (npy1) that was identified as an enhancer of the yuc1 yuc4 double mutants, which are partially auxin deficient (12). The npy1 yuc1 yuc4 triple mutants develop pin-like inflorescences and completely fail to form any flowers, a phenotype that resembles those of pin1, pid, and mp. We show that NPY1 belongs to a large family of proteins that includes the founding member NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3), which is a key component for blue-light-mediated phototropic responses (20, 21). We demonstrate that NPY1 is in the same pathway of PID and that NPY1 plays a critical role in auxin-regulated plant development.
Results and Discussion
Identification of a pin1-Like Arabidopsis Mutant.
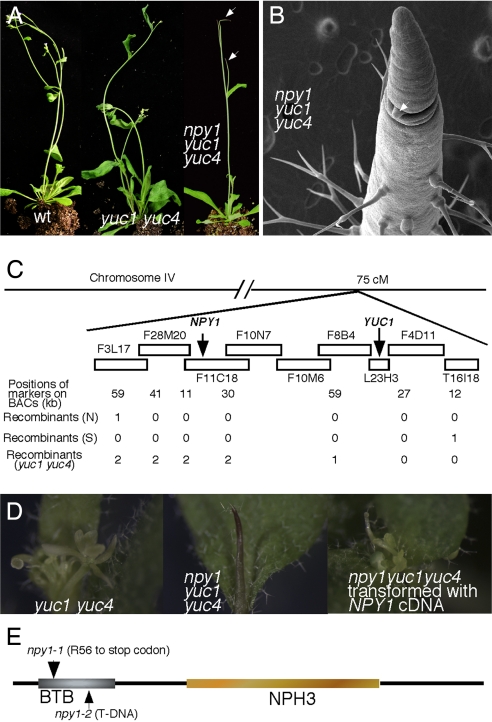
We carried out a genetic screen for enhancers of yuc1 yuc4 double mutants, which partially disrupt auxin biosynthesis and display dramatic defects in vascular and floral development (12) (Fig. 1A). A yuc1 yuc4 enhancer named npy1 was identified (Fig. 1A). The npy1 mutant in the yuc1 yuc4 background formed pin-shaped inflorescences and never produced any flowers (Fig. 1A). Electron microscopic analysis indicated that npy1 yuc1 yuc4 triple mutants failed to initiate flowers at the flanks of the inflorescence meristem (Fig. 1B). Lateral meristems can be initiated from the main inflorescence and lateral pin-like inflorescences were formed in npy1 yuc1 yuc4 triple mutants (Fig. 1 A and B). These lateral pin-like inflorescences also failed to form flowers (Fig. 1A). Compared with the previously described mutants with pin-like inflorescences, npy1 yuc1 yuc4 triple mutants appeared to have the strongest phenotypes in terms of flower initiation. Null alleles of mp, pid, and pin1 all produce some abnormal flowers, but npy1 yuc1 yuc4 did not produce any flowers (data not shown).
Fig. 1.
Identification and molecular cloning of npy1. (A) Mutations in NPY1 in yuc1 yuc4 background caused the formation of pin-like inflorescences. From left to right: WT, yuc1 yuc4, and npy1 yuc1 yuc4. Arrows point toward the pin-like inflorescences. (B) An electron micrograph of npy1 yuc1 yuc4. Note that no flowers were initiated from the inflorescence. Lateral meristems can be initiated from the main inflorescence in npy1 yuc1 yuc4 (arrow). (C) Molecular cloning of NPY1. (D) Complementation of npy1 with the At4g31820 cDNA. From left to right: yuc1 yuc4, npy1 yuc1 yuc4, and npy1 yuc1 yuc4 transformed with At4g31820 cDNA. Note that yuc1 yuc4 developed abnormal flowers, whereas npy1 yuc1 yuc4 never produced any flowers. Introduction of At4g31820 cDNA to npy1 yuc1 yuc4 led to yuc1 yuc4 phenotypes. (E) Domain structure of NPY1.
Transcription of PIN1, PID, and MP Are Not Affected in npy1 yuc1 yuc4.
Because npy1 yuc1 yuc4 displayed phenotypes similar to those of pin1, pid, and mp, we investigated whether the pin-like phenotypes of npy1 yuc1 yuc4 were caused by disruption of the expression of PIN1, or PID, or MP. RNA in situ hybridization analysis demonstrated that the three genes were still expressed in the tip of inflorescences of npy1 yuc1 yuc4 triple mutants [supporting information (SI) Fig. 6], indicating that the pin-like phenotypes of npy1 yuc1 yuc4 are not caused by disruption of PIN1, PID, or MP at the transcriptional level.
The Pin-Like Phenotypes of npy1 Depends on both yuc1 and yuc4 Mutations.
We next determined whether the formation of pin-like inflorescences depended on the presence of the yuc mutations. We first genotyped the pin-like mutants and found that they were all yuc1 yuc4 homozygous, suggesting that the pin-like phenotypes may depend on the presence of yuc1 yuc4. Because npy1 yuc1 yuc4 triple mutants never produced any flowers and were completely sterile, we recovered the sister plants that was npy1 heterozygous, yuc1 homozygous (yuc1−/−), and yuc4 heterozygous (yuc4+/−). Among the progenies from a single plant of npy1+/−yuc1−/−yuc4+/−, ≈12% formed pin-like inflorescences, and 12% displayed yuc1 yuc4 phenotypes. All of the plants with pin-like inflorescences were found to be yuc1 and yuc4 homozygous, suggesting that the pin-like phenotypes require simultaneous inactivation of YUC1, YUC4, and NPY1. The segregation ratios suggested that npy1 was recessive and was tightly linked to one of the YUC genes. Because progenies from plants with npy1+/−yuc1−/−yuc4+/− genotypes displayed both pin-like phenotypes and yuc1 yuc4 phenotypes with similar frequencies, we concluded that npy1 was tightly linked to yuc1. We also crossed npy1+/−yuc1−/−yuc4+/− to WT and analyzed the F2 population from an F1 plant that was heterozygous for all of the three mutations (yuc1, yuc4, and npy1). Among the 221 F2 plants that were yuc1 yuc4 homozygous, 220 plants formed pin-like inflorescences and only one displayed yuc1 yuc4 phenotypes, further demonstrating that npy1 is recessive and tightly linked to one of the yuc mutation.
Molecular Cloning of NPY1.
To further understand the role of NPY1 in auxin-mediated plant development, we identified the molecular lesion in npy1 mutant by map-based cloning (Fig. 1C). From an out-crossed F2 population, we used plants with pin-like inflorescences to locate the npy1 mutation. As we expected, only two linkages were identified: one is located between markers nga151 and nga225 on chromosome V (data not shown), and the other was at the bottom of chromosome IV (Fig. 1C). The linkage on chromosome V corresponds to the yuc4 mutation, whereas the linkage on chromosome IV is where the yuc1 and npy1 mutation are located (Fig. 1C). We used the F2 plants and some F3 plants with pin-like inflorescences to further narrow the interval on chromosome IV down to several BACs (Fig. 1C). We then took advantage of the rare recombination between yuc1 and npy1, which led to yuc1 yuc4 phenotypes with yuc1 yuc4 genotypes. Such a recombination further narrowed down the mapping interval of npy1 to ≈200 Kb.
Because npy1 alone did not display pin-like inflorescences, we hypothesized that NPY1 probably belongs to a gene family whose members may have overlapping functions. We first sequenced the candidate genes that belong to a gene family in the mapping interval and identified a C to T conversion in the gene At4g31820 in npy1 yuc1 yuc4 triple mutants, which converted the Arg-56 to a stop codon in At4g31820 (Fig. 1C). The pin-like inflorescence phenotypes of npy1 yuc1 yuc4 were rescued by expression of At4g31820 cDNA under the control of the 35S promoter, demonstrating that At4g31820 is NPY1 (Fig. 1D). Furthermore, analysis of a T-DNA allele of npy1 (npy1-2) where a T-DNA fragment was inserted in the second exon (22) provided additional evidence that At4g31820 is NPY1 (see below) (Fig. 1).
NPY1 Encodes a BTB-NPH3-Like Protein.
The predicted NPY1 protein belongs to a family that includes 31 other homologous proteins in Arabidopsis (SI Fig. 7). The founding member of this family NPH3 was isolated from a genetic screen for nonphototropic hypocotyl mutants in response to directional blue light (20, 21). Loss-of-function nph3 mutants do not bend toward blue-light stimuli (21). NPH3 forms a complex with PHOT1, a Ser/Thr protein kinase and the photoreceptor for blue-light-mediated phototropic responses (23–25). PHOT1 itself is also a member of the AGC kinase family that includes PID and 37 other Arabidopsis kinases (26) (SI Fig. 8). The AGC kinase is the collective name for cAMP-dependent protein kinase A, cGMP-dependent protein kinase G, and phospholipids-dependent protein kinase C.
NPY1 is a modular protein with a Bric-a-brac, Tramtrack, Broad-complex/Poxvirus Zinc finger (BTB-POZ) domain at the N terminus and a plant-specific NPH3 (nonphototropic hypocotyl 3) domain in the middle (Fig. 1E). Because PID and PHOT1 are homologous and pid mutants form pin-like inflorescences (26), the domain structure of NPY1 and its homology to NPH3 immediately lead us to suggest that NPY1 may work with PID to regulate Arabidopsis organogenesis.
NPY1 and PID Show Similar Expression Patterns.
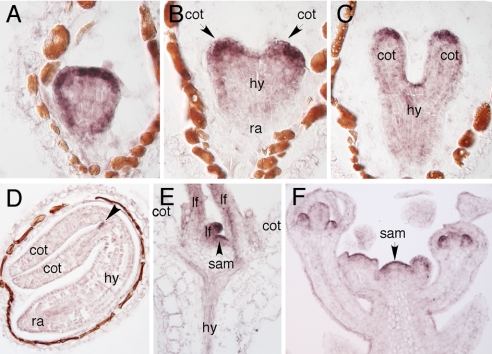
We carried out several experiments to investigate whether NPY1 and PID are in the same pathway in regulating auxin-mediated organogenesis. First, we analyzed the expression patterns of NPY1 by RNA in situ hybridization and found that NPY1 showed similar expression patterns to those of PID (14). NPY1 was expressed throughout the life cycle of Arabidopsis. In globular-stage embryos, NPY1 mRNA expression was detectable in the epidermis with the highest concentration at the apical region (Fig. 2A). At early heart stage, NPY1 expression was restricted to the two cotyledon primordia and the meristem (Fig. 2B). In the late heart to torpedo stages, NPY1 mRNA was further restricted to the meristem and the cotyledon tips (Fig. 2C). In mature embryos, the only place where NPY1 mRNA was detected was the apical meristem (Fig. 2D).
Fig. 2.
Analysis of the expression patterns of NPY1 by RNA in situ hybridization. NPY1 expression in a globular-stage embryo (A), an early heart-stage embryo (B), a late heart-stage embryo (C), a mature embryo (D), a light-grown seedling (E), and an inflorescence apex (F). Cot, cotyledon; hy, hypocotyl; ra, root meristem; sam, shoot apical meristem; lf, leaf.
During seedling development, NPY1 was highly expressed in young leaf primordia and the apical meristems (Fig. 2E). After transition to the reproductive phase, NPY1 mRNA was detected in the inflorescence meristem and at various stages of floral development (Fig. 2F). Furthermore, NPY1 expression also greatly overlaps with YUC1 and YUC4 expression domains (12, 19).
Synergistic Interactions Between npy1 and pid in Arabidopsis Organogenesis.
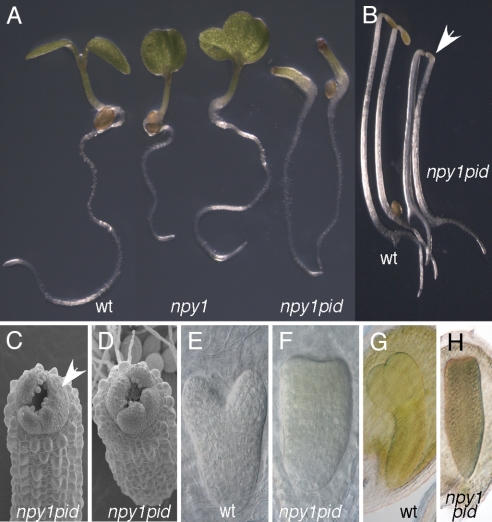
Because npy1 itself did not develop pin-like inflorescences, it is likely that other NPY1 homologs have overlapping functions with NPY1. PID is also a member of a gene family, and PID homologs likely also have overlapping functions with PID. Both npy1 and pid probably are only partial loss-of-function mutants because of the genetic redundancy. If NPY1 and PID participate in the same pathway, we expect that npy1 pid double mutants would display synergistic interactions. Disruption of NPY1 alone led to seedlings with abnormal cotyledons (Fig. 3A), but the frequency of such abnormal seedlings was very low (5 of 203 seedlings). However, pid mutants often produced seedlings with three cotyledons (data not shown). The double mutants of npy1-1 pid failed to form any cotyledons (Fig. 3A), a phenotype that was not observed in either npy1-1 or pid alone. The npy1-2 pid double mutants displayed phenotypes identical to those of npy1-1 pid. Data shown in this article were from the analysis of npy1-1. The no cotyledon phenotype of npy1 pid was fully penetrant, and the hypocotyls and roots did not display apparent defects (Fig. 3A). In the dark, npy1 pid had slightly shorter hypocotyls than WT controls (Fig. 3B). Interestingly, npy1 pid still had an apical hook, indicating that cotyledons are not required for the formation of an apical hook.
Fig. 3.
Genetic interactions between npy1 and pid. (A) Complete loss of cotyledons in npy1 pid double mutants. From left to right: WT, 2 seedlings of npy1, and npy1 pid. (B) Dark-grown npy1 pid seedlings. Note that npy1 pid did not have cotyledons, but it still retained the apical hook (arrow). The two seedlings at Left are WT, and the two at Right are npy1 pid. (C and D) Electron micrographs of npy1 pid seedlings grown in light. Note that the true leaves were initiated in npy1 pid, but normal phyllotaxis is disrupted. (E) A heart-stage WT embryo. (F) A heart-stage npy1 pid embryo. (G) A torpedo-stage WT embryo. (H) A torpedo-stage npy1 pid embryo.
Electron microscopic analysis of npy1 pid seedlings showed that cotyledons in npy1 pid were completely lacking (Fig. 3 C and D). The leaf-like structures were likely true leaves because of the presence of trichomes on the leaves (Fig. 3 C and D). Furthermore, dark-grown npy1 pid lacked such structures (data not shown). Although npy1 pid did not abolish the initiation of true leaves, the phyllotaxis of leaf formation appeared abnormal in npy1 pid (Fig. 3 C and D). The morphological defects of npy1 pid were initiated during embryogenesis. At early heart stage, when cotyledon primordia start to form in WT embryos, npy1 pid did not form cotyledon primordia (Fig. 3 E and F). The mature embryos of npy1 pid completely lacked cotyledons and instead assumed an appearance that appeared more cylindrical (Fig. 3 G and H).
Phenocopy npy1 pid Double Mutants by yuc1 yuc4 pid Triple Mutants.
We have shown that NPY1 and YUC1 YUC4 synergistically control the initiation of flowers and that npy1 yuc1 yuc4 triple mutants formed pin-like inflorescences (Fig. 1). We also demonstrated that NPY1 and PID displayed synergistic genetic interactions (Fig. 3). If PID and NPY1 work in the same pathway as sequence and genetic analysis suggested, we predict that pid and yuc should also display synergistic genetic interactions.
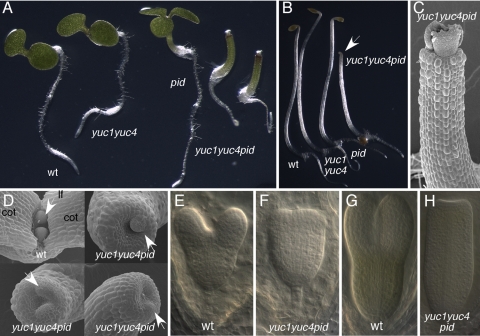
To test this hypothesis, we constructed yuc1 yuc4 pid triple mutants. As shown in Fig. 4A, the triple mutants yuc1 yuc4 pid in which all three mutations are null did not make any cotyledons, a phenotype that was not observed in either pid or yuc1 yuc4 alone. In yuc1 yuc4 pid triple mutants, the cotyledons were precisely deleted without affecting the formation of hypocotyls and roots (Fig. 4 A and B). Interestingly, yuc1 yuc4 pid seedlings and npy1 pid seedlings were morphologically indistinguishable. Both yuc1 yuc4 pid and npy1 pid lacked cotyledons and had an apical hook when grown in the dark (Figs. 3B and 4B). As in the npy1 pid double mutants, simultaneous disruption of YUC1, YUC4, and PID did not abolish the initiation of true leaves (Fig. 4C), but phyllotaxis was disrupted in yuc1 yuc4 pid (Fig. 4D). The earliest morphological deviation of yuc1 yuc4 pid from WT became obvious during early heart-stage development (Fig. 4 E to H).
Fig. 4.
Genetic interactions between pid and yuc1 yuc4 double mutants. (A) Disruption of cotyledon development in yuc1 yuc4 pid triple mutants. From left to right: WT, yuc1 yuc4, pid, two seedlings of yuc1 yuc4pid. (B) Effects of yuc1 yuc4 pid on seedling growth in the dark. From left to right: WT, yuc1 yuc4, pid, and yuc1 yuc4 pid triple mutants. (C) An electron micrograph of yuc1 yuc4 pid grown in light for 5 days. Note the lack of cotyledons in the seedling. (D) Electron micrographs of 3-day-old dark-grown seedlings. The Top Left is WT, and the other seedlings are yuc1 yuc4pid. Note that the apical region is concave shape in yuc1 yuc4 pid and that leaf primordia are initiated. In WT, two true leaf primordia are in opposite position and are similar in size. (E) A WT heart-stage embryo. (F) A yuc1 yuc4 pid heart-stage embryo. (G) A torpedo-stage WT embryo. (H) A torpedo-stage yuc1 yuc4 pid embryo. Cot, cotyledon; lf, leaf.
The pid Mutations Become Haploid-Insufficient in yuc1 yuc4 Background.
Strong genetic interactions among npy1, pid, and yuc1 yuc4 became even more obvious when we observed that pid was haploid-insufficient in yuc1 yuc4 or npy1 yuc1 backgrounds (SI Fig. 9). The pid mutant is known to be recessive in WT background, but in yuc1 yuc4, inactivation of one copy of PID was sufficient to cause the formation of pin-like inflorescences (SI Fig. 9). Heterozygous pid in npy1 yuc1 background also led to phenotypes similar to those of homozygous pid mutants (SI Fig. 9). Furthermore, we observed that yuc1−/−yuc4+/− formed pin-like inflorescences in pid-8, a weak pid mutant that never develops pin-like inflorescences (SI Fig. 9). The observations that pid is haploid-insufficient in yuc1 yuc4 or yuc1 npy1 suggest that YUC, PID, and NPY1 are in the same pathway.
Model for Auxin-Regulated Plant Development.
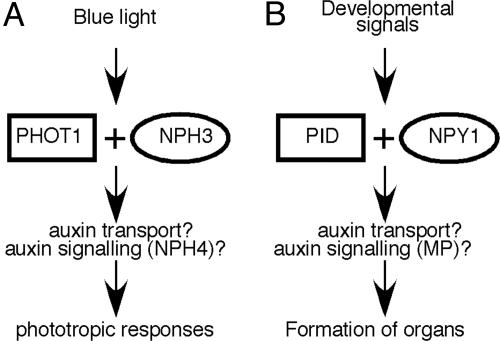
Our results lead us to conclude that NPY1 and PID act in the same pathway to control auxin-regulated plant development, a model that is derived on the basis of the phototropic response pathway (Fig. 5A). In response to blue-light stimuli, PHOT1 and NPH3, which are homologous to PID and NPY1, respectively, mediate phototropic responses possibly by regulating auxin transport and other downstream components (20, 23). We believe that NPY1 and PID probably also regulate auxin transport in response to developmental signals that have yet to be defined (Fig. 5B). Another similarity between the NPY1/PID pathway and phototropism pathway is that both require the involvement of an auxin response factor (ARF). Inactivation of ARF7/NPH4 leads to nonphototropic hypocotyls, whereas arf5/mp mutants form pin-like inflorescences (7, 27).
Fig. 5.
Model for NPY1 in auxin-regulated plant development. (A) PHOT1 and NPH3 in blue-light-mediated phototropism. (B) Regulation of plant development by NPY1 and PID. PHOT1 is homologous to the protein kinase PID, whereas NPH3 is a homolog of NPY1. Note that both PHOT1/NPH3 and PID/NPY1 pathways require the involvement of an auxin response factor. Inactivation of ARF7/NPH4 leads to nonphototropic hypocotyls, whereas disruption of ARF5/MP leads to the formation of pin-like inflorescence.
Although the developmental signal regulating the NPY1/PID pathway has not been defined, our data suggest that the signal may be auxin itself. Both PID and NPY1 display synergistic genetic interactions with yuc1 yuc4, which are defective in auxin biosynthesis (12) (Figs. 1 and 3). Furthermore, pid is haploid-insufficient in yuc1 yuc4, whereas yuc1 yuc4 is haploid-insufficient in pid-8 background (SI Fig. 9). Our hypothesis that auxin itself may regulate auxin transport via NPY1/PID is consistent with previous findings that auxin is able to regulate its own efflux (28). Alternatively, the genetic interactions between YUC1 YUC4 and NPY1/PID may also suggest that NPY1/PID regulates YUC-mediated local auxin biosynthesis.
NPY1 contains a BTB domain, which is known to interact with CUL3 and serve as an E3 ligase (29, 30), suggesting that NPY1 may also regulate protein degradation in response to developmental signals. Interestingly, the auxin efflux carrier PIN2 is reported to undergo proteolysis in response to gravity changes (31). It will be interesting to explore whether NPY1 is involved in targeted proteolysis of PIN proteins.
Among the 32 members of NPH3/NPY1 proteins in Arabidopsis, NPH3 (20, 21) and RPT2 (32) have been shown to participate in phototropism, and we have shown that NPY1 is involved in auxin-mediated plant development. Interestingly, NPH3, RPT2, and NPY1 all work with an AGC kinase (26). It remains to be investigated whether other PHOT1/PID-like AGC kinases in Arabidopsis also work with a specific NPH3/NPY1-like protein in response to a specific environmental or developmental signal.
Materials and Methods
The T-DNA insertion mutants for npy1-2 and pid were obtained from the Arabidopsis Biological Resource Center at Ohio. The mutant pid-8 (33) was a gift from Dr. E. Sundberg (Uppsala University, Uppsala, Sweden). The T-DNA mutants were genotyped according to the published protocols (22).
The insertion mutant for npy1-2 was the SALK_108406 line. Our DNA sequencing data showed that the T-DNA was inserted into the second exon, 1,300 bp downstream of the start codon of NPY1. Primers for genotyping npy1-2 are: LP, 5′-cctctggatattctaaactaggc-3′; RP, 5′- caaactccttgtaccggtcatc-3′; and the T-DNA-specific primer JMLB1.
The npy1-1 allele is genotyped with the above LP and RP primers. The PCR product is then digested with the enzyme BstI, which generates 900-bp and 400-bp bands for the mutant, and 1,300-bp band for WT.
The T-DNA line for PID was the SALK_049736 line from the Arabidopsis Biological Resource Center at Ohio. The T-DNA was inserted in the intron of the gene, 845 bp downstream of the start codon. Genotyping primers for pid were: 5′-atgttacgagaatcagacggtg-3′, 5′-tcaaaagtaatcgaacgccgctgg-3′, and JMLB1.
The pid-8 mutant was genotyped with a dCAPS marker: pid-8_LP, 5′-gacgtcattagtcggcgcaac-3′ and pid-8_RP, 5′-cgttcgttggtacgcatgaatacgtggctt-3′. The PCR fragment was digested with MseI.
Supplementary Material
Acknowledgments
We thank Drs. J. Chory, M. Chrispeels, J. Ecker, R. Schmidt, and M. Yanofsky for advice and comments on the manuscript; Dr. J. Long for technical assistance on analysis of embryos; Dr. E. York for SEM analysis; and F. Pierri, K. Saroca, L. Guerra, S. Lee, A. Fang, Y. Liu, M. Chang, Jin O, Y. Sun, M. Lewis for DNA preps and genotyping various mutants. This work is supported by National Institutes of Health Grant R01GM68631 (to Y.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708506104/DC1.
References
- 1.Leyser O. Annu Rev Plant Biol. 2002;53:377–398. doi: 10.1146/annurev.arplant.53.100301.135227. [DOI] [PubMed] [Google Scholar]
- 2.Hobbie L, Estelle M. Plant Cell Environ. 1994;17:525–540. doi: 10.1111/j.1365-3040.1994.tb00147.x. [DOI] [PubMed] [Google Scholar]
- 3.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 4.Kepinski S, Leyser O. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 5.Dharmasiri N, Dharmasiri S, Estelle M. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 6.Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 7.Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- 8.Bennett SRM, Alvarez J, Bossinger G, Smyth DR. Plant J. 1995;8:505–520. [Google Scholar]
- 9.Berleth T, Jurgens G. Development (Cambridge, UK) 1993;118:575–587. [Google Scholar]
- 10.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 11.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y, Dai X, Zhao Y. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, Koes R. Genes Dev. 2002;16:753–763. doi: 10.1101/gad.219502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen SK, Dagenais N, Chory J, Weigel D. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 15.Hardtke CS, Berleth T. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- 17.Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- 18.Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Y, Dai X, Zhao Y. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedmale UV, Liscum E. J Biol Chem. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- 21.Motchoulski A, Liscum E. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- 22.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 23.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 24.Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 25.Liscum E, Briggs WR. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogre L, Okresz L, Henriques R, Anthony RG. Trends Plants Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 27.Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jurgens G, Geldner N, Friml J. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- 29.Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma L, Vierstra RD. J Biol Chem. 2005;280:18810–18821. doi: 10.1074/jbc.M413247200. [DOI] [PubMed] [Google Scholar]
- 30.Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, Deng XW. Plant Cell. 2005;17:1180–1195. doi: 10.1105/tpc.105.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. Nat Cell Biol. 2006;8:249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- 32.Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. Plant Cell. 2004;16:887–896. doi: 10.1105/tpc.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E. Plant J. 2006;47:112–123. doi: 10.1111/j.1365-313X.2006.02775.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.