Abstract
RecQ helicases are involved in the processing of DNA structures arising during replication, recombination, and repair throughout all kingdoms of life. Mutations of different RecQ homologues are responsible for severe human diseases, such as Blooms (BLM) or Werner (WRN) syndrome. The loss of RecQ function is often accompanied by hyperrecombination caused by a lack of crossover suppression. In the Arabidopsis genome seven different RecQ genes are present. Two of them (AtRECQ4A and 4B) arose because of a recent duplication and are still nearly 70% identical on a protein level. Knockout of these genes leads to antagonistic phenotypes: the RECQ4A mutant shows sensitivity to DNA-damaging agents, enhanced homologous recombination (HR) and lethality in a mus81 background. Moreover, mutation of RECQ4A partially suppresses the lethal phenotype of an AtTOP3α mutant, a phenomenon that had previously been demonstrated for RecQ homologues of unicellular eukaryotes only. Together, these facts strongly suggest that in plants RECQ4A is functionally equivalent to SGS1 of Saccharomyces cerevisiae and the mammalian BLM protein. In stark contrast, mutants of the closely related RECQ4B are not mutagen-sensitive, not viable in a mus81 background, and unable to suppress the induced lethality caused by loss of TOP3α. Moreover, they are strongly impaired in HR. Thus, AtRECQ4B is specifically required to promote but not to suppress crossovers, a role in which it differs from all eukaryotic RecQ homologues known.
Keywords: Blooms syndrome, crossover suppression, Holliday Junction, Mus81, topoisomerase
Genes coding for RecQ helicases are present in all pro- and eukaryotes investigated. The data available so far from completed genome sequences indicate that the number of RecQ genes rises from organisms with low complexity to those of higher complexity. Single RecQ genes are present in Escherichia coli and Saccharomyces cerevisiae, and five to eight RecQ genes are found in mammals and plants, respectively (1, 2). Therefore, the function of RecQ helicases seems to have adapted to the complexity of genomes present in higher eukaryotes by increasing their number. Sequence duplications were followed by subsequent differentiation of known functions or possible acquisition of new ones. In most cases knockout of RecQ genes results in a hyperrecombination phenotype in various organisms, such as bacteria, yeasts, mammals, or plants (3–6). Mutations in the BLM, WRN, and RECQ4 genes are the cause of severe diseases, such as Blooms, Werner, and Rothmund–Thomson syndromes, respectively. Furthermore, a mutant of the mammalian RECQ5 gene shows a synergistic increase of sister chromatid exchange in a blm background (7, 8).
A prominent function of several RecQ helicases is the processing of double-holliday junctions (dHJs) that occur as intermediates during replication, DNA repair, or recombination and dissolve them in a manner which prevents deleterious crossover recombination (9–11). The respective RecQ homologue (e.g., BLM, SGS1, or RQH1) acts in concert with topoisomerase 3 (TOP3) in unicellular organisms, or TOP3α in multicellular eukaryotes (6, 12–17). As a third member of the complex, the RMI1 protein was characterized in mammals and yeast (10, 18, 19). Moreover, in the lower eukaryotes S. cerevisiae and Schizosaccharomyces pombe deletion of their respective RecQ homologue leads to partial suppression of the severe phenotypes caused by mutation of the TOP3 gene (14, 20–22).
Several RecQ helicase mutants are synthetically lethal in a combination with mutations in the endonuclease MUS81 (9, 23, 24). This inviability of the double mutants is most probably because both proteins act in parallel pathways resolving aberrant DNA structures arising during replication. When both genes are mutated, these structures accumulate, leading to cell death (9, 23).
Comparing the complete genome sequences of four different plants (Arabidopsis thaliana, Oryza sativa, Physcomitrella patens, and Populus trichocarpa), all in all, eight different RecQ-like gene types can be classified, seven of which are present in Arabidopsis (reviewed in ref. 1). So far, only one of these genes has been functionally characterized in planta: AtRECQ4A (3, 25). T-DNA insertion mutants were sensitive to DNA-damaging agents and showed an enhanced frequency of homologous recombination (HR) by using an assay specific for crossover events. Furthermore, expression of AtRECQ4A in yeast resulted in full suppression of the sgs1 mutant phenotype (3). Recently, we demonstrated that the recq4A mutant in combination with the mus81 mutation is lethal in Arabidopsis as has been shown for blm in Drosophila melanogaster, sgs1 in S. cerevisiae, and rph1 in S. pombe (9, 23–25). Interestingly, AtRECQ4A is one member of a recently duplicated gene pair in A. thaliana (26). The second ORF, AtRECQ4B, is nearly 70% identical to 4A on the amino acid level and shows most differences outside of the known domains of RecQ proteins. The aim of our present analysis was to elucidate the biological function of both genes, not only in regard to their role in HR, but also in respect to their genetic interactions with MUS81 and TOP3α.
Results
Sensitivity to Genotoxic Agents.
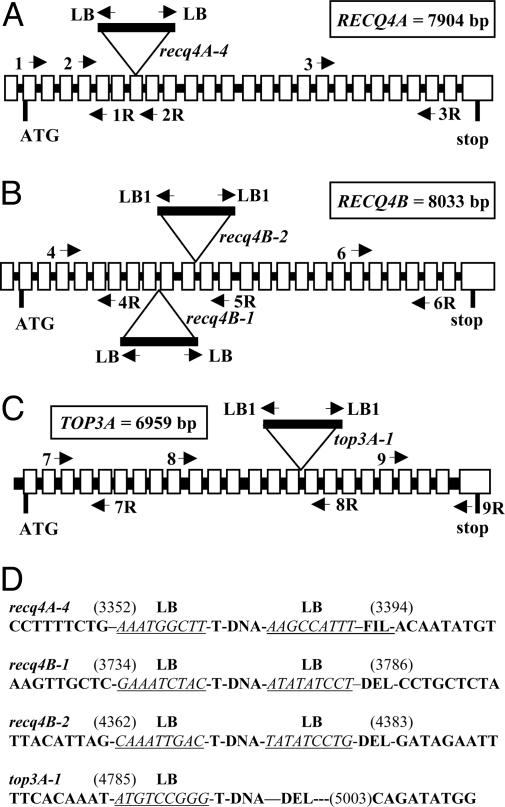
To elucidate the biological role of AtRECQ4B we characterized two different T-DNA insertion lines of AtRECQ4B (At1g60930): recq4B-1 (GABI_399C04) and recq4B-2 (SALK_011357) (27, 28). The sequences of the insertion sites were determined: the T-DNA of line 1 is inserted in the 8th intron (of a total of 24) with a genomic deletion of 51 bp in this area (Fig. 1 B and D). The T-DNA of line 2 is inserted in the 10th intron, accompanied by a genomic deletion of 20 bp (Fig. 1 B and D). Furthermore, we isolated a T-DNA insertion line (GABI_203C07) of AtRECQ4A (At1g10930) to compare mutants of both genes in the same ecotype. The T-DNA is inserted in the 7th exon (of 25 total exons), accompanied by a deletion of 40 bp of exon 7 sequence and an insertion of 19 bp of an ectopic filler sequence (Fig. 1 A and D). To distinguish it from the lines (3) published already, we refer to this line as Atrecq4A-4. All three lines described above harbored a tail-to-tail insertion of T-DNA with an LB on each end (Fig. 1 A and B). By RT-PCR full-length mRNA spanning the insertion sites could not be detected in any of the mutants (see supporting information (SI) Fig. 5 and SI Table 1 for details).
Fig. 1.
Molecular analysis of the T-DNA insertion lines. (A–C) The respective location of the T-DNA insertion in lines recq4A-4, recq4B-1 and 2, and top3α-1 is depicted. The schematically drawn genes of RECQ4A and RECQ4B contain 25 exons (gray boxes) and 24 introns (black lines) in the coding region and one additional exon and intron in the 5′ UTR, each. Both genes are interrupted in their helicase domains by the respective T-DNAs. The gene of TOP3α (At5g63920) contains 24 exons vs. 23 introns and the T-DNA is inserted in the 15th intron. Primers used are indicated by arrows. (D) Border sequence analysis of the insertion loci. The genomic sequences adjacent to each T-DNA insertion locus were determined by PCR and are depicted. In the case of top3α-1 on the right side, a deletion of ≈200 bp occurred. The genomic sequences are shown in bold, the respective left border sequence is in italics and underlined. FIL, filler sequence; DEL, deletion.
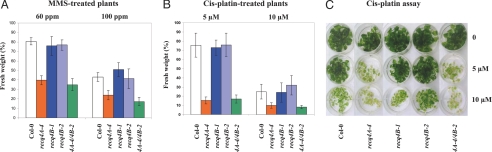
To analyze the mutagen sensitivity, we used a liquid medium assay and determined the weight of 3-week-old plantlets challenged by different concentrations of DNA-damaging agents. Whereas all mutants were as resistant as the wild type to mitomycin C and bleomycin (data not shown), Atrecq4A-4 was sensitive to methylmethanesulfonate (MMS) (3), and cis-platin (Fig. 2; SI Table 2). We could not detect any mutagen sensitivity at all in both Atrecq4B lines (Fig. 2). Additionally, we crossed recq4A-4 and recq4B-2 to characterize the phenotype of the double mutant. Recq4A-4/4B-2 exhibited a mutagen-sensitive phenotype that was indistinguishable from that of the single recq4A-4 mutant (Fig. 2). This demonstrates that AtRECQ4B, in contrast to AtRECQ4A, is not required for the repair of DNA damage induced by the applied mutagens.
Fig. 2.
Mutagen sensitivity assays with MMS or cis-platin. (A and B) Fresh weight of 10 plantlets at each mutagen concentration was determined and put into relation to the fresh weight of the untreated plantlets of the same line. The percentages were calculated from the relation of fresh weight at a given mutagen concentration to the fresh weight without mutagen. Each assay was performed at least four times as described, and the mean values including standard deviations are depicted (the detailed data are given in SI Table 2). recq4A-4 and the double mutant recq4A-4/4B-2 are sensitive to MMS and cis-platin, whereas both recq4B lines behave like the wild type. (C) A six-well assay plate showing a typical cis-platin assay revealing the enhanced sensitivity of recq4A-4 to this cross-linking agent. ppm, parts per million.
Homologous Recombination.
As a hallmark, mutants of eukaryotic RecQ helicases, such as WRN, BLM, or SGS1, show enhanced sister chromatid exchange (SCE) frequencies (6, 29–33). SCEs are a specific type of HR events based on crossovers between sister chromatids. To analyze the role of RECQ4A and RECQ4B in HR, we used the two different recombination substrates, 651 (34) and IC9C (35). Both lines harbor a transgene with nonfunctional overlapping parts of a β-glucuronidase gene (GUS) in an inverted but different spatial orientation (Fig. 3). Restoration of the marker is possible by intra- and interchromosomal recombination in the 651 line (36), whereas in the case of IC9C interchromosomal recombination by using the sister chromatid or the homologous chromosome is required (35). It is important to note that, in the case of line 651, a crossover is necessary to restore the function of the β-glucuronidase marker, whereas, in the case of IC9C, besides crossover (CO), gene conversion can also restore the marker. Each recombination event is represented visually as a blue sector on the plant. We determined the HR frequencies with and without induction by bleomycin. Whereas the application of bleomycin induces genomic double-strand breaks (DSBs), spontaneous recombination might be initiated by replication-associated DNA intermediates (like collapsed replication forks) that differ from classical DSBs. The HR frequencies of the reporter lines 651 (as described in ref. 3) and IC9C were enhanced in the recq4A-4 background (3- to 7-fold, respectively; Fig. 3). However, after induction of DSBs, such an enhancement in comparison with the wild type could not be detected anymore.
Fig. 3.
Recombination frequencies of untreated and bleomycin-treated recq4A and 4B insertion lines. (A) Mean value of at least five independent CO assays with use of the 651 background line that is capable of restoring the functional GUS gene either by intra- or intermolecular recombination (only the intramolecular mechanism of CO is shown schematically right of the diagram). recq4A-4 shows a 3-fold enhanced CO basic level and is poorly induced by bleomycin, whereas both recq4B lines have a 2- to 5-fold reduced basic level. The double mutant exhibits an intermediate phenotype. (B) Mean value of at least five independent HR assays with use of the IC9C background line (the mechanism of HR is restricted to an intermolecular reaction as shown schematically below the diagram). recq4A-4 shows a 7-fold enhanced HR level and is poorly induced by bleomycin, whereas both recq4B lines possess a 2- to 4-fold reduced basic level. The basic and induced HR levels of the double mutant recq4A-4/4B-2 are intermediate compared with the single lines. Because of the differing number of blue sectors per plant, the scales of the x axes vary. In the IC9C substrate line, much less HR occurs because only HR events using the sister chromatid or homologue as template lead to the restoration of the marker. The detailed data including the HR induction factor are given in SI Table 3.
In contrast, the HR level was significantly reduced in the recq4B lines in both substrate backgrounds with and without induction by using bleomycin (2- to 5-fold in line 651 and 2- to 4-fold in IC9C, respectively; Fig. 3). The double mutant recq4A-4/recq4B-2 showed an intermediate phenotype of HR for both substrate lines with and without bleomycin treatment (Fig. 3; SI Table 3). This intermediate phenotype indicates that RECQ4A and 4B are involved in different recombination pathways. HR, which is enhanced because of the loss of the RECQ4A gene, is clearly reduced in the double mutant but not to the same extent as in the single recq4B lines. This can be taken as a hint that a second HR pathway, independent of RECQ4B and possibly promoted by one of the other plant RecQ helicases, might be enhanced by the loss of RECQ4A and is responsible for the remaining events.
Genetic Interactions with AtMUS81 and AtTOP3α.
To detect a possible genetic interaction between AtRECQ4B and AtMUS81 we crossed Atrecq4B-2 with Atmus81-1. In stark contrast to recq4A-4, the recq4B-2/mus81-1 double mutant developed like the wild type, was fertile and exhibited the same mutagen-sensitive phenotype as the mus81 single mutant (data not shown) (25). Thus, it is obvious that RECQ4B performs a role not only different from RECQ4A in DNA repair and recombination, but also in respect to the resolution of aberrant replication intermediates.
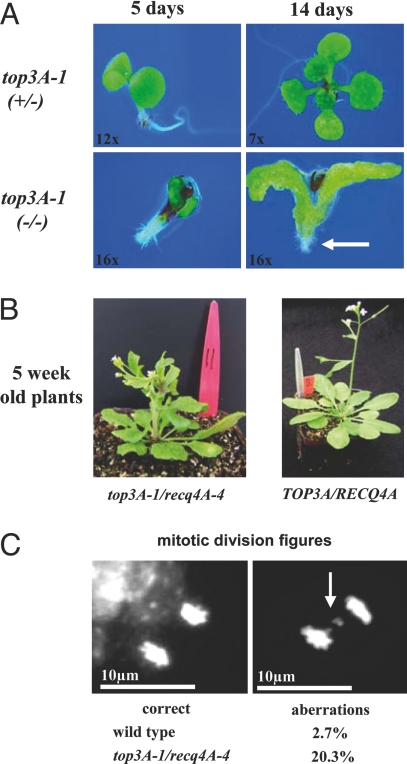
Because an intimate interaction of specific RecQ helicases with TOP3 or TOP3α has been demonstrated in several eukaryotes, we tested whether RECQ4A and/or 4B participate in such an interaction as well. For our study we used an Attop3α-1 mutant (SALK_139357, At5g63920) that harbors a T-DNA insertion in its 15th intron leading to a knockout of mRNA expression spanning the insertion site, whereas the mRNA level in front and behind the T-DNA remained unchanged (see SI Fig. 5). One border sequence of the T-DNA (the left border) was present, whereas the right border, together with 218 bp of the gene, were deleted in the mutant (Fig. 1 C and D). Attop3α-1 was severely impaired in growth and barely germinated on growth medium or in soil; the mutants had deformed cotyledons and were not able to form roots at all. In Fig. 4A the development of a top3α-1 seedling on an agar plate is shown in comparison with a heterozygous top3α-1 seedling. Yet, after 14 days only malformed cotyledons can be seen, whereas heterozygous seedlings had already developed four genuine leaves. The top3α-1 seedlings never reached larger sizes and slowly died. Seedlings heterozygous for the insertion developed like the wild type, showing no growth impairment and full fertility (Fig. 4 A Upper). Thus, the mutant exhibited a phenotype similar to the dramatic top3α phenotypes reported for other eukaryotes. Lethality was observed in mice and pleiotropic growth defects in Caenorhabditis elegans TOP3α mutants (12, 13).
Fig. 4.
Phenotype of the top3α-1 mutant and the double mutant top3α-1/recq4A-4. (A) Typical example of a homozygous top3α-1 seedling on agar plates shown at days 5 and 14 after germination. In comparison, a normally growing heterozygous seedling is shown above. The homozygous mutant developed neither a root (white arrow) nor genuine leaves, and even the cotyledons were malformed. (B) Typical example of a viable, double homozygous mutant plant (top3α-1/recq4A-4) in soil at 5 weeks of age. The plant shows adnated organs, demonstrates slower and dwarfed growth, and is finally sterile. (C) Examples of accurate (Left) and aberrant (Right) mitotic divisions in the double mutant (chromosome fragments are indicated by an arrow). In total, 256 mitotic figures of the control plant (Col-0) and 237 of the top3α-1/recq4A were analyzed, detecting 7 vs. 48 aberrant mitotic divisions, respectively.
Crossing the heterozygous top3α-1 plants with recq4A-4 or recq4B-2 resulted in strikingly different homozygous double mutants. Whereas the double mutant recq4B-2/top3α-1 clearly reproduced the lethal top3α-1 phenotype (data not shown), the recq4A-4/top3α-1 double mutant was viable and proliferated, demonstrating that recq4A possesses in Arabidopsis a suppressor function similar to the one of sgs1/rph1 in yeast. However, the mutation was not able to fully suppress the top3α phenotype, as the recq4A-4/top3α-1 plants show several abnormalities, such as adnated organs and dwarfism (Fig. 4 B Left) that finally result in sterility. Furthermore, the double mutant showed a very high level of mitotic distortions as determined by DAPI staining (Fig. 4C). In total, 256 mitotic divisions of a Col-0 wild-type plant and 237 of a top3A-1/recq4A-4 double mutant were analyzed. Col-0 possessed a level of 2.7% mitotic aberrations (7 of 256), whereas the double mutant exhibited 20.3% (48 of 237). We assume that these mitotic aberrations led to the observed growth defects of the top3A-1/recq4A-4 plants.
Discussion
AtRECQ4A: A Functional Homologue of SGS1 and BLM.
Seven different RecQ-like genes are present in the Arabidopsis genome and the question arises about which of the RecQs can be classified as the respective orthologues of human RecQs or SGS1 in yeast. One important aspect in the functional classification of RecQ homologues is whether their mutations induce defects in genome stability. The T-DNA mutant line recq4A-4 was sensitive to MMS and cis-platin and showed an enhanced HR level in both assay systems tested. This finding is in accordance with a role of RECQ4A in homology-based repair of DNA damage and can be found in RecQ mutants of other organisms, too (5, 29). Because cis-platin induces mostly intrastrand cross-links (CLs) and MMC induces mainly interstrand CLs, the lack of sensitivity to MMC might be taken as an indication that RECQ4A is involved specifically in repair events in which only one of the two DNA strands is blocked. This is in line with the “chicken foot” model, which has been put forward to explain the role of RecQ helicases during replicational repair (37). The fact that, in the absence of AtRECQ4A, the increase in HR was higher with the recombination substrate IC9C, which specifically detects interchromosomal and interchromatid recombination, is reminiscent of BLM cells that have enhanced numbers of SCEs (6, 29, 30, 32, 38), although we cannot formally exclude that the difference from line 651 is due to a position effect. We could not address SCE directly, because, due to its small chromosome sizes, SCE analysis in Arabidopsis was not set up. Interestingly, HR frequencies are not enhanced in the recq4A-4 background after treatment with bleomycin, indicating that RECQ4A is mainly involved in the resolution of aberrant DNA structures arising during replication but not in the repair of DSBs with two free ends.
Another important aspect in the functional classification of RecQ homologues is whether their mutations, in combination with mutations in other genes, cause new proliferation defects or suppress existing ones. A prominent example is the combination of mutations of certain RecQ homologues with the endonuclease MUS81 mutants leading to synthetic lethality. This was shown for S. cerevisiae for the combination sgs1/mus81 (24); for S. pombe, for the combination rqh1/slx3 (23); and for Drosophila blm/mus81 (9). Recently, we demonstrated that Atrecq4A shows synthetic lethality when combined with Atmus81 (25). These results indicate that the respective RecQ homologues are required to process aberrant replication intermediates that cannot be processed properly if MUS81 is mutated (39).
Mutations in the TOP3 or TOP3α gene result in proliferation defects and lethality in several eukaryotes (12–14). This phenotype is partially suppressed in S. cerevisiae and S. pombe if combined with a genetic mutation in the respective RecQ helicases SGS1 and RQH1, respectively (14, 21, 22). The top3α-1 mutant we used for crossings with recq4A-4 and recq4B-2 showed a very severe phenotype with lethality at the cotyledon stage. The mutant line recq4A-4 could partially suppress this phenotype; the double mutant is sterile and mitotically impaired but viable. Thus, to our knowledge, for the first time in a multicellular eukaryote the suppression of a top3α phenotype by the additional mutation of a RecQ helicase could be demonstrated.
In summary, we supply three lines of evidence that AtRECQ4A has a similar function in plants as SGS1 has in yeast and BLM has in mammals: (i) if mutated in RECQ4A, plants show an enhanced frequency of homologous crossovers; (ii) double mutants of RECQ4A and MUS81 are lethal in Arabidopsis (25); and (iii) mutation of RECQ4A in a top3α background changes the lethal phenotype to viability.
AtRECQ4B: A Surprisingly Different Sister.
In contrast to RECQ4A neither RECQ4B mutant showed any sensitivity to DNA-damaging agents. Moreover, also in all other aspects tested, RECQ4B differed from RECQ4A. The mutants of RECQ4B did not show lethality in a mus81 background, nor did they suppress the lethal top3α-1 phenotype. However, the most surprising finding was that RECQ4B has an antagonistic function in homologous recombination in comparison with RECQ4A: HR was not enhanced but reduced in the mutant background with both recombination substrates. Thus, AtRECQ4B is the first reported case of a eukaryotic RecQ homologue that is positively involved in homologous recombination. It is tempting to speculate about the different molecular roles of RECQ4A and RECQ4B: Together with TOP3α, RECQ4A seems to be involved in Holliday junction (HJ) resolution, which prevents CO recombination. In contrast, RECQ4B might be actively involved in the initiation or stabilization of recombination intermediates. One could imagine that RECQ4B with its helicase activity is involved in the formation of D-loop structures. This suggestion is in line with recent evidence that RecQ homologues might be actively involved in synthesis-dependent strand annealing (38, 40). Alternatively, RECQ4B might be able to stabilize formation of HJs by binding to these structures directly.
RECQ4A and 4B: Separation of Function During Plant Evolution?
The RECQ4A/RECQ4B gene pair can be found exclusively in dicotyledonous plants, whereas rice and the moss P. patens possess one RECQ4A orthologous gene only (1). Therefore, two putative scenarios can be envisaged about how the different functions of both gene products evolved: (i) after the duplication of the functional SGS1 homologue (ancestral RECQ4), one copy retained its function of CO suppression (RECQ4A), whereas the second (RECQ4B) became involved in promoting recombination, most probably by taking over the function of another unknown helicase; (ii) the ancestral protein RECQ4 possessed both functions (CO suppression and promotion) and the duplicated proteins specialized in different directions. We favor the second explanation because of different indications reported in the literature.
Early work on RecQ in E. coli and S. cerevisiae demonstrated that in both organisms RecQ (or SGS1) acts as a suppressor of illegitimate recombination and that even BLM can take over this function in a yeast sgs1 mutant (4, 41, 42). In a recent study, the group of Susan Rosenberg (43) was able to demonstrate that the RecQ helicase of E. coli is involved in the net accumulation of replication intermediates that might be resolved by HR. A combination of uvrD with ruvA, B, or C mutants in E. coli was shown to be inviable because of the presence of RecQ. The RuvAB complex is involved in branch migration and RuvC complex is involved in the resolution of HJs (44). UvrD is a DNA helicase required for mismatch repair and nucleotide excision repair, which resolves bimolecular recombination intermediates by removing RecA from single-stranded DNA (45, 46). Interestingly, the mutation of the unique bacterial RecQ helicase rescued the viability of the ruvA/uvrD cells (43). These results led the authors to suggest that, in general, RecQ homologues have two different functions: they might not only be involved in CO suppression, for example, by resolving of dHJs (the classical paradigm), but also in the promotion of recombinogenic DNA structures, such as dHJs (the new “second” paradigm). Whereas their study supplied first indications that the new paradigm might be valid for bacteria, we now clearly demonstrate that this paradigm can be applied to eukaryotes, too. We predict that in the long run other examples besides AtRECQ4B will be discovered in the eukaryotic kingdom.
Because of the duplicated RECQ4 gene, A. thaliana seems to be an ideal model system to further elucidate the molecular basis of both paradigmatic functions of RecQ helicases. It will be very interesting to define which parts of the highly homologous RECQ4A and B proteins are responsible for their antagonistic functions by complementation of the mutants with use of chimeric genes. Future analysis should also reveal whether RECQ4A and 4B have different biochemical properties in vitro, and if they interact with different factors in vivo to explain their antagonistic behavior.
Methods
See SI Tables 1–3 for details of the primers used and data of the sensitivity and HR assays.
Analysis of T-DNA Insertion Lines.
Seeds were obtained from the GABI (line GABI_203C07: At1g10930, recq4A-4; and GABI_399C04: At1g60930, recq4B-1) and SALK collections (line SALK_011357, recq4B-2, SALK_139357; At5g63920, top3α-1) (27, 28). Seeds derived from heterozygous plants were cultivated in soil, and PCR assays with primers flanking the T-DNA insertions were used to screen 3- to 4-week-old plants (Fig. 1 A and B; primers 2 and LB for recq4A-4; 5 and LB or LB1 for recq4B-1 and 2, respectively). Plants homozygous for the T-DNA insertion (or heterozygous in the case of top3α-1) were propagated further. The exact integration sites were determined by PCR with use of primer combinations specific for the left or right border of the respective T-DNA and genomic sequences within the respective gene (Fig. 1; primers 2, 2R, 5, 5R, 8, LB, and LB1). PCR products were purified and sequenced (GATC Biotech).
RNA Extraction and RT-PCR of the T-DNA Insertion Lines.
RNA from young Arabidopsis plantlets was isolated by using the RNeasy Plant Mini Kit from Qiagene according to the instructions of the manufacturer. Reverse transcription and RT-PCR were performed according to the SMART protocol from Clontech by using 2 μg of total RNA. The cDNA produced was used for different PCRs with primers 1 to 9R to evaluate the mRNA level of the respective interrupted gene in front of, spanning, and behind the T-DNA insertion sites (SI Fig. 5 and SI Table 1).
Mutagen Assays.
Homozygous mutant plants from all three lines, the double mutant and Col-0 wild type were sterilized by using 4% NaOCl solution and plated on a germination medium (GM). After 7 days the small seedlings were transferred into six-well plates containing 5 ml of pure liquid GM per well, or liquid GM with different amounts of the respective mutagen methylmethanesulfonate or cis-platin. Ten seedlings were transferred to each well. After 13 days in the respective mutagen solution, the seedlings were taken out and pressed on paper towels to remove any excess liquid. Finally, the weight of the seedlings was determined by using a fine scale balance. All experiments were performed at least four times, and for each experiment the mean value of each line was compared with the wild-type mean value given for each mutagen concentration (Fig. 2 A–C; SI Table 2).
Homologous Recombination Assay.
Homozygous plants for the T-DNA insertion in the respective gene were used for crossings with the two HR-reporter lines 651 and IC9C (34, 35). After crossing and propagation of the heterozygous F1 generation, the F2 generation was screened by PCR for double-homozygous plants and also for plants, in which the respective homozygous wild-type gene (RECQ4A or 4B) and the homozygous reporter construct locus were combined. The latter plants were used as internal controls for the recombination assays. For HR assays, the seedlings were treated in the same manner as for the mutagen assays, but instead of transferring them to six-well plates after 7 days, 30–35 seedlings each were transferred into halved Petri dishes containing 10 ml of either pure liquid GM, or liquid GM with 5 μg of bleomycin, respectively. After 5 additional days in liquid culture the seedlings were transferred to a staining solution (47). After 2 more days, the seedlings were incubated in 70% ethanol for 12 h, and subsequently, the number of blue sectors on each plant was determined by using a binocular microscope. The HR assays were repeated independently at least five times, and the mean values were calculated (Fig. 3; SI Table 3).
DAPI Staining of Mitotic Anaphase Figures.
Inflorescences were collected and incubated in a mixture of methanol and acetic acid in a ratio of 3 to 1. In this fixative the plant material can be stored at −20°C until preparation, which is done, at the earliest, the next day and, at the latest, after a few weeks.
For usage, the plant material was washed in 0.01 M citrate buffer (4 ml of 0.1 M citric acid and 6 ml of 0.1 M sodium citrate, 90 ml of distilled water, pH 4.8) for 5 min and then incubated for 30 min in citrate buffer containing 2% cellulase and 0.2% pectinase (Sigma Aldrich) at 37°C. This digestion was processed in a moist chamber, followed by washing the inflorescences twice for 5 min with citrate buffer.
Buds with a maximum size of 0.5 mm were prepared in 15 μl of acetic acid (45%). After adding a coverslip, the pollen grains were gently squeezed out of the dissected anthers by gentle, manual pressure. To enable fast removal of the coverslip with the help of a razor blade, the slides were quick-frozen in liquid nitrogen for 30 s. The air-dried preparation was then stained with Vectashield mounting medium containing 1.5 μg/ml DAPI (48). By using a fluorescence microscope (Zeiss Axio Imager M1) with an appropriate DAPI filter, mitotic anaphase stages were detected and counted, both accurate stages and stages showing misarranged DNA fragments or chromosomes, respectively.
Supplementary Material
Acknowledgments
We thank Manfred Focke and Shaun Lampi for their thorough reading of the manuscript and Sabine Zeiler for technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grants HA5055/1-1 and PU137/8-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705998104/DC1.
References
- 1.Hartung F, Puchta H. J Plant Physiol. 2006;163:287–296. doi: 10.1016/j.jplph.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Opresko PL, Cheng WH, Bohr VA. J Biol Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 3.Bagherieh-Najjar MB, de Vries OM, Hille J, Dijkwel PP. Plant J. 2005;43:789–798. doi: 10.1111/j.1365-313X.2005.02501.x. [DOI] [PubMed] [Google Scholar]
- 4.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watt PM, Hickson ID, Borts RH, Louis EJ. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Hickson ID. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, Enomoto T. Mol Cell Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson-Schlitz D, Engels WR. Proc Natl Acad Sci USA. 2006;103:16840–16845. doi: 10.1073/pnas.0607904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, Lee MH, Ryu SS, Kim JH, Koo HS. Genes Cells. 2002;7:19–27. doi: 10.1046/j.1356-9597.2001.00496.x. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Wang JC. Proc Natl Acad Sci USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad F, Stewart E. Mol Genet Genomics. 2005;273:102–114. doi: 10.1007/s00438-005-1111-3. [DOI] [PubMed] [Google Scholar]
- 16.Hu P, Beresten SF, van Brabant AJ, Ye TZ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Hum Mol Genet. 2001;10:1287–1298. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 18.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. EMBO J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin A, Wang SW, Toda T, Norbury C, Hickson ID. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maftahi M, Han CS, Langston LD, Hope JC, Zigouras N, Freyer GA. Nucleic Acids Res. 1999;27:4715–4724. doi: 10.1093/nar/27.24.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartung F, Suer S, Bergmann T, Puchta H. Nucleic Acids Res. 2006;34:4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartung F, Plchova H, Puchta H. Nucleic Acids Res. 2000;28:4275–4282. doi: 10.1093/nar/28.21.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 28.Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 29.Adams MD, McVey M, Sekelsky JJ. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 30.Kusano K, Johnson-Schlitz DM, Engels WR. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 31.McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S, Doherty KM, Brosh RM., Jr Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan H, McCane J, Toczylowski T, Chen C. J Cell Biol. 2005;171:217–227. doi: 10.1083/jcb.200502077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swoboda P, Gal S, Hohn B, Puchta H. EMBO J. 1994;13:484–489. doi: 10.1002/j.1460-2075.1994.tb06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molinier J, Ries G, Bonhoeffer S, Hohn B. Plant Cell. 2004;16:342–352. doi: 10.1105/tpc.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuermann D, Molinier J, Fritsch O, Hohn B. Trends Genet. 2005;21:172–181. doi: 10.1016/j.tig.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Hickson ID, Davies SL, Li JL, Levitt NC, Mohaghegh P, North PS, Wu L. Biochem Soc Trans. 2001;29:201–204. doi: 10.1042/0300-5127:0290201. [DOI] [PubMed] [Google Scholar]
- 38.McVey M, Larocque JR, Adams MD, Sekelsky JJ. Proc Natl Acad Sci USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabre F, Chan A, Heyer WD, Gangloff S. Proc Natl Acad Sci USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinert BT, Rio DC. Nucleic Acids Res. 2007;35:1367–1376. doi: 10.1093/nar/gkl831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmon FG, Kowalczykowski SC. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magner DB, Blankschien MD, Lee JA, Pennington JM, Lupski JR, Rosenberg SM. Mol Cell. 2007;26:273–286. doi: 10.1016/j.molcel.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharples GJ, Ingleston SM, Lloyd RG. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flores MJ, Bidnenko V, Michel B. EMBO Rep. 2004;5:983–988. doi: 10.1038/sj.embor.7400262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flores MJ, Sanchez N, Michel B. Mol Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt-Puchta W, Orel N, Kyryk A, Puchta H. Methods Mol Biol. 2004;262:25–34. doi: 10.1385/1-59259-761-0:025. [DOI] [PubMed] [Google Scholar]
- 48.Bleuyard JY, Gallego ME, White CI. Plant Mol Biol. 2004;56:217–224. doi: 10.1007/s11103-004-2812-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.