Abstract
Plants activate distinct defense responses depending on the lifestyle of the attacker encountered. In these responses, salicylic acid (SA) and jasmonic acid (JA) play important signaling roles. SA induces defense against biotrophic pathogens that feed and reproduce on live host cells, whereas JA activates defense against necrotrophic pathogens that kill host cells for nutrition and reproduction. Cross-talk between these defense signaling pathways has been shown to optimize the response against a single attacker. However, its role in defense against multiple pathogens with distinct lifestyles is unknown. Here we show that infection with biotrophic Pseudomonas syringae, which induces SA-mediated defense, rendered plants more susceptible to the necrotrophic pathogen Alternaria brassicicola by suppression of the JA signaling pathway. This process was partly dependent on the cross-talk modulator NPR1. Surprisingly, this tradeoff was restricted to tissues adjacent to the site of initial infection; A. brassicicola infection in systemic tissue was not affected. Even more surprisingly, tradeoff occurred only with the virulent Pseudomonas strain. Avirulent strains that induced programmed cell death (PCD), an effective plant-resistance mechanism against biotrophs, did not cause suppression of JA-dependent defense. This result might be advantageous to the plant by preventing necrotrophic pathogen growth in tissues undergoing PCD. Our findings show that plants tightly control cross-talk between SA- and JA-dependent defenses in a previously unrecognized spatial and pathogen type-specific fashion. This process allows them to prevent unfavorable signal interactions and maximize their ability to concomitantly fend off multiple pathogens.
Keywords: biotroph, cross-talk, jasmonic acid, necrotroph, salicylic acid
In their natural environment, plants are under continuous biotic stress caused by different attackers, including bacteria, fungi, viruses, and insects. Plant pathogens can generally be divided in two categories: biotrophs and necrotrophs. Biotrophs are pathogens that penetrate or establish close contacts with host cells for growth and reproduction in their life cycle. Consequently, biotrophs cause minimal damage to the plant, although symptoms usually occur as a result of nutrient depletion (1, 2). Necrotrophs, in contrast, depend on dead host tissue for nutrients and reproduction. They often secrete enzymes and toxins that degrade and kill host cells to make nutrients available (1, 2). Besides pathogens with these contrasting infection strategies, there are also those pathogens that are biotrophic in one stage of the infection cycle and necrotrophic in another stage of the infection cycle.
To fend off pathogens with different infection strategies, plants have evolved complex defense mechanisms. Each plant genome encodes hundreds of R proteins that play a pivotal role in defense against biotrophs (2). R proteins allow rapid recognition of the pathogen to trigger the hypersensitive response, which includes generation of an oxidative burst and programmed cell death (PCD), thereby rendering the pathogen avirulent. Besides this pathogen-specific R protein-mediated resistance, there are general resistance mechanisms against biotrophic pathogens. These mechanisms include local and systemic synthesis of salicylic acid (SA) (3, 4), which is a potent inducer of a large set of pathogenesis-related (PR) genes (5). The concerted action of PR gene products, some of which encode proteins with antimicrobial activity (6), is thought to establish resistance against biotrophs. Mutants that fail to accumulate SA (such as sid2) or are insensitive to SA (such as npr1) have enhanced general susceptibility to biotrophs (7–14).
The R protein-mediated hypersensitive response and SA-mediated basal resistance are normally ineffective against necrotrophic pathogens. Instead, necrotrophic infection often results in the rapid accumulation of jasmonic acid (JA), which activates a set of PR genes distinct from those induced by SA (15, 16). These genes, such as PDF1.2, HEL, and CHI-B, encode proteins with potent antifungal activities (17, 18). Mutations that disrupt JA signaling result in enhanced susceptibility to necrotrophic pathogens (19). In addition to JA, the secondary indole metabolite, camalexin, is important for defense against necrotrophs because the camalexin-deficient pad3 mutant also shows increased susceptibility to necrotrophs (20, 21). Thus, resistance against necrotrophs is exerted mainly through two defense mechanisms: JA signaling and camalexin synthesis, although other yet unidentified mechanisms also may play a role.
Interestingly, SA- or JA-dependent signaling pathways are not always activated exclusively in response to biotrophs or necrotrophs (2). For example, the biotrophic bacterial leaf-pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 can simultaneously trigger synthesis of both SA and JA (22). This pathogen produces a JA-mimicking phytotoxin, coronatine, which can induce a set of JA-responsive genes (23–25). Because coronatine/JA-insensitive coi1/jai1 mutant plants exhibit elevated expression of SA-responsive PR genes and enhanced resistance to Pst DC3000 (26, 27), coronatine is thought to increase pathogen virulence partly by diversion of the plant's SA-dependent defenses. However, in WT Arabidopsis plants, JA levels taper off soon after SA starts to accumulate, whereas in NahG plants, in which SA fails to accumulate because of the expression of salicylate hydroxylase, high JA levels persist (22). This finding suggests that, in response to a pathogen that can induce synthesis of both SA and JA, cross-talk is used by the plant to adjust the response in favor of the more effective pathway (i.e., the SA-mediated pathway). This cross-talk has been shown to be mediated by NPR1 because it is required for SA-mediated activation of PR gene expression, as well as suppression of JA synthesis and JA-responsive gene expression (22).
Cross-talk between different plant defense signals has been described not only for single attackers, but also for simultaneous invasion by pathogens and herbivorous insects (28, 29). Experiments using chemical elicitors or mutants impaired in SA/JA signaling have suggested that cross-talk between SA and JA may be the underlying mechanism for resistance tradeoffs observed in plants when challenged by both a microbial pathogen and an herbivorous insect. Treatment of plants with inducers of the SA signaling pathway had a detrimental effect on resistance against herbivorous insects (30, 31). Moreover, genetically engineered plants with a silenced phenyl-ammonia-lyase gene have reduced levels of SA, but higher levels of JA, compared with WT. Accordingly, resistance against tobacco mosaic virus was lost, whereas resistance to an herbivorous insect was boosted (32). Although such experiments clearly demonstrate the potential costs of turning on or off a specific defense pathway by using chemical inducers or mutations, they do not simulate a plant in its natural environment. Moreover, the molecular basis and the role of cross-talk in resistance against pathogens with different infection strategies are unknown. Here we investigated how plants coordinate their defenses when infected by both biotrophic and necrotrophic pathogens, and we explored the underlying molecular mechanism. We show that infection by a biotrophic pathogen significantly compromises resistance against a necrotroph. In this case, SA is the potent inhibitor of JA-dependent defense against necrotrophs. Interestingly, tradeoffs between biotrophs and necrotrophs are spatially limited to local or adjacent tissues and arise only from SA-dependent basal defense, but not from R protein-mediated defense. These data demonstrate that plants tightly regulate defense tradeoffs by previously unrecognized control mechanisms to cope with multiple biotic threats.
Results
Exogenous SA Suppresses JA-Dependent Defenses Against Alternaria brassicicola.
Previous experiments have shown that SA can suppress JA-responsive genes (22). To test whether this result leads to compromised JA-dependent defense against necrotrophic pathogens, we examined infection by A. brassicicola. The signals involved in defense against this necrotrophic pathogen are well studied. In WT Arabidopsis, resistance against A. brassicicola is established by two distinct mechanisms: (i) production of camalexin (21), and (ii) synthesis of JA and subsequent activation of a large set of defense genes (33). Expression profiling performed in the WT and camalexin-deficient pad3 mutant has shown that the JA-dependent defenses against A. brassicicola are not affected by the pad3 mutation (33). Therefore, by using pad3 mutant plants, it is possible to examine the sole effect of JA-dependent defenses against A. brassicicola. Mutant pad3 plants were treated with SA, methyl-JA, or a combination of SA and methyl-JA and subsequently infected with A. brassicicola. As shown in Fig. 1A, methyl-JA-treated plants developed smaller lesions and produced fewer A. brassicicola spores, compared with the control plants, indicating that methyl-JA successfully induced defense as reported previously (19). However, in the presence of SA, methyl-JA-induced restriction of fungal growth was compromised, indicating that treating plants with SA can indeed diminish JA-mediated resistance.
Fig. 1.
Effect of SA on JA-mediated defenses against the necrotroph A. brassicicola. (A) In planta-formed spores per lesion 4 dpi of mutant pad3 plants with A. brassicicola. Before inoculation, plants were treated with a solution of 10 mM MgSO4 containing either 1 mM SA, 50 μM methyl-JA, or a combination of both. Asterisks indicate statistically significant differences compared with control treatment (Tukey–Kramer ANOVA test; α = 0.05, n = 3). (B and C) In planta-formed spores per lesion (B) and lesion size 4 dpi of WT Col-0 plants with A. brassicicola (C). Before inoculation plants were treated with a solution of 10 mM MgSO4 supplemented with or without 1 mM SA. The absolute percentage of diseased leaves after SA treatment was 77.0% in this experiment and was always >50.0% in three replicate experiments. (D) SA-responsive PR-1 and JA-responsive PDF1.2 gene expression in untreated and SA-treated Col-0 plants at different dpi of A. brassicicola. Note that day 0 is shown only once because it is identical for untreated and SA-treated plants. To check for equal loading, blots were stripped and hybridized with a gene-specific probe for ubiquitin (UBQ). (E and F) Lesion size on right leaf halves of Col-0 plants 5 days after challenge inoculation with A. brassicicola. Two days before challenge inoculation, left halves of leaves were pressure-infiltrated with 10 mM MgSO4 or biotrophic virulent Pst DC3000 (107 cfu/ml). The absolute percentage of leaves diseased with A. brassicicola was 80.0%. Error bars in graphs represent SE. An asterisk indicates statistically significant differences compared with the control (Student's t test; α = 0.05).
A. brassicicola is unable to infect WT plants because of both camalexin synthesis and rapid activation of JA-dependent defenses. In the JA-insensitive coi1 mutant, resistance was lost (19). We hypothesized that SA-mediated suppression of JA signaling may have a similar effect on resistance as the coi1 mutation. To test this hypothesis, we treated WT Arabidopsis thaliana Col-0 plants with SA and subsequently infected the same plants with A. brassicicola. As shown in Fig. 1 B and C, the WT plants were resistant to this pathogen as indicated by the lack symptoms and fungal growth. Accordingly, JA-dependent expression of the plant defensin gene PDF1.2 was highly activated within 24 h of infection and lasted up to 3 days postinoculation (dpi) (Fig. 1D). Thus, there was a correlation between the restriction of pathogen growth and activation of JA-responsive genes. In contrast to the control plants, SA-treated WT plants developed spreading lesions with extensive sporulation of the pathogen (Fig. 1 B and C). Consistently, SA treatment resulted in activation of SA-responsive PR-1 gene expression, but a complete blockage of JA-responsive PDF1.2 gene expression (Fig. 1D). Together these data indicate that SA is a potent inhibitor of JA-induced defense against necrotrophic A. brassicicola, resulting in increased host susceptibility.
Biotroph-Induced SA Accumulation Leads to Inhibition of JA-Dependent Defenses Against A. brassicicola.
Because exogenous-applied SA suppresses JA-dependent defense against A. brassicicola, it is plausible that endogenous SA, produced upon biotrophic pathogen infection, exerts a similar inhibitory effect. The virulent pathogen Pst DC3000 strongly induces the accumulation of SA and activates PR gene expression (22). Therefore, we pressure-infiltrated leaf halves with 10 mM MgSO4 or Pst DC3000 and subsequently challenged the other leaf halves with A. brassicicola. As shown in Fig. 1 E and F, although the control WT plants were completely resistant to A. brassicicola, Pst DC3000-inoculated plants displayed severe spreading lesions. Similarly, A. brassicicola produced 2- to 5-fold more spores on Pst DC3000-inoculated pad3 plants, compared with the control plants treated with 10 mM MgSO4 [supporting information (SI) Fig. 5 A and B]. We then examined whether the increased susceptibility to A. brassicicola was indeed because of Pst DC3000-induced endogenous SA by using the SA synthesis mutant sid2. We found that in mutant sid2 plants, the Pst DC3000-induced susceptibility to A. brassicicola was abolished, indicating that endogenous SA is required to suppress defense against A. brassicicola (Fig. 2A). Similar results were obtained with the pad3 sid2 double-mutant plants (SI Fig. 5A). RNA gel blot analysis on A. brassicicola-infected tissue showed that Pst DC3000 infection, although causing a strong induction of the SA-dependent PR-1 gene, significantly repressed A. brassicicola-induced expression of the JA/ethylene-responsive genes PDF1.2, HEL, and CHI-B (Fig. 2B). Interestingly, Pst DC3000 infection did not suppress the expression of LOX2, which encodes an important enzyme in JA biosynthesis, suggesting that SA-mediated suppression of JA signaling was not the result of decreased expression of at least one key JA biosynthesis enzyme (Fig. 2B).
Fig. 2.
Biotroph infection locally suppresses JA-mediated defenses against necrotrophic A. brassicicola through SA and NPR1. (A) Percentage of spreading A. brassicicola lesions on WT, sid2, and npr1 plants. Left halves of leaves were pressure-infiltrated with 10 mM MgSO4 alone or with biotrophic virulent Pst DC3000 (107 cfu/ml). After 2 days, the right halves of these leaves were challenge-inoculated with A. brassicicola. Error bars indicate SE. An asterisk indicates statistically significant differences compared with the control (Student's t test; α = 0.05, n = 30). (B) RNA gel blot analysis of SA-responsive PR-1 and JA/ethylene-responsive PDF1.2, HEL, CHI-B, and LOX2 gene expression in pad3 plants at different dpi with A. brassicicola. At −2 dpi, plants were infected with Pst DC3000 and, at 0 dpi, challenge inoculated with A. brassicicola. Only A. brassicicola-inoculated leaf halves were collected for RNA extraction. To check for equal loading, blots were stripped and hybridized for constitutively expressed ubiquitin (UBQ). (C) RNA gel blot analysis of JA/ethylene-responsive PDF1.2, HEL, and CHI-B gene expression in pad3 and pad3 npr1 plants at different dpi with A. brassicicola. To check for equal loading, rRNA was stained with ethidium bromide.
It has been shown previously that SA exerts its inhibitory effect on JA-mediated gene expression through the action of the regulatory protein NPR1 (22). Therefore, we analyzed whether NPR1 also is required for tradeoffs between SA-dependent defense against biotrophs and JA-dependent defense against necrotrophs. In contrast to the WT, Pst DC3000 did not cause increased susceptibility to A. brassicicola in npr1 mutant plants (Fig. 2A). We found similar results with mutant pad3 npr1 plants. Compared with pad3 plants, Pst DC3000 infiltration of pad3 npr1 plants caused only a moderate increase in A. brassicicola infection (33% increase in pad3 npr1 vs. 70% in pad3) (SI Fig. 5B), implying that SA-mediated repression of JA-dependent defenses was partly lost. This finding corresponded to a partial loss of repression of JA/ethylene-responsive PDF1.2, HEL, and CHI-B in pad3 npr1, compared with pad3 (Fig. 2C). Together these findings clearly indicate that biotroph-induced SA accumulation exerts a strong negative effect on JA-dependent defenses against necrotrophs partly through the regulatory molecule NPR1.
Spatial Control of Tradeoffs Between Defenses Against Biotrophs and Necrotrophs.
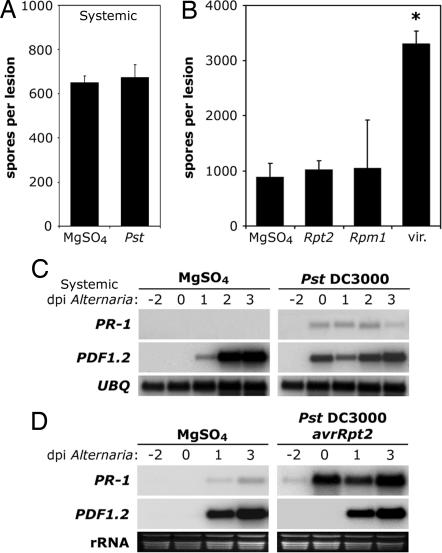
Our experiments clearly show that tradeoffs occur between defenses against biotrophs and necrotrophs when both pathogens are in close proximity to each other (e.g., on the same leaf). To investigate whether tradeoffs between defenses also occur when biotrophic and necrotrophic pathogens are separated on different leaves, we pressure-infiltrated three lower leaves of pad3 plants with 10 mM MgSO4 or Pst DC3000 2 days before challenge inoculating three upper leaves with A. brassicicola. Surprisingly, Pst DC3000 inoculation had no significant effect on the growth of A. brassicicola (Fig. 3A). We examined the expression of SA- and JA-responsive genes in the A. brassicicola-infected tissue. As shown in Fig. 3C, Pst DC3000 infection resulted in weak systemic activation of SA-responsive PR-1 and JA-responsive PDF1.2 gene expression (0 dpi). The subsequent A. brassicicola challenge inoculation induced a second expression peak of PDF1.2 at 2 dpi. Compared with the control plants, Pst DC3000-inoculated plants were slightly attenuated for A. brassicicola-induced PDF1.2 gene expression (Fig. 3C), indicative of modest cross-talk between SA and JA signaling pathways. These data demonstrate that spatial separation of a biotrophic and necrotrophic pathogen results in a low level of antagonism between the SA and JA signaling pathways, which is not sufficient for significant tradeoffs in resistance.
Fig. 3.
Systemic SA signaling and R protein-mediated resistance to biotrophs do not suppress JA-dependent defense against necrotrophic A. brassicicola. (A) In planta-formed spores per lesion 5 days after A. brassicicola challenge-inoculation of control (MgSO4) or Pst DC3000-infected mutant pad3 plants. Two days before challenge inoculation, three lower leaves per plant were pressure-infiltrated with 10 mM MgSO4 alone or with biotrophic virulent Pst DC3000 (107 cfu/ml). Next, three systemic intact upper leaves were challenge-inoculated with A. brassicicola. Error bars indicate SE. (B) In planta-formed spores per lesion 4 days after A. brassicicola challenge-inoculation of pad3 plants that were previously treated with 10 mM MgSO4, virulent Pst DC3000 (vir.) or Pst DC3000 carrying the avirulence genes avrRpt2 or avrRpm1. Plants received half-leaf pathogen inoculations as described in the legend of Fig. 2. Error bars indicate SE. Asterisk indicates statistically significant differences compared with the control (Tukey–Kramer ANOVA test; α = 0.05, n = 3). (C) RNA gel blot analysis of systemic expression of SA-responsive PR-1 and JA-responsive PDF1.2 genes from plants described in A. To check for equal loading, blots were stripped and hybridized for constitutively expressed ubiquitin (UBQ). (D) RNA gel blot analysis of SA-responsive PR-1 and JA-responsive PDF1.2 gene expression from plants described in B. To check for equal loading, rRNA was stained with ethidium bromide.
R Protein-Mediated Resistance to Biotrophs Does Not Suppress Defense Against Necrotrophs.
Besides SA-dependent basal defense, plants exhibit R protein-mediated resistance against biotrophs that carry avirulence genes. We investigated whether SA produced during R protein-mediated PCD against biotrophs can also suppress JA-dependent defense against necrotrophs. We pressure-infiltrated leaf halves with either 10 mM MgSO4 or Pst DC3000/avrRpt2, carrying the avrRpt2 avirulence gene, and subsequently challenged the other leaf halves with A. brassicicola. Surprisingly, in contrast to the virulent strain, avirulent Pst DC3000/avrRpt2 inoculation did not result in enhanced susceptibility to A. brassicicola (Fig. 3B), indicating that a tradeoff was absent. RNA gel blot analysis revealed that infection with avirulent Pst DC3000/avrRpt2 strongly up-regulated PR-1 gene expression in the adjacent tissue, demonstrating that SA signaling pathway was fully activated (Fig. 3D). However, this high level of SA signaling had little effect on the A. brassicicola-induced JA-responsive PDF1.2 gene expression. To investigate whether the lack of tradeoff was specific to Pst DC3000/avrRpt2, we tested a Pst DC3000 strain carrying another avrRpm1 avirulence gene. Fig. 3B shows that Pst DC3000/avrRpm1 also failed to enhance susceptibility to A. brassicicola, indicating that the absence of plant defense tradeoffs between avirulent strains of Pst DC3000 and A. brassicicola was because of a general mechanism. Perhaps R protein-mediated resistance activates an unknown plant signal that blocks SA-mediated inhibition of JA signaling.
Discussion
Defense responses in plants are regulated by complex interconnecting signaling pathways, in which the signaling molecules SA and JA play important roles (28, 34, 35). Cross-talk between SA and JA signaling pathways may help fine-tune defense responses against a single pathogen according to its mode of infection (22, 26–28). However, little is known about cross-talk between defense signaling pathways in response to attack by multiple pathogens. Here we investigated whether tradeoffs occur between SA- and JA-dependent resistance against the well characterized biotrophic and necrotrophic pathogens Pst DC3000 and A. brassicicola, respectively.
Our data provide strong evidence that exogenous SA is a potent suppressor of JA-mediated defenses against necrotrophs, resulting in enhanced pathogen performance (Fig. 1). Similar observations have been reported for JA-mediated plant defenses against insects. Tomato plants treated with BTH, a mimic of SA, were attenuated for JA-mediated defenses and displayed enhanced susceptibility for the herbivores Spodoptera exigua and Helicoverpa zea (30, 31). Besides exogenous application, pathogen-induced accumulation of endogenous SA also has been shown to suppress defense against certain insects (29). For example, tobacco mosaic virus-induced SA signaling in tobacco was associated with increased herbivory by the tobacco hornworm, Manduca sexta (36). Moreover, cucurbit scab fungus-induced SA signaling reduced resistance to both a chewing and a sucking insect in cucumber (37). Whereas biological tradeoffs between SA-inducing pathogens and certain insects are evident, a direct link to cross-talk between SA and JA signaling pathways is lacking. Furthermore, a similar tradeoff between pathogens with different lifestyles has been hypothesized, but not tested. Our results show that biotrophic Pst DC3000-induced endogenous SA exerts a robust negative effect on JA/ethylene-responsive gene expression and A. brassicicola resistance (Fig. 2 A and B and SI Fig. 5A). Moreover, the suppressive effect of SA can partly be attributed to the regulatory protein NPR1 because a mutation in NPR1 partially restored resistance to A. brassicicola (Fig. 2 A and C and SI Fig. 5B). This finding is consistent with the previous one that NPR1 is a key modulator of SA/JA cross-talk when these compounds were applied exogenously (22). We now demonstrated that this antagonism between SA and JA signaling pathways is the underlying mechanism for biological tradeoff between resistance to biotrophs and necrotrophs.
In plants, a local infection often induces a systemic defense response that is effective against pathogens with a similar infection strategy (38, 39). However, in its natural environment, plants also may simultaneously encounter pathogens with different infection strategies. Therefore, antagonism between SA and JA signaling pathways, which is used to fine-tune defense against a single pathogen, may be detrimental in systemic defense against pathogens with opposing lifestyles. To test this possibility, we directly compared tradeoffs in local versus systemic tissues. We demonstrated that, contrary to local tissues where a successful infection by a biotroph reduces resistance to a necrotroph, in systemic tissues tradeoff between biotroph and necrotroph resistance is negligible (Fig. 3A).
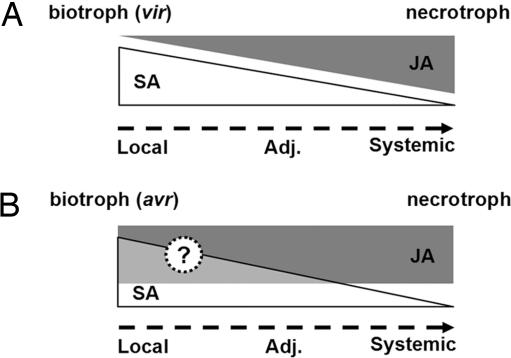
Interestingly, moderate SA-mediated repression of JA-responsive PDF1.2 gene expression was still detected in systemic tissues even in the absence of a resistance tradeoff (Fig. 3C). Perhaps in systemic tissues the SA/JA cross-talk is counteracted by the systemic accumulation of the bacterial produced phytotoxin coronatine (40). Coronatine is a mimic of JA that activates a battery of JA-responsive genes (24, 25) and has been shown to promote Pst DC3000 virulence by overcoming SA-mediated defense (41). We tested this hypothesis by using the Pst DC3000 cmaA/cfa6 double mutant (42) and found that coronatine deficiency in this double mutant did not affect cross-talk in systemic tissue at the level of gene expression or disease resistance (S.H.S. and X.D., unpublished data). Therefore, it is more plausible that tradeoff requires high concentrations of SA. Indeed, it was recently reported that high SA concentrations antagonized JA-induced gene expression, whereas low levels of SA were less effective in this respect or even had synergistic effects on JA signaling (43). We propose that tradeoff between biotroph and necrotroph resistance requires a certain threshold level of SA relative to JA. In tissues close to the site of initial infection, levels of SA are high and tradeoff occurs, whereas in systemic tissues the relative levels of SA compared with JA drop below the threshold and tradeoff is diminished (Fig. 4A).
Fig. 4.
Models for tradeoffs between plant defenses against biotrophic and necrotrophic pathogens. (A) Proposed model for the spatial regulation of tradeoff between defenses against virulent (vir) biotrophs and necrotrophs. Virulent biotroph infection induces strong SA signaling in local and adjacent tissues, which tapers off with increasing distance from the site of infection. As a result of cross-talk, this gradient of SA signaling from local to systemic tissues is inverse-correlated to JA-mediated defenses launched against necrotrophs. In systemic tissues, SA signaling is relatively low, compared with JA signaling, and biological tradeoffs are diminished. (B) Proposed model for interaction between R protein-mediated resistance to biotrophs and JA-mediated resistance to necrotrophs. R protein-mediated resistance to avirulent (avr) biotrophs also induces a gradient of SA signaling from local to systemic tissues. Because of an unidentified factor, this SA signaling is not sufficient to suppress JA-mediated defenses against necrotrophs.
Surprisingly, this concentration gradient hypothesis cannot explain the lack of tradeoff in adjacent tissue when two different avirulent Pst DC3000 strains were used instead of the isogenic virulent strain Pst DC3000 (Fig. 3 B and D). Avirulent Pst DC3000 strains can induce SA signaling to high levels both locally as well as systemically (16, 44), but this is evidently not sufficient to suppress JA-mediated defense against A. brassicicola. This may be explained by the fact that R protein-mediated resistance is associated with the concomitant engagement of both SA and JA signaling pathways (45). In tomato, R protein-mediated resistance to aphids induced simultaneous expression of SA- and JA-dependent defenses (46). Moreover, avirulent Pst DC3000 strains robustly activated the expression of JA biosynthesis enzymes and induced high levels of both SA and JA in Arabidopsis (16, 47). In fact, the JA level in Pst DC3000/avrRpt2-inoculated plants was 4-fold higher, compared with that induced by the virulent strain (22). Biologically, this finding suggests that R protein-mediated resistance against a biotroph does not compromise resistance to a necrotroph (Fig. 4B). Accordingly, in our experiment, resistance to Pst DC3000/avrRpt2 had no detrimental effect on resistance to A. brassisicola. This finding also may explain the observation made by Cui et al. (48) that inoculation of Arabidopsis with an avirulent P. syringae strain caused enhancement, rather than repression, of resistance to the herbivore Trichoplusia ni.
The lack of cross-talk by R protein-mediated defense is unexpected, but may have biological significance. Whereas induction of PCD is highly effective against biotrophs, it may render plants more attractive to necrotrophic pathogens. It has been shown previously that PCD induced by an avirulent P. syringae strain made the plant tissue more susceptible to the necrotroph Botrytis cinerea (49). Our data suggest that, during the R protein-mediated response, plants can deploy a mechanism to counteract SA/JA cross-talk and thereby prevent this enhanced susceptibility to necrotrophs from spreading to the neighboring tissues.
In summary, our study shows that cross-talk between SA and JA is more tightly controlled than we first anticipated. When a virulent biotrophic pathogen infects, tradeoff with necrotroph resistance may be necessary to maintain basal resistance against the biotroph. However, tradeoff does not spread to distant tissues. In addition, there appears to be an active mechanism that prevents SA/JA cross-talk during R protein-mediated PCD probably to ensure that this defense mechanism is not hijacked by necrotrophs, which thrive on dead cells. This spatial and pathogen type-specific control of cross-talk highlights the complexity and sophistication of the plant defense network to simultaneously cope with multiple pathogens.
Materials and Methods
Cultivation of Plants and Pathogens.
Seeds of WT A. thaliana (Col-0), mutant sid2-2 (50), npr1-1 (11), pad3-1 (20), pad3-1 sid2-2, and pad3-1 npr1-1 plants were grown on soil (Metro Mix 200; Grace-Sierra, Milpitas, CA) for 2 weeks. Next, 2-week-old seedlings were transplanted to individual pots and allowed to grow for another 2 weeks before treatment.
The virulent bacterial leaf pathogen Pst DC3000 and avirulent strains carrying the avirulence genes avrRpt2 or avrRpm1 (51) were grown overnight in liquid King's B and prepared as described previously (22). A. brassicicola strain MUCL20297 was grown on PDA agar for 2 weeks at room temperature. Spores were harvested in 10 ml of sterile water and filtered through two layers of Miracloth. Subsequently, spores were collected by centrifugation and washed once with sterile water.
Chemical Treatments and Pathogen Bioassays.
Three leaves of 4-week-old plants were pressure-infiltrated or sprayed with a solution containing 10 mM MgSO4 and either 1 mM SA (Sigma–Aldrich, St. Louis, MO), 50 μM methyl-JA (Sigma–Aldrich), or a combination of both 24 h before pathogen challenge.
For local tripartite assays, the left halves of three leaves from 4-week-old plants were pressure-infiltrated with virulent or avirulent Pst DC3000 carrying avrRpt2 or avrRpm1 at a final concentration of 107 cfu/ml in 10 mM MgSO4. After 2 days of infection, the right halves of the same leaves were inoculated with a 3-μl drop of water (for infection of the pad3 mutant) or 0.5% potato dextrose agar (for infection of the WT) containing A. brassicicola at 106 spores per milliliter. For systemic tripartite assays, three lower leaves of 4-week-old plants were pressure-infiltrated with virulent Pst DC3000 at a final concentration of 107 cfu/ml in 10 mM MgSO4. After 2 days of infection, three upper leaves were inoculated with a 3-μl drop of water containing A. brassicicola at 106 spores per milliliter. At 3–5 days after A. brassicicola inoculation, three batches of 30 right leaf halves or 15 whole leaves per treatment for local or systemic tripartite assays, respectively, were collected for determination of newly formed spores. Spores were detached from the leaves by vigorous shaking in 10 ml of 0.1% Tween 20. The spore suspension was filtered through two layers of Miracloth and centrifuged at 3,200 × g for 15 min. After resuspension in 100 μl of 0.1% Tween 20, spores were counted in a hemacytometer. All bioassays were repeated at least two times with similar results.
RNA Analysis.
RNA extraction, electrophoresis, and hybridization to gene-specific probes were performed as described previously (11, 22).
Supplementary Material
Acknowledgments
We thank Dr. Corné Pieterse (Utrecht University, Utrecht, The Netherlands) and Dr. Jane Glazebrook (University of Minnesota, St. Paul, MN) for the A. brassicicola strain and the pad3 mutant, Dr. Barbara Kunkel (Washington University, St. Louis, MO) for the Pst DC3000 cmaA/cfa6 double mutant; and Natalie Spivey and Drs. Nisha Philip, Yasuomi Tada, Karolina Mukhtar, and Thomas Mitchell-Olds for their useful suggestions and for critically reading the manuscript. This work was supported by a National Institutes of Health grant (to X.D.).
Abbreviations
- dpi
days postinoculation
- SA
salicylic acid
- JA
jasmonic acid
- PCD
programmed cell death
- Pst
Pseudomonas syringae pv. tomato.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708139104/DC1.
References
- 1.Hancock JG, Huisman OC. Annu Rev Phytopathol. 1981;19:309–331. [Google Scholar]
- 2.Glazebrook J. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 3.Malamy J, Carr JP, Klessig DF, Raskin I. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 4.Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- 5.Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- 6.Van Loon LC, Van Strien EA. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- 7.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessman H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 8.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 9.Lawton KA, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Mol Plant–Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- 10.Nawrath C, Métraux J-P. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao H, Bowling SA, Gordon AS, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney TP, Friedrich L, Ryals JA. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glazebrook J, Rogers EE, Ausubel FM. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah J, Tsui F, Klessig DF. Mol Plant–Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 15.Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux J-P, Van Loon LC, Dicke M, et al. Mol Plant–Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 17.Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazebrook J, Ausubel FM. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF. Plant J. 1999;19:163–171. doi: 10.1046/j.1365-313x.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 22.Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, et al. Plant Cell. 2003;15:760–770. [Google Scholar]
- 23.Bender CL, Alarcon-Chaidez F, Gross DC. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uppalapati SR, Ayoubi P, Weng H, Palmer DA, Mitchell RE, Jones W, Bender CL. Plant J. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 25.Thilmony R, Underwood W, He SY. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 26.Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. Plant J. 2001;26:509–522. doi: 10.1046/j.1365-313x.2001.01050.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 28.Beckers GJM, Spoel SH. Plant Biol. 2006;8:1–10. doi: 10.1055/s-2005-872705. [DOI] [PubMed] [Google Scholar]
- 29.Stout MJ, Thaler JS, Thomma BPHJ. Annu Rev Entomol. 2006;51:663–689. doi: 10.1146/annurev.ento.51.110104.151117. [DOI] [PubMed] [Google Scholar]
- 30.Stout MJ, Fidantsef AL, Duffey SS, Bostock RM. Physiol Mol Plant Pathol. 1999;54:115–130. [Google Scholar]
- 31.Thaler JS, Fidantsef AL, Duffey SS, Bostock RM. J Chem Ecol. 1999;25:1597–1609. [Google Scholar]
- 32.Felton GW, Korth KL, Bi JL, Wesley SV, Huhman DV, Mathews MC, Murphy JB, Lamb C, Dixon RA. Curr Biol. 1999;9:317–320. doi: 10.1016/s0960-9822(99)80140-7. [DOI] [PubMed] [Google Scholar]
- 33.Van Wees SCM, Chang H-S, Zhu T, Glazebrook J. Plant Physiol. 2003;132:606–617. doi: 10.1104/pp.103.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reymond P, Farmer EE. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel BN, Brooks DM. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 36.Preston CA, Lewandowski C, Enyedi AJ, Baldwin IT. Planta. 1999;209:87–95. doi: 10.1007/s004250050609. [DOI] [PubMed] [Google Scholar]
- 37.Moran PJ. Oecologia. 1998;115:523–530. doi: 10.1007/s004420050550. [DOI] [PubMed] [Google Scholar]
- 38.Ton J, Van Pelt JA, Van Loon LC, Pieterse CMJ. Mol Plant–Microbe Interact. 2002;15:27–34. doi: 10.1094/MPMI.2002.15.1.27. [DOI] [PubMed] [Google Scholar]
- 39.Durrant WE, Dong X. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 40.Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. Proc Natl Acad Sci USA. 2005;102:1791–1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks DM, Bender CL, Kunkel BN. Mol Plant Pathol. 2005;6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- 42.Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, Kunkel BN. Mol Plant–Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- 43.Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C. Plant Physiol. 2005;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Wees SCM, Luijendijk M, Smoorenburg I, van Loon LC, Pieterse CMJ. Plant Mol Biol. 1999;41:537–549. doi: 10.1023/a:1006319216982. [DOI] [PubMed] [Google Scholar]
- 45.Bostock RM. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 46.De Ilarduya OM, Xie Q, Kaloshian I. Mol Plant–Microbe Interact. 2003;16:699–708. doi: 10.1094/MPMI.2003.16.8.699. [DOI] [PubMed] [Google Scholar]
- 47.Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Proc Natl Acad Sci USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui J, Jander G, Racki LR, Kim PD, Pierce NE, Ausubel FM. Plant Physiol. 2002;129:551–564. doi: 10.1104/pp.010815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govrin EM, Levine A. Curr Biol. 2000;10:751–757. doi: 10.1016/s0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 50.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Nature. 2002;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 51.Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.